Abstract
Descriptions of recently evolved genes suggest several mechanisms of origin including exon shuffling, gene fission/fusion, retrotransposition, duplication-divergence, and lateral gene transfer, all of which involve recruitment of preexisting genes or genetic elements into new function. The importance of noncoding DNA in the origin of novel genes remains an open question. We used the well annotated genome of the genetic model system Drosophila melanogaster and genome sequences of related species to carry out a whole-genome search for new D. melanogaster genes that are derived from noncoding DNA. Here, we describe five such genes, four of which are X-linked. Our RT-PCR experiments show that all five putative novel genes are expressed predominantly in testes. These data support the idea that these novel genes are derived from ancestral noncoding sequence and that new, favored genes are likely to invade populations under selective pressures relating to male reproduction.
Keywords: adaptation, comparative genomics, lineage-specific, de novo gene
Understanding the genetic basis of adaptation remains a key priority for evolutionary biologists. Most adaptation likely results from modification of ancestral genetic function. Such modifications include coding sequence substitutions (1) and the origination of novel genes by partial or complete duplication of preexisting genes (2). The contribution of more radical “de novo” genetic changes to adaptive divergence, such as the recruitment of noncoding DNA into coding function, remains an open question. Although there are some rare examples that support partial recruitment of noncoding DNA into new genes (3, 4), there is no evidence thus far for novel genes derived primarily from ancestrally noncoding DNA. Such de novo genes would be difficult to identify for two reasons. First, novel gene discovery, which often occurs by serendipitous discovery of lineage-specific exon duplication (5, 6), biases against de novo gene identification. Second, if novel genes evolve rapidly under directional selection and/or if the associated ancestral noncoding DNA evolves rapidly under low functional constraint, there may be only a brief evolutionary window during which a new gene and its noncoding ancestor can be identified.
Results and Discussion
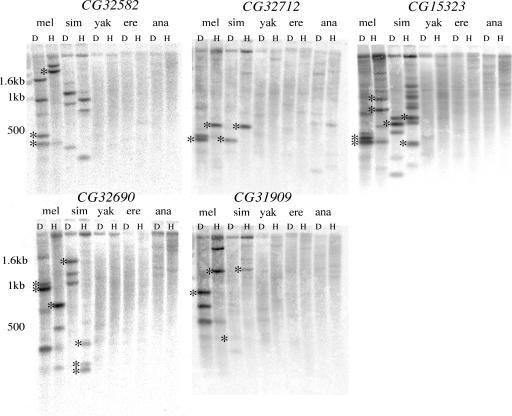
We took advantage of the recently assembled Drosophila genomes to carry out an analysis of lineage-specific de novo genes using the annotated model system genome of Drosophila melanogaster and the genome sequences of its close relatives. We generated a preliminary list of candidate genes from annotated D. melanogaster genes that returned poor hits in an automated blastn analysis against the genomes of Drosophila yakuba, Drosophila erecta, and Drosophila ananassae (see Methods). This search should exclude novel genes that are primarily composed of D. melanogaster-specific duplications of preexisting functional exons present in the common ancestor of all four species. The list of D. melanogaster and/or Drosophila simulans-specific genes was reduced by retaining only genes with empirical support [i.e., EST or complete cDNA sequence (http://flybase.org)], with the exception of one gene, CG32712, which we experimentally confirmed with RT-PCR (see below). This list was further reduced to five genes by requiring high-quality syntenic alignments of the flanking regions of the candidate D. melanogaster lineage-specific genes with the corresponding regions in D. yakuba, D. erecta, and D. ananassae (Fig. 5, which is published as supporting information on the PNAS web site). Those genes whose syntenic alignments revealed the absence of the focal D. melanogaster gene, rather than a highly diverged ortholog or a gap in the genome of assembly of the close relative, were retained for experimental confirmation by using D. melanogaster probes in a low-stringency Southern blot analysis. Southern blot analysis of the five candidate genes was consistent with the computational prediction that these D. melanogaster genes are absent from D. yakuba, D. erecta, and D. ananassae (Fig. 1). Hybridization to multiple bands in D. melanogaster and/or D. simulans is explained by the presence of paralogous DNA (see below). Weak hybridization of the D. melanogaster CG32712 probe to D. ananassae genomic DNA (Fig. 1) is likely due to the presence of homologous sequences in D. ananassae (see coordinates 4439–4765 of Fig. 5g). RT-PCR experiments on D. ananassae provided no evidence that this homologous sequence is transcribed (data not shown). Given the conservative nature of our analysis (only the top candidates were investigated), five is a minimum number of de novo D. melanogaster genes.
Fig. 1.
Southern blot analysis of putative novel genes in D. melanogaster (mel), D. simulans (sim), D. yakuba (yak), D. erecta (ere), and D. ananassae (ana). D, Dde; H, HhaI. Asterisks indicate predicted band sizes of focal genes. There are no predicted band sizes for CG32582 in D. simulans because of a gap in the assembly.
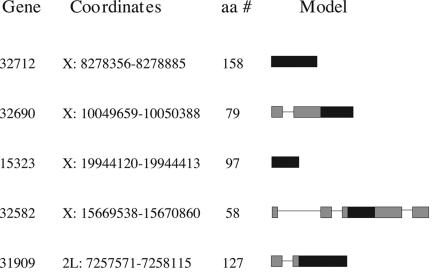
Although these lineage-specific genes evolved recently (2–5 million years ago), our sequence data from North American and African population samples suggest that all five genes are fixed in D. melanogaster. For each gene, all sequenced alleles possess an intact ORF that is homologous to the annotated ORF. Of the five genes, four occur on the X chromosome and one occurs on 2L (Fig. 2). The excess of X-linked genes is significant (binomial probability = 0.013). RT-PCR data from RNA isolated from whole adult females and adult male reproductive tracts of D. melanogaster revealed that all five genes exhibit testis-biased expression (Fig. 3). The multiple bands associated with CG32582 correspond to four alternative splice variants (data not shown). pfam analysis (www.sanger.ac.uk/Software/Pfam) of the five predicted proteins revealed no identifiable protein domains.
Fig. 2.
List of novel genes, chromosome number, coordinates, amino acid number, and gene models. Black boxes indicate coding regions. Gray boxes indicate untranslated regions.
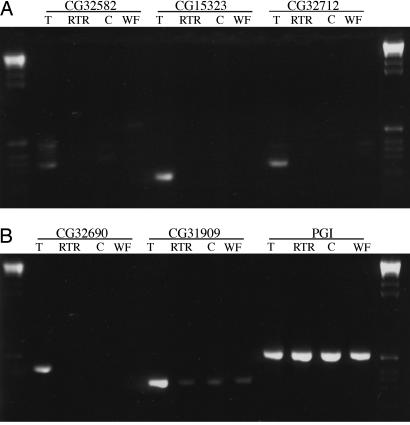
Fig. 3.
RT-PCR analysis of five putative novel genes in D. melanogaster. T, testes; RTR, reproductive tract remainder (male reproductive tract minus testes); C, male carcass (adult male minus reproductive tract); WF, whole female.
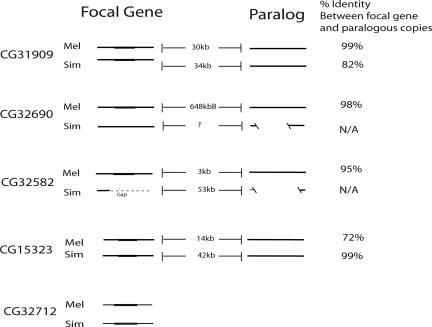
The repetitive hybridization patterns for all Southern blot probes (Fig. 1, CG32172 probe excepted) motivated an investigation into the possibility that these novel genes were related to paralogous D. melanogaster sequence. Using blastn, we found evidence of paralogous sequence (significant second best blast hits, P < e−13) located between 25 kb and 650 kb away from the focal gene for the same four genes (Fig. 4); we found evidence of more than six paralogous regions for CG15323. Moreover, focal genes and their corresponding paralogous sequence show high sequence similarity (Fig. 6, which is published as supporting information on the PNAS web site). These paralogous sequences are annotated as intergenic or intronic in D. melanogaster (http://flybase.org). Although RT-PCR experiments showed that a subset of these paralogous sequences is transcribed at a low level (Fig. 7, which is published as supporting information on the PNAS web site), none possess an ORF similar to that of the corresponding focal gene. These findings suggest that recent, intrachromosomal duplication is associated with the origin of four of the five de novo genes. We cannot determine whether the focal gene or the paralogous sequence is ancestral. However, blat searches revealed that D. yakuba lacks not only the focal gene but also sequence homologous to the paralogous, noncoding DNA regions of D. melanogaster (data not shown). The absence of both the focal genes and their associated paralogous sequences from D. yakuba supports the hypothesis that all five genes evolved from noncoding DNA in the common ancestor of D. melanogaster and D. simulans.
Fig. 4.
Paralogous regions associated with novel genes. Parallel solid lines indicate homology among flanking regions of focal genes. Filled rectangular box indicates gene region. Distances between focal gene and paralogous copy are indicated. CG15323 seems to be part of a larger, noncoding “family” of copies (only most closely related sequence is shown). CG32712 returned no significant nonself blast hits. Homologous sequence for all other genes as well as the paralogous copies was found in D. simulans, suggesting that most duplications occurred along the lineage leading to D. melanogaster and D. simulans. D. simulans contains a region homologous to the D. melanogaster gene CG32690 but has no homologous ORF. This D. simulans sequence, using blat, points to chromosome X_random, making unclear the distance between two D. simulans copies. Extensive indel variation among paralogous copies of the CG32690 gene and CG32582 region disallowed calculation of percentage identity. D. simulans sequence homologous to the CG32582 gene region occurs in a currently unassembled region in D. simulans genome.
For the three genes with complete ORFs in D. simulans, we analyzed within and between species silent and replacement variation using a McDonald–Kreitman test (7). CG32712 and CG31909 significantly deviate from neutrality whereas CG15323, although not significant, has a dN/dS (nonsynonymous:synonymous substitution rate) that is >1. These results are consistent with adaptive protein divergence (Table 1). This conclusion is further supported by the finding that replacement substitution rates (dN) are generally high relative to the genomic average (8) (Table 2, which is published as supporting information on the PNAS web site). These data are consistent with previous reports that novel proteins often experience adaptive protein evolution subsequent to their origin (5, 9, 10).
Table 1.
McDonald–Kreitman test of combined North American and African population samples
| Gene | Synonymous |
Nonsynonymous |
G value | ||
|---|---|---|---|---|---|
| Fixed | Polymorphic | Fixed | Polymorphic | ||
| 32712 | 11 | 17 | 17 | 9 | 3.74* |
| 15323 | 11 | 4 | 36 | 9 | 0.285 |
| 31909 | 12 | 9 | 45 | 11 | 4.04* |
| Total | 34 | 30 | 98 | 29 | 9.018** |
∗, 0.01 < P < 0.05;
∗∗, 0.001 < P < 0.01.
The testis-biased expression pattern for all five genes suggests that previous conclusions regarding the importance of adaptive evolution for male reproduction-related proteins also may apply to the origin of completely novel genetic functions. Unlike previously described novel genes with sex-specific expression such as Jingwei (11) and Hydra (H.-P. Yang, personal communication), the genes described here probably evolved from noncoding sequence rather than from functional elements that, in the parent copy, exhibited sex-biased expression. Thus, for these de novo genes, it is unlikely that recruitment into male function is related to the ancestral function.
The predominance of X-linkage among the five genes was unexpected. Previous reports suggested that male-biased genes, including novel genes generated by retrotransposition (12), are underrepresented on the X chromosome in Drosophila (13). Multiple explanations have been offered for this pattern, including an evolutionary advantage of avoiding X-inactivation during spermatogenesis (12) and sexual antagonism driving demasculinization and germ-line inactivation of the X chromosome (14). Our analysis suggests that young de novo genes tend to be both male-biased and X-linked, which casts doubt on the hypothesis that X-linkage of such genes is strongly disfavored by natural selection. Moreover, the preponderance of X-linked de novo genes suggests that mutations generating such genes either occur more often on the X chromosome or fix more readily on the X chromosome compared with similar mutations on the autosomes. It is notable that CG15323 is located in polytene band 19, which also happens to contain Sdic (4) and Hydra (H. P. Yang, personal communication), two other novel, testes-expressed genes. This observation raises the intriguing possibility that certain genomic regions have properties that favor origination of novel, testis-expressed genes.
A D. melanogaster whole-genome tiling array experiment has revealed widespread transcription of intergenic DNA (15). Our RT-PCR experiments support the idea that significant amounts of noncoding DNA may be transcribed at a low level. Such transcription could increase the probability of de novo gene evolution. In particular, promiscuous transcription of the testis, at least in mammals (16), and hypertranscription of the male X in Drosophila testis (17) suggest the possible contribution of the testis environment to origination of de novo genes. Intergenic DNA harbors large numbers of ORFs, few of which are functional genes (18). Although for most such ORFs expression is likely deleterious, transcription of an intergenic ORF might occasionally be beneficial, resulting in recruitment of noncoding DNA into novel function (19). Such phenomena may be more likely to occur in transcriptional domains associated with functions under directional selection, such as male reproduction. Interestingly, polytene band 19 (see above) seems to overlap a chromosomal domain of testis transcription (20).
The vast majority of D. melanogaster genes are found in other fly species and, indeed, other animal genomes (21). Furthermore, these genes shared among taxa tend to be highly conserved at the amino acid level. These aspects of conservation reflect the fact that most basic cellular and developmental functions are conserved across animals. Despite the relative rarity of lineage-specific genes, the possibility that they may play a disproportionately important role in lineage-specific adaptations and species incompatibilities could be revealed by functional analysis. Spermatogenesis-related phenotypes such as sperm competition or meiotic drive are interesting possibilities. Previous studies of Drosophila have revealed rapid sequence (22–25) and transcriptome (26) evolution in genes associated with male reproduction. Our results suggest that male reproductive function is associated with yet another aspect of genome divergence: de novo gene evolution from ancestral noncoding DNA.
Methods
Informatic Strategy.
Local blast databases were constructed for D. ananassae, D. erecta, D. melanogaster, D. simulans, and D. yakuba using the assemblies available as of November 2004. A total of 19,572 mRNA sequences corresponding to 13,449 D. melanogaster genes were extracted from the FlyBase database. tblastn was used to compare these D. melanogaster mRNA sequences against the D. ananassae, D. erecta, and D. yakuba genomes; a similar analysis was carried out against the D. melanogaster transposable element collection. Genes that returned an e-value <0.000001 against all sequence sets were removed from the list of possible candidate novel genes. This analysis yielded a collection of 77 genes that had either weak similarity or no similarity to species other than D. melanogaster or D. simulans. To enrich for bona fide D. melanogaster genes, as opposed to incorrect gene predictions, we retained only the 66 genes corresponding to a D. melanogaster cDNA or EST. The average number of ESTs or cDNAs per transcript was 2.5.
These 66 genes were then ranked by an index that weighted each gene by similarity to other species. The index was generated by multiplying the e-values from a D. melanogaster tblastx to D. yakuba, D. erecta, D. ananassae, and D. melanogaster transposable elements. The higher the value, the more probable the candidate. Consequently, weaker similarity to several species other than D. melanogaster and D. simulans reduced the rank whereas absence of matches to any species other than D. melanogaster or D. simulans increased the rank. Short genes are penalized in such an analysis. The 18 highest-ranked candidates were selected for further analysis based on quality of D. melanogaster annotation and a re-blast using the entire gene rather than just the transcripts.
Syntenic Alignments.
Syntenic alignments to the D. yakuba genome were made for the 18 D. melanogaster candidates. Candidates that were located in gaps in the D. yakuba genome or corresponded to small, highly diverged D. yakuba orthologs were removed from the analysis. The D. yakuba assembly for the remaining genes was confirmed by using PCR and sequencing of D. yakuba genomic DNA from strain Tai18E2. In all cases, the PCR and sequence data supported the genome assembly. Candidates that were absent from D. yakuba based on syntenic alignments were subjected to syntenic alignments in D. erecta and D. ananassae. D. melanogaster genes that corresponded to gaps in the syntenically aligned regions of these other species were further investigated by Southern blot analysis as described below.
Experimental Confirmation.
We used Southern blot analysis to determine whether our computationally produced list of D. melanogaster candidate, lineage-specific genes was (i) absent from D. yakuba, D. erecta, and D. ananassae and (ii) single copy in D. melanogaster. Genomic DNA (2–2.5 μg) was purified from D. melanogaster (y;cnbw;sp), D. simulans (w501), D. yakuba (Tai18E2), D. erecta (Tucson Stock Center), and D. ananassae (Tucson Stock Center). These DNAs were digested with DdeI and HhaI (Invitrogen), run on 1.5% agarose gels, and Southern blotted to Nytran Nylon membranes (Whatman). Each of five blots was probed by one of the five candidate genes. The probes were 32P-labeled (Stratagene Prime-it II random primer labeling kit) D. melanogaster PCR products (for probe locations, see Fig. 5). Blots were hybridized at 55°C overnight and subjected to three low-stringency washes (one for 2 min at 55°C and two for 10 min each at room temperature) with a solution composed of 40 mM NaPi, 0.001 M EDTA, and 0.01% SDS.
Assessing Presence/Absence and Population Genetics of Lineage-Specific Genes.
Sequence data from a population sample of all five lineage-specific genes were obtained from inbred D. melanogaster lines (http://dpgp.org) and Malawi isofemale D. melanogaster (B. Ballard, University of Iowa, Iowa City). For genes with complete ORFs in D. simulans, sequence data were obtained from a population sample that included both inbred lines from Wolfskill Orchard, CA (D.J.B.) and isofemale lines from Harare, Zimbabwe (C. Aquadro, Cornell University, Ithaca, NY). Most data were obtained by sequencing directly off the PCR product; however, in the few cases of residual heterozygosity, PCR products were cloned in PCR-4 vector (Invitrogen), and individual colonies were sequenced. Population genetic parameters and tests of neutrality were calculated in dnasp 4.0 (27). Polarized analyses were not possible because these genes do not occur in D. yakuba. Sequence data for this paper have been submitted to GenBank under accession numbers DQ657247–DQ657347.
Patterns of Expression.
Sex-biased and male reproductive organ-biased expression was investigated in D. melanogaster by using reverse transcriptase PCR. Thirty males (line 301A; T. MacKay, North Carolina State University, Raleigh) were dissected in RNA Later (Ambion, Austin, TX). Tissues collected were (i) testes/seminal vesicles, (ii) reproductive tract remainder (including external genitalia, ejaculatory bulb, accessory glands, ducts, and connective tissue), and (iii) remaining male carcass. Total RNA was isolated from each of these tissues and from whole females (n = 13) by using mirVana miRNA Isolation Kit using the Total RNA Isolation protocol (Ambion), followed by a reverse transcription reaction on 500 ng of RNA using SuperScript III Reverse Transcriptase reagents (Invitrogen). RT-PCR on heteroduplex was carried out by using the following primer pairs: CG32690 forward (F): GTTACAGCTACATTGCCGACGAA, reverse (R): ATCCAAATCAACGCAGTATCAAT; CG32582 F: AACCGAGTCCCAACAATAAAATCT, R: ATCCCAAAACCGAGTCGTAAGAAC; CG32712 F: CGCATCTTAGCCGGCAGGAGTTA, R: GGCGGTGTTCAGGGCGATGTA; CG15323 F: CCAGGAGGCGATCGAATAACAG, R: CCAGGAGGCGATCGAATAACAG; CG31909 F: AATCGGAACTTCAGAACCAACTACG, R: AGCGTCTACTTCATCCAGTA. RT-PCRs were then run on 1% agarose gels. The Pgi locus (F: AGAACCGCGCCGTCCTCCAC, R: GACCGCCCACCCAATCCCAAAAA) was used as a positive control and as an indicator of genomic contamination (RT-PCR primers flank an intron, thereby allowing detection of genomic DNA contamination; none was observed).
Investigation of Paralogous Regions.
We performed blast on the coding sequence of each lineage-specific gene back to the D. melanogaster genome. Sequences longer than 60 bp and with an e-value <10−7 were considered significantly similar. Evidence for larger-scale duplication was investigated by using blast on longer stretches of sequence (that included the focal coding sequence) back to the D. melanogaster genome. We used University of California, Santa Cruz blat to assess the presence/absence of such sequences in the genome of D. simulans.
Supplementary Material
Acknowledgments
A. Holloway, C. Langley, M. Hahn, B. Oliver, and three anonymous reviewers provided useful comments. This work was supported by National Science Foundation (NSF) Grant DEB-0327049 and National Institutes of Health Grant GM071926. M.T.L. was supported by an NSF Graduate Research Fellowship. A.D.K. was supported by a Howard Hughes predoctoral fellowship.
Footnotes
References
- 1.Bustamante C. D., Fledel-Alon A., Williamson S., Nielsen R., Hubisz M. T., Glanowski S., Tanenbaum D. M., White T. J., Sninsky J. J., Hernandez R. D. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 2.Long M., Betran E., Thorton K., Wang W. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 3.Chen L., Devries A. L., Cheng C. H. Proc. Natl. Acad. Sci. USA. 1997;94:3817–3822. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurminsky D. I., Nurminskaya M. V., De Aguiar D., Hartl D. L. Nature. 1998;396:572–575. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 5.Long M., Langley C. H. Science. 1993;260:91–95. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 6.Wang W., Brunet F. G., Nevo E., Long M. Proc. Natl. Acad. Sci. USA. 2002;99:4448–4453. doi: 10.1073/pnas.072066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald J. L., Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 8.Begun D. J. Mol. Biol. Evol. 2002;19:201–203. doi: 10.1093/oxfordjournals.molbev.a004072. [DOI] [PubMed] [Google Scholar]
- 9.Jones C. D, Begun D. J. Proc. Natl. Acad. Sci. USA. 2005;102:11373–11378. doi: 10.1073/pnas.0503528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betran E., Long M. Genetics. 2003;164:977–988. doi: 10.1093/genetics/164.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W., Zhang J., Alvarez C., Llopart A., Long M. Mol. Biol. Evol. 2000;17:1294–1301. doi: 10.1093/oxfordjournals.molbev.a026413. [DOI] [PubMed] [Google Scholar]
- 12.Betrán E., Thornton K., Long M. Genome Res. 2002;12:1854–1859. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parisi M., Nuttall R., Naiman D., Bouffard G., Malley J., Andrews J., Eastman S., Oliver B. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C.-I., Xu E. Y. Trends Genet. 2003;19:243–247. doi: 10.1016/s0168-9525(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 15.Stolc V., Gauhar Z., Mason C., Halasz G., van Batenburg M. F., Rifkin S. A, Hua S., Herreman T., Tongprasit W., Barbano P. E., et al. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt E. E. Curr. Biol. 1996;6:768–769. doi: 10.1016/s0960-9822(02)00589-4. [DOI] [PubMed] [Google Scholar]
- 17.Gupta V., Parisi M., Sturgill D., Nuttall R., Doctolero M., Dudko O. K., Malley J. D., Eastman P. S., Oliver B. J. Biol. 2006;5:3.1–3.22. doi: 10.1186/jbiol30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yandell M., Bailey A. M., Misra S., Shu S. Q., Wiel C., Evans-Holm M., Celniker S. E., Rubin G. M. Proc. Natl. Acad. Sci. USA. 2005;102:1566–1571. doi: 10.1073/pnas.0409421102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begun D. J., Lindfors H. A., Thompson M. E., Holloway A. K. Genetics. 2006;172:1675–1681. doi: 10.1534/genetics.105.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutanaev A. M., Kalmykova A. I., Shevelyov Y. Y., Nurminsky D. I. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- 21.Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Begun D. J., Whitley P., Todd B. L., Waldrip-Dail H. M., Clark A. G. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson W. J., Clark A. G., Waldrip-Dail H. M., Wolfner M. F., Aquadro C. F. Proc. Natl. Acad. Sci. USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z., Hambuch T. M., Parsch J. Mol. Biol. Evol. 2004;21:2130–2139. doi: 10.1093/molbev/msh223. [DOI] [PubMed] [Google Scholar]
- 25.Richards S., Liu Y., Bettencourt B. R., Hradecky P., Letovsky S., Nielsen R., Thornton K., Hubisz M. J., Chen R., Meisel R. P., et al. Genome Res. 2005;15:1–18. doi: 10.1101/gr.3059305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meiklejohn C. D., Parsch J., Ranz J. M., Hartl D. L. Proc. Natl. Acad. Sci. USA. 2003;100:9894–9899. doi: 10.1073/pnas.1630690100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rozas J., Sánchez-DelBarrio J. C., Messegyer X., Rozas R. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.