Abstract
Analyses of whole-genome sequences and experimental data sets have revealed a large number of DNA sequence motifs that are conserved in many species and may be functional. However, methods of sufficient scale to explore the roles of these elements are lacking. We describe the use of protein arrays to identify proteins that bind to DNA sequences of interest. A microarray of 282 known and potential yeast transcription factors was produced and probed with oligonucleotides of evolutionarily conserved sequences that are potentially functional. Transcription factors that bound to specific DNA sequences were identified. One previously uncharacterized DNA-binding protein, Yjl103, was characterized in detail. We defined the binding site for this protein and identified a number of its target genes, many of which are involved in stress response and oxidative phosphorylation. Protein microarrays offer a high-throughput method for determining DNA–protein interactions.
Keywords: proteomics, transcription factor, yeast
A fundamental problem in biology is to identify cis-regulatory DNA sequence elements and the proteins that bind to them. Such information is necessary for uncovering gene regulatory networks that control cellular and developmental processes. Genome-wide approaches have revealed many DNA sequence elements that may regulate gene expression: comparison of genome sequences of related organisms has identified thousands of evolutionarily conserved DNA sequence motifs (1–3); comparison of the sequences adjacent to coregulated sets of genes of an organism often reveals shared sequence motifs (4–7). Verifying functionality of these sequences and identifying the proteins that bind to them remains a significant challenge.
Several methods have recently been developed to map globally the DNA-binding sites of transcription factors. The SELEX method enables in vitro selection of the optimal binding site of a transcription factor (8), although applying it genome-wide may be difficult. In the chromatin immunoprecipitation “(ChIP)-chip” method, chromatin bound by a transcription factor of interest is immunoprecipitated, and the associated DNA is identified by using it to probe a genomic DNA microarray, thereby identifying the targets of the transcription factor (9, 10). Two related methods are direct probing of a DNA microarray with a DNA-binding protein and capture of genomic DNA in vitro with a DNA-binding protein, followed by its identification by probing a DNA microarray (“DIP chip”) (11, 12). Although these methods have achieved considerable success, their resolution is comparatively low because they identify relatively large segments of DNA bound by a protein. Pinpointing the binding site within these segments requires inference (usually by computational analysis). Indeed, the DNA sequences recognized by over half of the predicted DNA-binding proteins in yeast remain to be identified.
Although these methods promise comprehensive identification of the targets of a known transcription factor, they are not able to do the converse: identify the binding protein that recognizes a sequence motif of interest. Thus, these approaches are unable to take advantage of the thousands of conserved functional DNA sequence elements that have been predicted from a variety of studies and whose DNA-binding proteins are unknown (2, 4, 5, 7, 13, 14). One method that potentially offers this capability is the one hybrid method for identifying proteins that bind to a particular sequence in vivo (15), but its application on the whole-genome scale may be difficult.
To fill this void, we have developed a high-throughput method for identifying sequences recognized by DNA-binding proteins that uses an array of transcription factors. Oligonucleotides containing evolutionarily conserved DNA sequence motifs were used to probe an array of ≈300 known or potential transcription factors from Saccharomyces cerevisiae. We identified numerous protein–DNA interactions and characterized the DNA sequence recognized by a previously uncharacterized DNA-binding protein. This method should be applicable to any organism.
Results
Development of Protein Arrays for Assaying DNA-Binding Activity.
We first tested whether proteins arrayed on a surface could be used to detect specific protein–DNA interactions by arraying a few transcription factors (Rap1, Abf1, and Swi6), whose binding sites are well defined, along with two proteins that do not bind to DNA (Cmd1 and Cmk1). This miniarray was probed with a Cy3-labeled oligonucleotide containing three copies of the canonical binding site of Rap1, prepared as described in Fig. 1A. Multiple copies of the Rap1 recognition sequence were incorporated into the probe to increase the local concentration of binding sites. A Cy5-labeled probe with two base pair changes in the central invariant nucleotides of the binding sites was used in parallel to test the specificity of binding (Fig. 1B; see Materials and Methods).
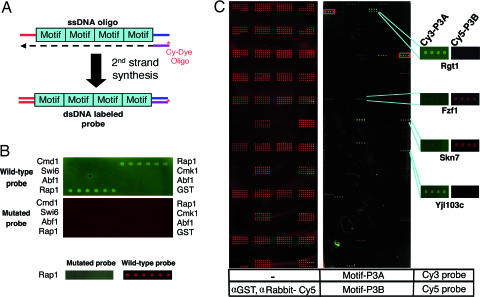
Fig. 1.
Probing the transcription factor microarray. (A) Probes were made by extending a universal primer labeled at its 5′ end with a fluorophore on an oligonucleotide template containing conserved sequence motifs. Because the length of the sequence motifs varies and we kept the length of the oligonucleotide probes constant, three or four copies of a motif are present in each probe. (B) Rap1 protein binds to a probe containing Rap1-binding sites. Each protein depicted on the right-hand and left-hand sides was spotted six times on the nitrocellulose surface and probed with an oligonucleotide containing three Rap1-binding sites (ACACCCAT/GCA) (labeled with Cy3, shown in green) and a probe containing three Rap1-binding sites with two nucleotide changes (ACACttAT/GCA) (labeled with Cy5, shown in red). Probing with reciprocally labeled probes is depicted below C. (C) Yeast transcription factor microarrays probed with fluorescent DNA probes. The GST-fused transcription factors purified from yeast (see Materials and Methods) were spotted (in quadruplicate) on each slide and probed with Cy5-labeled anti-GST (Left) or a pair of probes (Right). Examples of specific DNA binding are enlarged at the right. Yjl103c binds specifically to P3A but not P3B.
The proteins were arrayed on a variety of different surfaces and probed under different conditions (see Materials and Methods). Conditions were identified in which Rap1 bound to the wild-type probe but not to the mutant probe, regardless of the fluorophore used to label the probes (Fig. 1B). These probes did not bind to any other DNA-binding proteins on the array or to the non-DNA-binding proteins, indicating that binding is specific. In all of our preliminary experiments, we tested a total of seven proteins with binding sites of known sequence: Rap1, Zap1, Ume6, Yap1, Abf1, Swi6, and Mbp1. The first five of these proteins bound to probes containing their known binding sites (Fig. 1 and data not shown).
Survey of Proteins That Bind to Conserved Sequences Using a Transcription Factor Array.
To identify proteins that bind to specific DNA sequence motifs, we produced a microarray of 282 known or potential DNA-binding proteins, chosen based on their Gene Ontology (GO) designation as transcription factors, their homology to known DNA-binding domains, or their association with an in vitro DNA-binding activity (16). Most of the proteins known to bind DNA nonspecifically, such as chromatin-binding proteins and subunits of the general transcription machinery, were excluded from the array. The proteins were expressed in yeast cells as fusions to GST, purified by glutathione affinity chromatography (17), and spotted on microscope slides (Fig. 1C). The concentration of protein applied to each spot varied from ≈0.2 to 4 ng/μl.
The transcription factor array was probed with 40 Cy3-labeled double-stranded DNA oligonucleotides containing, in total, 75 DNA sequence motifs previously identified by their evolutionary conservation (1) (see Table 1, which is published as supporting information on the PNAS web site). Each oligonucleotide probe contained three or four copies of the sequence motif to be tested (Fig. 1A). We were able to represent the 75 sequence motifs in 40 oligonucleotides by careful design of the junctions between the repeated sequence motifs. To distinguish between specific DNA–protein interactions and nonspecific interactions, the array was probed with a second set of mutant Cy5-labeled probes that contained two base pair changes in the conserved sequence motifs (Fig. 1C and Table 1). Because sequence motifs are relatively short, judicious design of the mutant probes meant that two base pair changes in each copy of the sequence motif changed the sequence of most motifs represented in the oligonucleotide. These changes also create sequence motifs absent in the wild-type probes. For example, Fzf1, which recognizes TATCGTAT (18), binds to the two mutant probes (P3B (Fig. 1C) and P30B (see Table 2, which is published as supporting information on the PNAS web site), because they contain the sequences TATCG and TATGGTGT. These sequences are not represented in the corresponding probes (P3A and P30A) that serve as the variants of the P3B and P30B probes.
Twenty-three proteins on the array appear to bind DNA nonspecifically, because they bound to most probes with approximately equal affinity (Fig. 1C, shown in yellow), and to a double-stranded oligonucleotide consisting of the universal sequences that flank the motifs in each probe. These proteins, which included several known nonspecific DNA-binding proteins, such as Nph6A/B and Htz1, were excluded from further analysis. Some proteins bind more strongly to the wild-type DNA probe (Fig. 1C, shown in green); others bind more strongly to the mutant probe (Fig. 1C, shown in red). Sixty-two proteins on the array bound to at least one probe (Table 2).
Many Specific DNA–Protein Interactions Can Be Detected.
We identified a total of 211 specific DNA–protein interactions with the 80 probes (40 pairs of probes). Thirty-five probes did not interact specifically with any proteins on the array; 9 probes had only one specific DNA-protein interaction; 15 probes bound to between 5 and 22 different proteins. This latter result is not surprising, because concatenation of motifs creates multiple binding sites that can be recognized by different proteins.
Among the 211 specific DNA–protein interactions detected, 80 involved proteins with previously characterized binding sites (30 total proteins), including Met-31 and Met-32, which have been shown to bind as a heterodimer (19). For 17 of these proteins, their characterized/canonical binding sites are present in at least one bound probe, a minimal positive rate (17 of 30, or 57%), because we avoided including known binding sites in the probes as much as possible. Surprisingly, the putative recognition sequence was not apparent in the probes that bound to 13 previously characterized proteins. Perhaps the sequences recognized by these proteins are not well defined, or perhaps they recognize more than one sequence.
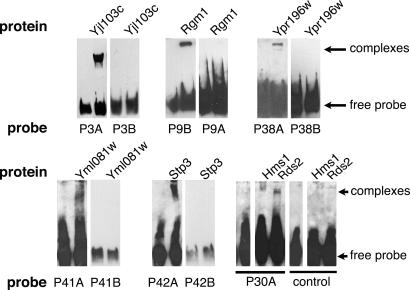
We further analyzed eight of these proteins whose DNA-binding sites are not known: Yjl103, Rgm1, Ypr196, and Rds2, each of which bound a single probe; Stp4, which bound two probes; Stp3 and Hms1, which bound four probes; and Yml081, which bound five probes. Stp3 and Yml081 bound to probes containing sequences similar to their respective binding sites, predicted by using the model described by Benos et al. (20) (G. Stormo, personal communication).
To verify the specific DNA interactions of the eight proteins, we purified the proteins, incubated them with their corresponding probes, and subjected them to EMSA. We were able to detect specific binding of the appropriate probes to seven of the eight proteins (Fig. 2), confirming that their binding sites are contained within the probe sequence.
Fig. 2.
EMSA of seven proteins that showed specific DNA binding on protein microarrays. Only one probe of each probe pair (Left) binds specifically to the protein. There are two or three base pair differences in each motif in each pair of probes. P30A binds to both Hms1 and Rds2. P38A is used as a control to show that binding is specific to P30A. See Materials and Methods for details.
Yjl103 Binds to CGGN8CGG.
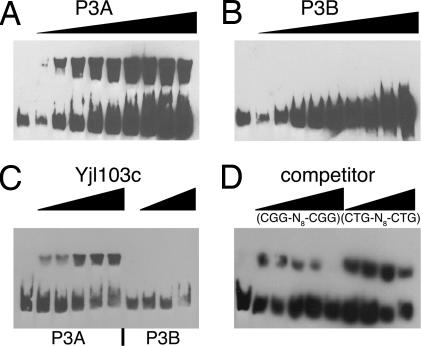
One protein–DNA interaction was studied in detail. Yjl103 is a member of the zinc cluster family of transcription factors. Several members of this family, which includes Gal4, have been well characterized (21, 22). All defined binding sites for this family of proteins consist of CGG repeats, with the recognition sequences for each protein differing in the orientation (direct, inverted, or convergent) and spacing of the CGG sequences. Yjl103 binds to a probe containing two overlapping copies of a direct repeat of CGG separated by eight nucleotides (CGGN8CGG). None of the other 39 probe pairs contains this sequence.
Yjl103 binds to its specific probe and not to the mutant probe in gel shift assays (Fig. 3). Binding was competed with a sequence containing a single copy of CGGN8CGG; an oligonucleotide containing the sequence CTGN8CTG did not compete for binding.
Fig. 3.
EMSA of Yjl103c. (A and B) A constant concentration of Yjl103c (5 μM) incubated with increasing amounts of labeled probes P3A (wild-type binding site) (A) and P3B (mutant binding site) (B), respectively. Probe concentrations increase from 60 to 600 pM. (C) Constant concentration of probes P3A and P3B (250 pM) with increasing amounts of Yjl103c. Protein increases from 0.7 μM to 8.5 μM. (D) Competition with unlabeled DNA: Increasing amounts of cold competitor DNA are added to the reaction with constant concentration of Yjl103c (1.6 μM) and labeled probe P3A (250 pM). Cold competitor is added at effective excess of labeled probe of 10-fold, 50-fold, 100-fold, and 800-fold.
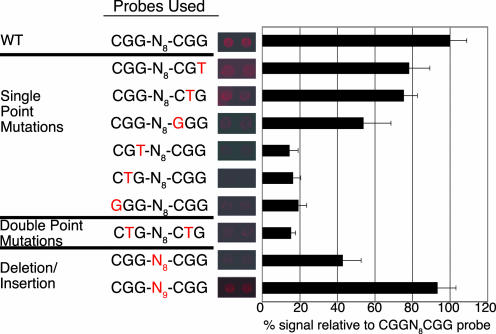
The protein chip assay was used to further elucidate the binding site for Yjl103. A Yjl103-GST fusion protein was purified from yeast, immobilized on a surface and incubated with a panel of probes containing variants of the CGGN8CGG sequence (Fig. 4). The first CGG appears to be required for binding, but the latter two residues in the second CGG appear to be less important, because substitutions in either of these positions reduce, but do not abolish, binding. Yjl103 binds in vitro to both CGGN8CGG and CGGN9CGG. This result is somewhat surprising because other members of this family of DNA-binding proteins appear to have a strict requirement for a specific spacing of the CGG repeats. The inclusion of metal chelators (EGTA and, especially, EDTA) during the probing severely impaired the binding of Yjl103 to DNA, suggesting that zinc is important for its DNA-binding activity (data not shown).
Fig. 4.
Yjl103c binds to CGGN8CGG and CGGN9CGG. Oligonucleotides containing variations of the putative binding site of Yjl103c were used to probe the transcription factor microarrays. Binding intensity, relative to the wild-type probe, is plotted on the right (average of three to five independent probings with each sequence).
Yjl103 Binds Upstream of Genes with CGGN8/9CGG That Are Involved in Energy Utilization.
To identify targets of Yjl103 and thereby gain clues to its function, we compared the gene-expression profile of a wild-type strain to those of strains that overexpress or are deleted for YJL103C. More than 500 genes were differentially expressed between the wild-type and the YJL103C-overexpressing strains (approximately half of these were up-regulated by Yjl103 overexpression). These genes are enriched for proteins involved in carbon compound and carbohydrate metabolism (P = 3.73 × 10−5) and also for proteins involved in stress response (P = 4.79 × 10−5), two roles previously suggested for Yjl103 (23). We found 131 genes that were expressed differently in the YJL103C deletion mutant compared with the wild-type strain (23 of them were among the 551 genes affected by YJL103C overexpression), approximately two-thirds of which are up-regulated in the deletion mutant. Thirty five of the >500 genes whose expression was altered by YJL103C overexpression and 7 of the 131 genes whose expression was altered by deletion of Yjl103C contain CGGN8CGG or CGGN9CGG in their promoters. This result is not significantly more than would be expected by chance, but this may be because we have not yet found the optimal conditions for inducing Yjl103 function.
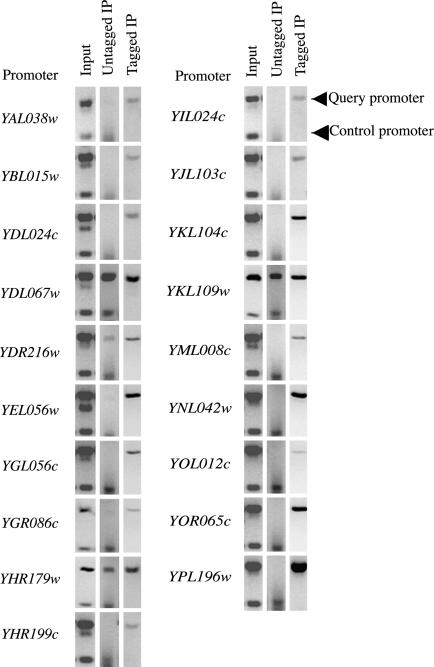
To determine whether Yjl103 binds in vivo to any of these genes whose expression is altered in strains lacking or overexpressing Yjl103 and that contain a CGGN8CGG sequence motif upstream, several of them were tested for Yjl103 binding by using ChIP. As shown in Fig. 5, 19 of 22 genes were enriched in immunoprecipitates from a Yjl103::c-myc-tagged strain relative to controls. Thus, Yjl103 associates with CGGN8CGG targets in vivo as well as in vitro.
Fig. 5.
ChIP assay for Yjl103 binding. Chromatin was crosslinked to proteins, Yjl103 tagged with a 13-myc epitope was precipitated with anti-myc antibody, and the precipitated DNA was released from protein and detected by PCR (as described in Materials and Methods) using primers specific for sequences upstream of the indicated 19 genes (query promoter) and primers specific for the GAL4 promoter (control promoter) that amplify a 150-bp fragment.
Discussion
Although a large number of potentially functional DNA sequence motifs have been identified from gene expression and sequence conservation studies, no facile method for identifying the proteins that bind to them has been available. Here, we describe implementation of protein arrays for this purpose. Using a microarray of yeast transcription factors, we were able to detect many known and previously unidentified DNA–protein interactions. Nucleotide substitutions in the known binding sites completely abolished binding of a protein, providing validation for the assay. In this way, we were able to define the proteins that bind to several sequence motifs and discover a previously unknown DNA-binding specificity.
Many of the proteins that bound to our probes bound to multiple probes, suggesting that they bind to DNA nonspecifically. For example, Phd1 bound 11 probes with no common sequence among them. Although many of these proteins probably bind DNA nonspecifically in vitro, such as Nhp6A and Nhp6B, others, like Phd1, seem to bind specific sequences in vivo (24). It is therefore likely that these proteins use additional cofactors to achieve sequence-specific binding.
The transcription factor arrays were probed with oligonucleotides containing multiple copies of the sequence motifs. Thus, each probe may contain several overlapping binding sites capable of being recognized by several proteins. The pattern of binding by each transcription factor can often be deconvoluted by examining the different probes each one binds. For example, Hms1 bound three probes, each of which contained the sequence ACCACA. Probes that bound to Yml081 also contained sequences similar to its predicted binding motif (20). In other cases, it is necessary to determine the exact sequence bound by the particular factor. One possible solution to this problem is to separate defined sequence motifs with random nucleotides, which would keep other binding sites at single copy and retain multiple copies of the intended binding site in the probe (Table 1).
Many transcription factors bind to DNA as heterodimers. It is noteworthy that we detected binding of both Met-31 and Met-32 to a probe containing the binding site of this heterodimer. We expect that other heterodimeric DNA-binding proteins purified from yeast extracts will similarly be associated with their partner protein(s). It should also be possible to carry out the binding reactions in the presence of another protein, or perhaps in the presence of a nuclear extract, to recreate heterodimers not present on the array. Combinations of proteins could also be spotted on the array, providing a matrix of all possible heterodimers.
We characterized in detail the binding site of a previously uncharacterized protein, Yjl103, a member of the Zn cluster family of transcription factors, whose bindings sites are variations of CGG repeats. We defined the binding site of Yjl103 to be two direct repeats of CGG separated by eight or nine nucleotides (CGGN8CGG or CGGN9CGG). It is somewhat surprising that the spacing of the CGG repeats is variable, because the binding sites of nearly all members of this family of DNA-binding proteins have rigid spacing requirements. In fact, it is the spacing of the CGG repeats (and their orientation) that determines the specificity of DNA binding of each protein. Perhaps Yjl103 forms a complex with other proteins that modify its sequence spacing requirement. Gene-expression profiling identified several genes differentially regulated when Yjl103 is overexpressed or deleted. Yjl103 binds in vivo upstream of 19 of 22 of the genes we tested, and all of them contain the CGGN8CGG sequence motif. The known or predicted functions of the proteins encoded by these genes are enriched in carbon compound and carbohydrate metabolism, consistent with the proposed role of Yjl103 in energy utilization (23).
In yeast, a very well characterized organism, the sequences recognized by only approximately half of its 200 or more transcription factors are known. Protein array technology offers the possibility for high-throughput analysis of all transcription factors with many probes under a variety of conditions and should bring the catalog of transcription factor binding sites within our reach. Application of this technology to mammals, with ≈1,000–1,500 transcription factors, would require only a modest increase in the scale of the analysis. Thus, it should be possible to determine cis-regulatory sequences and the proteins that bind to them across the genome, which is the first step in decoding the regulatory networks of an organism.
Materials and Methods
Strains.
Transcription factors fused to GST were expressed in a pep4 (protease-deficient) strain (Y258) (17). The yjl103c mutant was from the yeast gene-deletion collection (25). All other strains were derivatives of S228c (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0/met15Δ0/MET LYS/lys2Δ0) or the filamentous strain L5321 (MATa leu2::hisG ura3–52). Yjl103 was expressed from its own promoter and tagged at its C terminus with 13 copies of the myc epitope by integrating into the chromosome 13Myc-KanMX, as described in ref. 26.
Probe Preparation.
Probes were made by a fill-in reaction with TaqDNA polymerase using a universal oligonucleotide labeled with Cy3 or Cy5 (for protein chips) or biotin (for EMSA). Probes were purified, concentrated, and quantified by acrylamide gel electrophoresis and a NanoDrop apparatus (NanoDrop Technologies, Wilmington, DE).
Transcription factors tagged on their N termini with GST-His6 (17) were overexpressed in yeast cells and purified from 100-ml cultures grown to midlog phase in 1% yeast extract and 2% peptone and induced for 5 h with 2% galactose. Proteins were purified from cell extracts in 96-well deep-well plates by using glutathione beads (GE Healthcare) as described in ref. 17.
Protein Microarrays.
The GST-tagged transcription factors were arrayed into 384 microwell plates and printed on FAST slides (8 pads, 16 pads, or single pad slides; Schleicher & Schuell) in duplicate, triplicate, or quadruplicate. In pilot experiments, nickel-coated (XENOSLIDE N; Xenopore, Hawthorne, NJ) and aldehyde-coated (SMAI; Telechem International, Sunnyvale, CA) slides were also tested. We chose the FAST slides because of their higher capacity for protein.
The protein microarrays were probed (in duplicate) with labeled oligonucleotides by using the following protocol. Printed slides were blocked for 1 h with 3% BSA in hybridization buffer [25 mM Hepes (pH 8.0)/50 mM KCl/0.5% Triton X-100/2 mM MgCl2/1 mM PMSF/3 mM DTT and protease inhibitors (Complete; Roche)] and then probed for 90 min with 40 nM fluorescently labeled double-stranded DNA oligonucleotides (see Table 1 for list of oligonucleotides) in hybridization buffer at 4°C, washed three times in cold hybridization buffer, and air dried. The slides were scanned with a GenePix 4000 scanner (Axon Instruments, Union City, CA). Proteins whose signal was reproducibly above background levels (n ≥ 2 slides) and specific for the wild-type probes were classified as putative targets and tested further as described below. The conditions for binding were chosen based on extensive experimentation with a wide variety of conditions for binding of probes to Rap1 and Zap1, including different buffers (Tris·HCl and Tris·borate and Hepes), at a variety of concentrations (25–150 mM), at pHs between 7.0 and 8.0, with different salts (KCl and NaCl), at several different concentrations (25–150 mM), and at different temperatures. Nonspecific carrier DNA (salmon-sperm DNA and poly dI-dC) was omitted from the final protocol because it increased the background signal of labeled probe to the nitrocellulose surface. Glycerol in the binding buffer >20% smeared the slides; 10% glycerol seemed optimal. Neither Triton X-100 nor Tween 20 detergents (0.1–10%) had an observable effect on binding of probes to the array. Binding of probes to the array increased with probe concentration to ≈50 nM, after which increased background binding to the nitrocellulose surface of the slides was observed.
EMSAs.
Binding reactions were carried out according to the manufacturer’s recommendations (Pierce Light Shift Chemiluminescence kit) in 20 μl of 50 nM KCl, 25 mM Hepes (pH 8), 10% glycerol, and 0.1% Triton X-100. Probe concentrations varied from 60 to 600 pM. Protein concentrations varied from 0.7 to 8.5 μM. The reactions were incubated for 20 min at room temperature, followed by 10 min on ice before 5 μl of 5% Ficoll loading dye was added, and loaded onto 8-cm × 7-cm 8% acrylamide gels prerun at 100 V for 1 h. The gels were run at 4°C and 100 V until the bromophenol blue dye had migrated two-thirds of the way down the gel. Nucleic acids were transferred to nylon membranes and visualized according to the manufacturer’s recommendations.
DNA Microarrays.
DNA microarrays were printed with 6,388 oligonucleotides manufactured by QIAGEN Operon that represent virtually all S. cerevisiae ORFs. The oligonucleotides were resuspended to a concentration of 40 μM in 3× SSC with 0.75 M betaine and were printed in duplicate on Epoxy slides (MWG Biotech).
RNA Preparation.
Cells were grown to log phase in yeast peptone 2% raffinose medium and induced with galactose for 5 h. RNA extraction, labeling, and hybridization were done as described in ref. 27.
Data Analysis.
The scanned array images were analyzed by using the default settings in genepix pro 4.0. For each spot on the array, the median of the pixel-by-pixel ratios of the two channel intensities (with median background intensity subtracted) was calculated, and the two-step mixed-model ANOVA was applied to the log-transformed values (28) and used to normalize the expression differences between spots that were due to factors in which we were not interested: log2(Yijkm) = μ + Gi + Tj + Ak(ij) + GTij + ϕ(ijk)m, where Yijkm is the median of ratios for each spot, Gi is the average genotype effect (overexpressed or δ-strain), Tj is the average treatment effect (wild-type or modified strain), GTij is the average genotype times treatment-interaction effect (wild-type overexpressed strain, wild-type δ-strain, modified overexpressed strain, modified δ-strain), Ak(ij) is the average array effect, which is nested within the genotype by treatment-interaction effect, and ϕ(ijk)m is the residual.
A second ANOVA model was applied to each gene separately by using the residual ϕ(ijk)m from each spot as a response variable: ϕgijkm = γg + γ Ggi + γTgj + γ GTgij + γ Agk(ij) + ε(gijk)m, where ϕgijkm is the residual from the first ANOVA model for each spot, γg is the average gene expression for each gene g, Ggi is the gene expression due to genotype i, Tgj is the gene expression due to treatment j, GTgij is the gene expression due to genotype i interacting with treatment j, Agk(ij) is the gene expression due to array effect, and ε(gijk)m is the residual.
Genes that showed differential expression between wild-type (GT10) and overexpressed (GT11) strains were selected based on the criteria: γGT10 − γGT11 ≠ 0 at α = 0.05. Genes that showed differential expression between wild-type (GT00) and δ- (GT01) strains were selected based on similar criteria: γGT00 − γGT01 ≠ 0 at α = 0.05. To select genes that show differential expression between the overexpressed and δ-strains, several filters were applied. First, genes that satisfied γGT01 − γGT11 ≠ 0 at α = 0.05 were kept. Next, genes that satisfied γGT00 − γGT10 ≠ 0 at α = 0.05 were filtered out. Last, we filtered out genes that did not show any significance for the genotype by treatment-interaction effect.
ChIP.
The strain expressing Yjl103 tagged at its C terminus with 13 copies of the myc epitope from its native gene on chromosome 10 and the corresponding wild-type strain were inoculated at an OD600 of 0.2 and grown in yeast peptone dextrose medium overnight and reinoculated into fresh medium at an OD600 of 0.2. The strains were grown for 4 h at 30°C with shaking before heated medium was added to bring the temperature of the cultures to 37°C. After 20 min of shaking at 37°C, cells were fixed by addition of formaldehyde to a final concentration of 1%. Proteins were precipitated by using 9E10 anti-c-myc antibody (Santa Cruz Biotechnology), and the associated DNA was liberated, purified, amplified, and labeled with Cy3 and Cy5 fluorophores and used to probe a DNA microarray of intergenic regions of the yeast genome, as described in ref. 29. Twenty-two gene promoters that showed an increased hybridization signal relative to the signal obtained with probe prepared from the immunoprecipitate of a strain without a myc-tagged protein and whose genes were differentially expressed in strains missing or overexpressing Yjl103 (determined by a gene-expression profiling experiment as described above) were chosen for further analysis by conventional ChIP, preformed as described above, except that 40 ng of the liberated DNA was amplified (30 cycles at 95°C for 1.5 min, 57°C for 2 min, and 72°C for 3 min, with a final extension at 72°C for 10 min) in a 50-μl reaction with Mango Taq (Bioline, Randolph, MA). Two sets of primers were used in each reaction. Primers to query promoters were designed to generate a product of ≈500 base pairs and were added to a final concentration of 2 nM; primers to the GAL4 promoter generated a product of 150 base pairs and were added to a final concentration of 0.4 nM.
Supplementary Material
Acknowledgments
We thank Justin Gerke, Jay Gertz, Josh Witten, and Xiaowei Zhu for help with data analysis and Jon Armstrong and Ken Nelson for help in printing the protein microarrays. This work was supported by National Institutes of Health Grants GM063803 (to M.J.) and CA77808 (to M.S.).
Abbreviation
- ChIP
chromatin immunoprecipitation.
Footnotes
Conflict of interest statement: M.S. consults for Invitrogen.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B. A., Johnston M. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 2.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 3.Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., et al. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes J. R., Cheng J. F., Ventress N., Prabhakar S., Clark K., Anguita E., De Gobbi M., de Jong P., Rubin E., Higgs D. R. Proc. Natl. Acad. Sci. USA. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatesh B., Yap W. H. BioEssays. 2005;27:100–107. doi: 10.1002/bies.20134. [DOI] [PubMed] [Google Scholar]
- 6.Stein L. D., Bao Z., Blasiar D., Blumenthal T., Brent M. R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., et al. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cliften P. F., Hillier L. W., Fulton L., Graves T., Miner T., Gish W. R., Waterston R. H., Johnston M. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- 8.Roulet E., Busso S., Camargo A. A., Simpson A. J., Mermod N., Bucher P. Nat. Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 9.Horak C. E., Luscombe N. M., Qian J., Bertone P., Piccirrillo S., Gerstein M., Snyder M. Genes Dev. 2002;16:3017–3033. doi: 10.1101/gad.1039602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 11.Mukherjee S., Berger M. F., Jona G., Wang X. S., Muzzey D., Snyder M., Young R. A., Bulyk M. L. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Noll D. M., Lieb J. D., Clarke N. D. Genome Res. 2005;15:421–427. doi: 10.1101/gr.3256505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T., Stormo G. D. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 14.Gertz J., Riles L., Turnbaugh P., Ho S. W., Cohen B. A. Genome Res. 2005;15:1145–1152. doi: 10.1101/gr.3859605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deplancke B., Dupuy D., Vidal M., Walhout A. J. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall D. A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., et al. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 18.Avram D., Leid M., Bakalinsky A. T. Yeast. 1999;15:473–480. doi: 10.1002/(SICI)1097-0061(199904)15:6<473::AID-YEA388>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 19.Blaiseau P. L., Isnard A. D., Surdin-Kerjan Y., Thomas D. Mol. Cell. Biol. 1997;17:3640–3648. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benos P. V., Lapedes A. S., Stormo G. D. J. Mol. Biol. 2002;323:701–727. doi: 10.1016/s0022-2836(02)00917-8. [DOI] [PubMed] [Google Scholar]
- 21.Liang S. D., Marmorstein R., Harrison S. C., Ptashine M. Mol. Cell. Biol. 1996;16:3773–3780. doi: 10.1128/mcb.16.7.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellauer K., Rochon M. H., Turcotte B. Mol. Cell. Biol. 1996;16:6096–6102. doi: 10.1128/mcb.16.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng Y., He T., Wu Y., Vanka P., Yang G., Huang Y., Yao H., Brown S. J. Int. J. Mol. Med. 2005;15:123–127. [PubMed] [Google Scholar]
- 24.Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., Snyder M. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Veronneau S., Dow S., Lucau-Danila A., Anderson K., Andre B., et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 26.Longtine M. S., McKenzie A., III, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Dudley A. M., Aach J., Steffen M. A., Church G. M. Proc. Natl. Acad. Sci. USA. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfinger R. D., Gibson G., Wolfinger E. D., Bennett L., Hamadeh H., Bushel P., Afshari C., Paules R. S. J. Comput. Biol. 2001;8:625–637. doi: 10.1089/106652701753307520. [DOI] [PubMed] [Google Scholar]
- 29.Iyer V. R., Horak C. E., Scafe C. S., Botstein D., Snyder M., Brown P. O. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.