Abstract
Ionizing radiation (IR) can induce apoptosis via p53, which is the most commonly mutated gene in human cancers. Loss of p53, however, can render cancer cells refractory to therapeutic effects of IR. Alternate p53-independent pathways exist but are not as well understood as p53-dependent apoptosis. Studies of how IR induces p53-independent cell death could benefit from the existence of a genetically tractable model. In Drosophila melanogaster, IR induces apoptosis in the imaginal discs of larvae, typically assayed at 4–6 hr after exposure to a LD50 dose. In mutants of Drosophila Chk2 or p53 homologs, apoptosis is severely diminished in these assays, leading to the widely held belief that IR-induced apoptosis depends on these genes in Drosophila. In this article, we show that IR-induced apoptosis still occurs in the imaginal discs of chk2 and p53 mutant larvae, albeit with a delay. We demonstrate that this phenomenon is a true apoptotic response because it requires caspase activity and the chromosomal locus that encodes the pro-apoptotic genes reaper, hid, and grim. We also show that Chk2- and p53-independent apoptosis is IR dose-dependent and is therefore probably triggered by a DNA damage signal. We conclude that Drosophila has Chk2- and p53-independent pathways to activate caspases and induce apoptosis in response to IR. This work establishes Drosophila as a model for p53-independent apoptosis, which is of potential therapeutic importance for inducing cell death in p53-deficient cancer cells.
Keywords: apoptosis, DNA damage
Programmed cell death, or apoptosis, is crucial for normal organismal development and eliminating damaged cells. Defects in the apoptotic response can allow cells with damaged or incomplete genetic information to live and contribute to tumor development. Two tumor suppressors, the protein kinase checkpoint kinase 2 (Chk2) and the transcription factor p53, play key roles in inducing apoptosis when DNA is damaged by genotoxic agents such as ionizing radiation (IR). The importance of Chk2 and p53 in maintaining genome integrity is highlighted by the fact that mutations in chk2 and p53 are associated with human disease. Mutations in p53 have been found in at least half of all human cancers (1), and somatic mutations in chk2 have been found in a variety of sporadic tumors (2). In addition, inherited germ-line mutations in either chk2 or p53 have been associated with a familial multicancer syndrome called Li–Fraumeni syndrome (3–5).
Mouse models of Chk2 and p53 demonstrate the requirement for these molecules in stimulating apoptosis in certain tissues and in preventing tumor development. Thymocytes from both Chk2- and p53-null mice are defective in IR-induced apoptosis (6, 7). The majority of p53-null mice develop tumors by 6 months of age (8), and although Chk2-null mice do not develop spontaneous tumors at a high rate, they are highly sensitive to developing tumors when treated with the chemical carcinogen 7,12-dimethylbenz[a]anthracene (6).
The molecular mechanisms by which Chk2 and p53 induce apoptosis have been much studied. In response to IR, Chk2 becomes activated via phosphorylation on threonine-68 by ataxia telangiectasia mutated (ATM) (9–11). Activated Chk2 then phosphorylates p53 on serine-20, disrupting the interaction of p53 with Mdm2 and leading to increased stability of p53 (12, 13). ATM can also phosphorylate p53 directly on serine-15, contributing to transcriptional activation of p53 (14–16). p53 is thought to stimulate apoptosis by transcriptional activation of Bax, Noxa, and PUMA, which then stimulate the release of Diablo/Smac and cytochrome c from mitochondria, inhibit inhibitor of apoptosis proteins (IAPs), and subsequently activate caspases (17).
Mammalian cells lacking p53 can also undergo apoptosis when treated with chemical genotoxins, IR, or UV. p53-independent apoptosis that occurs in response to topoisomerase inhibitors occurs via another p53-family member, p73 (18). In contrast, mechanisms for radiation-induced, p53-independent cell death are poorly understood.
Homologs of Chk2 and p53 exist in Drosophila melanogaster and have conserved roles in regulating IR-induced apoptosis; imaginal discs from Dmchk2 mutant larvae are defective in IR-induced apoptosis, at least at 4 hr after exposure to x-rays (19, 20). Like Dmchk2 mutants, Dmp53 mutants are also defective in IR-induced apoptosis (20–22), demonstrating a conserved role for p53 in flies. As in mammalian systems, DmChk2 appears to regulate Dmp53 (20, 23). Dmp53 is thought to stimulate apoptosis by up-regulating transcription of the proapoptotic genes reaper (rpr), head involution defective (hid), and sickle (skl) (20, 24), which encode functional orthologs of Diablo/Smac.
Contrary to the established thought that DmChk2 and Dmp53 (to be called Chk2 and p53, hereafter) are required for IR-induced apoptosis, we show here that cell death does indeed occur in the imaginal discs of irradiated chk2 and p53 mutant larvae but with a significant temporal delay compared with wild type (WT). This Chk2- and p53-independent cell death is probably a bona fide apoptotic response because it requires the activity of activated caspases and the chromosomal locus that encodes the proapoptotic genes rpr, hid, and grim, which encode another Smac/Diablo ortholog. p53-independent cell death that requires another p53-family member, p73, has been described in mammalian systems (18, 25). Because Dmp53 is the only known p53 family member in Drosophila, the Chk2- and p53-independent cell death we document in this article is likely to rely on a previously uncharacterized mechanism. Systematic studies of IR-induced, p53-independent cell death in a genetically tractable model are thought to be unfeasible because work in Caenorhabditis elegans and Drosophila had led to the view that IR-induced apoptosis depends on p53 (20–22, 26). Our work establishes Drosophila as a model for p53-independent apoptosis, which provides a powerful genetic tool for identifying novel regulators of IR-induced, p53-independent apoptosis that may be clinically relevant.
Results
Radiation-Induced Cell Death Occurs in chk2 and p53 Mutants.
Exposure of Drosophila larvae to IR such as x-rays or γ-rays results in increased apoptosis in imaginal discs. This response is routinely assayed at 4–6 hr after irradiation by staining with acridine orange (AO), a vital dye that is excluded from live cells. chk2- and p53-null mutants fail to show an increase in apoptosis by this assay, leading to the conclusion that Chk2 and p53 are essential for IR-induced apoptosis (19, 20, 23). In these experiments, larvae were irradiated with 4,000 R, which allows >95% of pupae to eclose into adults. Approximately 50% of these adults die soon thereafter, however, such that 4,000 R represents the LD50 for larvae. We used this standard dose of 4,000 R in our experiments, except where otherwise indicated.
We found that radiation-induced cell death, as detected by AO staining, was indeed diminished or absent in chk2 and p53 mutants at 6 hr after irradiation. When a time course was performed, however, we detected robust increases in AO staining in irradiated chk2 and p53 mutants compared with nonirradiated controls, albeit with a delay (Fig. 1). The response was reproducibly detectable by 18 hr after irradiation and persists for at least an additional 12 hr.
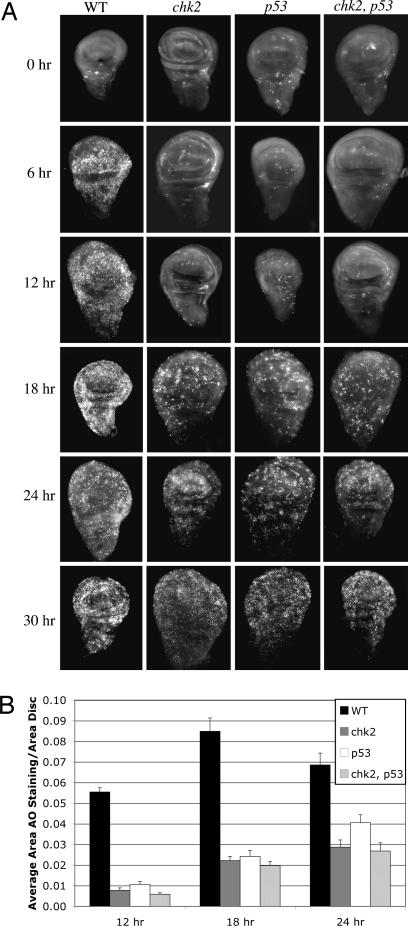
Fig. 1.
Time course of cell death in chk2 and p53 single and chk2, p53 double mutants. (A) Ninety-six- to 120-hr-old third-instar larvae were irradiated with 4,000 R of x-rays. Imaginal discs were dissected at various times after irradiation and stained with AO to detect dead cells. Only wing imaginal discs are shown. Genotypes are indicated for WT, mnkp6 (chk2), and p535A-1-4 (p53) mutants and mnkp6, p535A-1-4 (chk2, p53) double mutants. (B) The fractional area of each disk stained with AO was quantified for 12-, 18-, and 24-hr time points. Error bars represent SEM. The data are from 10 to 21 discs per genotype per time point.
As mentioned above, larvae that are single mutants in either chk2 or p53 show a lack of apoptosis at 4–6 hr after irradiation. In the embryo, Drosophila p53 is phosphorylated in response to IR, and this phosphorylation requires Chk2 (20). These data led to the idea that p53 acts downstream of Chk2 to activate radiation-induced apoptosis in the larvae. This idea, however, had not been rigorously tested, for example in comparative analysis of double and single mutants. That is, if Chk2 and p53 act in the same pathway for the apoptotic response, chk2, p53 double-mutant larvae should behave as each single mutant. If Chk2 and p53 act in distinct pathways, the phenotype of a double mutant should be more severe than that of single mutants, for example showing further delay in apoptosis in the former. We found that quantified AO stain was similar between chk2 single mutants and the double mutant at all time points assayed (Fig. 1B). AO stain in p53 mutants was also similar to that of chk2 mutants and chk2, p53 double mutants at 12 hr and importantly at 18 hr after irradiation when Chk2-/p53- independent cell death begins. At 24 hr after irradiation, AO stain in p53 mutants was only slightly (≈30%) higher than in the other two genotypes. These data are consistent with Chk2 and p53 acting in a linear pathway for IR-induced apoptosis. This allows us to define Chk2-/p53-independent cell death as cell death seen in either chk2 or p53 mutants.
Caspase Activation Accompanies Chk2-/p53-Independent Cell Death.
In Drosophila, AO is specific for apoptotic cells and does not stain necrotic cells (27). Nonetheless, we wanted to confirm that cell death in chk2 and p53 mutants was due to apoptosis. To this end, we stained discs with a commercially available antibody to activated caspase 3, which cross-reacts with the activated Drosophila caspase drICE (28). We found that drICE was indeed activated after irradiation in chk2 mutants and p53 mutants, although the latter did not show activation as robust as in chk2 mutants (Fig. 2). We refrained from using the TUNEL assay in these experiments because it can, in principle, detect damaged DNA induced by radiation.
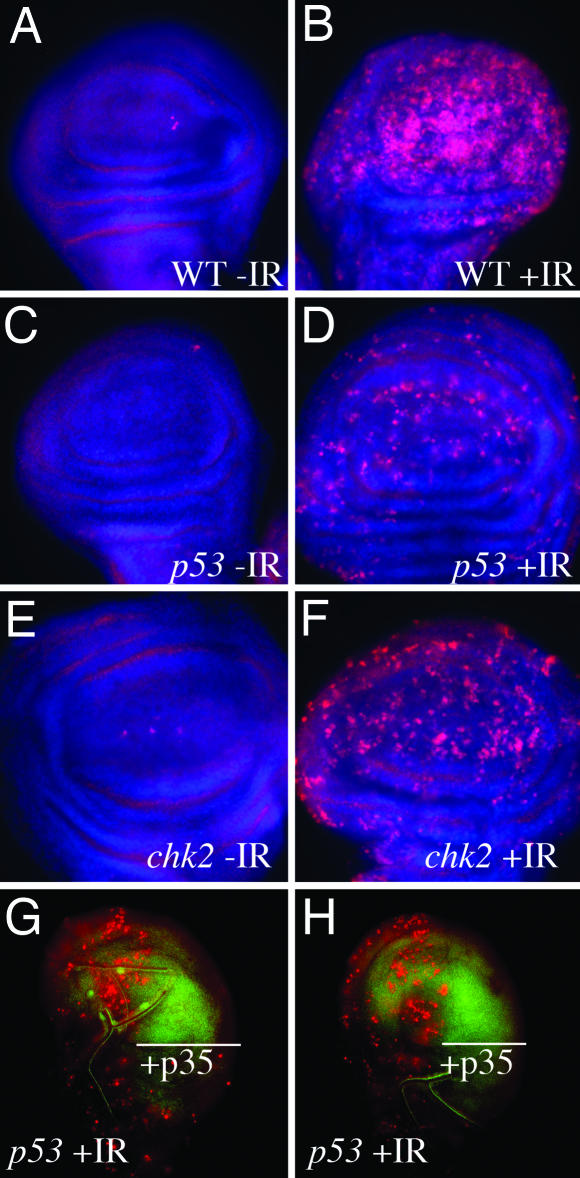
Fig. 2.
Role of caspases in Chk2-/p53-independent cell death. (A–F) Ninety-six- to 120-hr-old third-instar larvae were irradiated with 0 (−IR) or 4,000 R (+IR) of x-rays. Imaginal discs were dissected 24 hr after irradiation, fixed, and stained with Hoechst 33258 (blue) and an antibody against activated caspase 3 (red). Only the wing pouch portions of wing imaginal discs are shown. Genotypes are indicated for WT, mnkp6 (chk2), and p535A-1-4 (p53) mutants. Twenty-one to 34 wing discs are examined for each genotype. Representative examples are shown. (G–H) Ninety-six- to 120-hr-old third-instar larvae of the genotype en-Gal4>UAS-p35 UAS-GFP, p53 were irradiated with 4,000 R of x-rays (+IR). Imaginal discs were dissected 24 hr after irradiation and stained with AO (red) to detect dead cells. The GFP signal (green) indicates the area where engrailed drives p35 expression. Eighteen wing discs were examined. Two representative discs are shown in G and H to demonstrate reproducibility.
Furthermore, we found that inhibition of caspase activity blocked p53-independent cell death. Baculovirus protein p35 is a broad caspase inhibitor that blocks the activity of effector caspases at a step downstream of their proteolytic activation (29). When p35 was expressed in the posterior compartment of p53 mutant imaginal discs under control of the engrailed (en) promoter and the GAL4-UAS system, cell death in these compartments resembled those of unirradiated controls (wing imaginal discs at 24 hr after irradiation are shown in Fig. 2). We conclude that p53-independent cell death requires caspase activity.
The Apoptosis Locus Is Required for Chk2-Independent Cell Death.
rpr, hid, grim, and skl are clustered on chromosome III (to be called the apoptosis locus). Protein products of these genes inhibit DIAP1, an inhibitor of caspases (30). H99 is a chromosomal deficiency that removes rpr, hid, and grim; homozygotes die as embryos, thus genes of the apoptosis locus are essential for normal fly development (31). Previous work showed that imaginal discs from H99 heterozygous larvae have extremely low levels of IR-induced cell death, again assayed at 4–6 hr after irradiation, leading to a model in which products of proapoptotic genes must accumulate above a certain threshold to induce apoptosis (20). Analysis over a time course can lead to a more complete understanding of the process than what is achieved by analysis of a single time point. To better understand the importance of gene dose in radiation-induced cell death, we performed a time course of AO staining in imaginal discs from H99 heterozygotes.
Cell death is reduced in H99 heterozygotes at 6 hr after irradiation, in agreement with previous work. By 18 and 24 hr after irradiation, however, cell death in H99 heterozygotes appears as robust as WT at similar stages (Fig. 3; compare with Fig. 1). This result is still consistent with the threshold model because the accumulation of proapoptotic gene products would be slower in a hemizygote, but a threshold could still be reached after a delay.
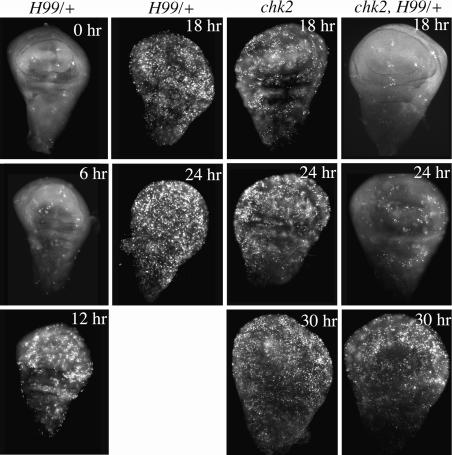
Fig. 3.
Requirement of the proapoptotic locus for Chk2-independent cell death. Ninety-six- to 120-hr-old third-instar larvae were irradiated with 4,000 R of x-rays. Imaginal discs were dissected at various times after irradiation and stained with AO to detect dead cells. Genotypes are indicated for H99/+ and mnkp6 (chk2) mutants and mnkp6, H99/+ (chk2, H99) double mutant. Five to 18 discs were examined per genotype per time point.
To determine whether the proapoptotic genes encoded by H99 play a role in Chk2-/p53-independent cell death, we analyzed chk2 mutants that also lack a copy of the apoptosis locus. chk2 homozygous mutants that are also H99 heterozygous display significantly lower levels of AO staining at 18 and 24 hr after irradiation compared with chk2 mutant controls (Fig. 3), indicating that the apoptosis locus is required for proper timing of Chk2-independent cell death. By 30 hr after irradiation, however, we observe a substantial increase in cell death in chk2, H99/+ double mutants. We surmise that, in the absence of Chk2 and p53, proapoptotic genes uncovered by H99 are activated, but more slowly. The H99 gene products accumulate but require a longer time to reach a threshold necessary for apoptosis.
To address this idea experimentally, we measured the level of proapoptotic transcripts by real-time PCR (RT-PCR; Fig. 4 and Table 1). We examined time points before and including the onset of Chk2/p53-dependent cell death (2 and 4 hr after irradiation) and before and including the onset of Chk2/p53-independent cell death (12 and 18 hr after irradiation). Imaginal discs from WT larvae showed an increase in rpr, hid, and skl transcripts, to varying degrees and at various times after irradiation. chk2 mutants accumulated rpr and hid transcripts, albeit with a delay, and for rpr, to lower levels than in WT. p53 mutants accumulated hid and skl transcripts also to lower levels and with a longer delay. grim transcripts also accumulated in chk2 mutants but, interestingly, not in WT or p53 mutants. Lower levels of transcripts could result in slower accumulation of proteins and a longer time before a threshold is reached.
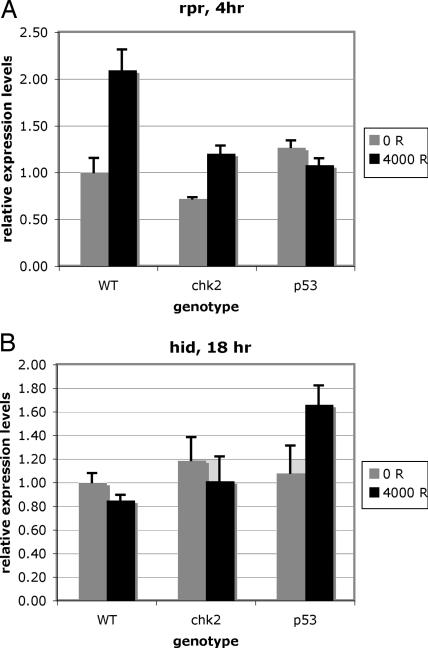
Fig. 4.
Expression of proapoptotic genes in chk2 and p53 mutants. mRNA levels were quantified by RT-PCR of cDNA derived from total RNA isolated from imaginal discs of irradiated (4,000 R) or control (0 R) larvae. For each experiment, RT-PCRs were run in triplicate and normalized to the average of actin and α-tubulin controls, and the average and the standard deviation were calculated and normalized to the values for unirradiated WT (0 R). (A) Sample data from an experiment in which rpr levels show a statistically significant (P < 0.05) increase in WT and chk2 larvae but not p53 larvae at 4 hr after irradiation. (B) Sample data from an experiment in which hid levels show a statistically significant (P < 0.05) increase in p53 larvae but not WT or chk2 larvae at 18 hr after irradiation.
Table 1.
Summary of RT-PCR data from three to four different experiments on WT, chk2, and p53 mutants
| Gene | Time, hr |
|||
|---|---|---|---|---|
| 2 | 4 | 12 | 18 | |
| rpr | ||||
| WT | 3/3* | 3/3* | 4/4* | 2/3* |
| [5.4, 3.6, 4.7] | [2.9, 1.9, 2.1] | [2.1, 1.4, 1.7, 1.4] | [1.4, 1.8] | |
| chk2 | 0/3 | 2/3* | 0/4 | 1/3 |
| [1.7, 1.7] | ||||
| p53 | 0/3 | 0/3 | 0/4 | 1/3 |
| hid | ||||
| WT | 2/3* | 1/3 | 2/4 | 1/3 |
| [1.4, 1.7] | ||||
| chk2 | 0/3 | 2/3* | 0/4 | 0/3 |
| [1.6, 1.7] | ||||
| p53 | 0/3 | 0/3 | 0/4 | 2/3* |
| [1.6, 1.5] | ||||
| skl | ||||
| WT | 3/3* | 2/3* | 2/4 | 1/3 |
| [2.5, 2.8, 3.5] | [1.6, 2.3] | |||
| chk2 | 1/3 | 0/3 | 0/4 | 1/3 |
| p53 | 0/3 | 0/3 | 0/4 | 2/3* |
| [1.4, 2.1] | ||||
| grim | ||||
| WT | 1/3 | 0/3 | 0/4 | 0/3 |
| chk2 | 0/3 | 2/3* | 0/4 | 0/3 |
| [1.8, 2.1] | ||||
| p53 | 0/3 | 0/3 | 0/4 | 1/3 |
Times shown represent duration after irradiation, in hours. The first line in each cell is the number of times a statistically significant increase was seen relative to unirradiated controls/number of experiments (e.g., 3/3). Fold increases are given in brackets.
*Conditions in which at least two of three or three of four experiments showed an increase in expression level of the pertinent gene.
p53-Independent Cell Death Depends on IR Dose.
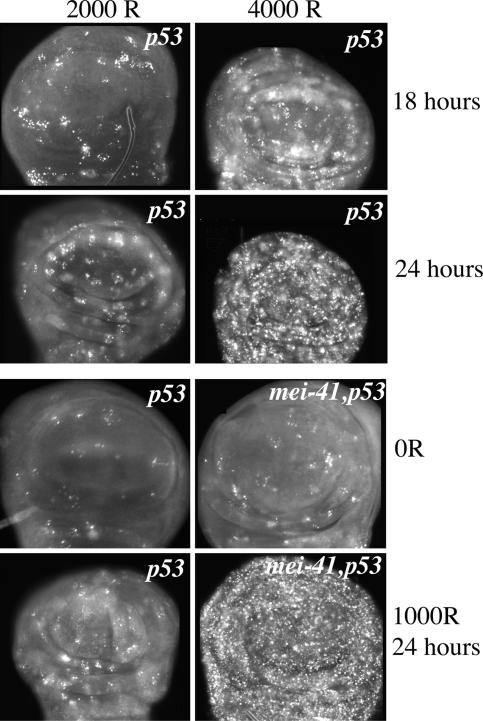
We observed more AO-stained cells in imaginal discs from p53 mutant larvae that had been irradiated with higher doses. For example, the abundance of AO-stained cells at 4,000 R outnumbers that at 2,000 R (shown for p53 mutants at 18 and 24 hr after irradiation in Fig. 5). Because we expect higher doses of x-rays to produce more damage, these data indicate that the amount of p53-independent cell death correlates with the amount of damage and suggests that p53-independent cell death occurs due to damage caused by x-rays.
Fig. 5.
Dose dependence of p53-independent cell death. Ninety-six- to 120-hr-old third-instar larvae were irradiated with 1,000, 2,000, or 4,000 R of x-rays. Imaginal discs were dissected at 18 or 24 hr after irradiation and stained with AO to detect dead cells. Only the wing pouch region of wing imaginal discs is shown. Genotypes are indicated for p535A-1-4 (p53) mutants and mei-4129D, p535A-1-4 (mei-41, p53) double mutants. Eleven to 17 discs were examined per genotype per time point.
We found that mutational loss of mei-41 (Drosophila ATR) exacerbated p53-independent cell death. For example, 1,000 R of radiation, a dose that produced little p53-independent cell death, produced robust AO staining in mei-41, p53 double mutants (Fig. 5). The fact that p53-independent cell death occurs in mei-41 mutants indicates that the Drosophila ATR homolog is not required for this response. mei-4129D mutants are deficient in cell-cycle checkpoints and the ability to repair DNA (32). Deficient checkpoints and repair should result in more damage at a given dose of radiation. This phenomenon could explain why mei-41, p53 double mutants show increased p53-independent cell death compared with p53 single mutants at the same radiation dose.
Radiation Sensitivity of chk2, p53, and H99/+ Mutants.
Major cellular responses to IR include the activation of cell-cycle checkpoints, DNA repair, and apoptosis. In previous work, we attempted to dissect the contribution of each response to organismal survival after irradiation. We did so by comparing the radiation sensitivity of mutants that are defective in one or two of the three pathways. For instance, mutants in Drosophila Chk1 (grapes) are defective in cell-cycle checkpoints but survive radiation as well as the WT, leading us to conclude that cell-cycle regulation by checkpoints is dispensable for radiation survival (32).
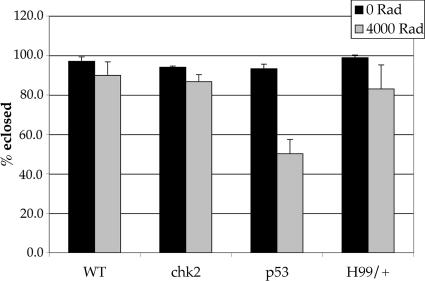
We began to analyze mutants in chk2 and p53 to determine the contribution of apoptosis to radiation survival. However, because chk2 and p53 mutants do exhibit IR-induced apoptosis, as shown here, we cannot unequivocally determine the contribution of this response to survival. Nonetheless, we found that chk2 mutants and H99 heterozygotes survive radiation as well as the WT (Fig. 6). Therefore, neither Chk2-/p53-dependent apoptosis nor the normal timing of apoptosis after irradiation is critical for radiation survival. p53 mutants, however, are moderately radiation-sensitive, with only 50% eclosing after irradiation with 4,000 R. This difference in radiation sensitivity between chk2 and p53 mutants might be explained by differences in the roles that Chk2 and p53 play in regulating transcription in response to irradiation (20).
Fig. 6.
Radiation sensitivity of chk2, p53, and H99/+ mutants. Ninety-six- to 120-hr-old third-instar larvae were irradiated with 4,000 R of x-rays. The number of empty and full pupae cases was scored for up to 10 days after irradiation. Eclosion is the number of empty pupae cases expressed as a percentage of total pupae formed. All irradiated larvae formed pupae in these experiments.
Discussion
Our results show that the Drosophila homologs of Chk2 and p53 are required, not for induction of apoptosis, but for timely induction of apoptosis in response to irradiation. Radiation-induced cell death still occurs in chk2 and p53 mutants, albeit with a delay. Four lines of evidence support the idea that this delayed cell death is apoptosis rather than necrosis: (i) it is detected by staining with AO, which has been shown to stain apoptotic but not necrotic cells (Fig. 1) (27); (ii) it accompanies activation of caspases, a hallmark of apoptosis but not necrosis (Fig. 2 A–F); (iii) it requires caspase activity, which is required for apoptosis but not necrosis (Fig. 2 G–H); and (iv) it requires the chromosomal locus encoding the proapoptosis genes rpr, hid, and grim, whose protein products are required to inhibit DIAP1 and activate caspases (Fig. 3) (30). These results indicate that there is a Chk2-/p53-independent pathway that commits damaged cells to apoptosis and utilizes many of the same downstream components as the Chk2-/p53-dependent apoptosis pathway.
Two lines of evidence presented here support the idea that DNA damage is the signal that induces Chk2-/p53-independent apoptosis after exposure to ionizing radiation. First, the amount of Chk2-/p53-independent apoptosis appears to increase with IR dose. This dose dependence suggests that the amount of DNA damage is what induces Chk2-/p53-independent apoptosis but does not rule out the contribution of other damages that result from IR. Second, we observed higher levels of Chk2-/p53-independent apoptosis when the ability to repair DNA was compromised, as in mei-41, p53 double mutants (Fig. 5). Collectively, these data suggest that DNA damage caused by x-rays induces Chk2-/p53-independent apoptosis.
IR-induced apoptosis in chk2 and p53 mutants shows a temporal delay. IR-induced apoptosis is also delayed in H99 heterozygotes, possibly because H99 heterozygotes contain half the gene dose of the proapoptotic Smac/Diablo orthologs and it may take longer for the proapoptotic gene products to accumulate to the point of an apoptosis-stimulating threshold. We found that IR induced increase in the transcripts of rpr and hid, two of the H99-encoded genes, still occurred in chk2 (rpr and hid) and p53 (hid) mutants, but to lower levels (for rpr) and after a delay. Therefore, apoptosis may be delayed in chk2 and p53 mutants because proapoptotic gene products take longer to accumulate to a threshold in the absence of Chk2 or p53 regulation. Our data showing that IR-induced apoptosis is further delayed in a chk2, H99/+ double mutant, compared with a chk2 single mutant (Fig. 3), support this claim. Furthermore, our results suggest the existence of at least another signaling pathway that does not operate through Chk2 or p53, but nonetheless links the same signal (DNA damage) to a similar outcome (accumulation of H99-encoded gene products).
RT-PCR experiments revealed interesting differences in the identity and onset of induction of proapoptotic genes in chk2 and p53 mutants. rpr and hid are induced at 4 hr after irradiation in chk2 mutants, whereas hid and skl are induced between 12 and 18 hr after irradiation in p53 mutants. We do not understand the basis for these differences. More detailed time courses as well as deletion analysis of the H99 locus to determine the contribution of each proapoptotic gene to Chk2-/p53-independent apoptosis needs to be performed to address these issues.
The data presented here establish D. melanogaster as a model for studying p53-independent apoptosis. p53 is the most commonly mutated gene in human cancers. Loss of p53 poses an immense clinical problem because p53-deficient cancer cells no longer stimulate p53-dependent apoptosis in response to radiation or genotoxic chemotherapy drugs. In this scenario, p53-independent apoptotic pathways become key for inducing cancerous cells to die because they provide potential therapeutic targets. In mammals, a p53-independent apoptosis pathway that is mediated by p73, another member of the p53 family, has been identified (25, 33). In Drosophila, Dmp53 is the only known p53 family member. Therefore, the p53-independent apoptosis we have identified and characterized in this article is likely to represent a previously unknown process. An important goal in the future will be to dissect the Chk2-/p53-independent pathway that links DNA damage to the proapoptotic genes of the H99 locus.
We have tested and eliminated several candidates as regulators of Chk2-/p53-independent cell death. Mei-41 (ATR) is not required for Chk2-/p53-independent cell death because mei-41, p53 double mutants actually exhibit more cell death than p53 alone (Fig. 5). Recent work showed that ectopic induction of eiger, a TNF ligand homolog, can induce apoptosis in Drosophila (20, 34, 35). We found that Chk2-/p53-independent cell death still occurred in p53, eiger double mutants (data not shown), suggesting that the TNF pathway is not involved in the induction of cell death characterized here. Work in mammalian cells showed that overexpression of c-Myc can induce p53-independent apoptosis (36). We found that Chk2-/p53-independent apoptosis still occurred in Dmyc, p53 double mutants (data not shown), indicating that Dmyc is not required for this response.
A classical genetic screen may identify components of the Chk2-/p53-independent apoptosis pathway, as well as testing more candidates, such as the transcription factor de2f1, grapes (DmChk1), DmATM, and genes required for autophagy. Autophagic cell death, in which a cell lyses itself, occurs during Drosophila metamorphosis to lyse polyploid tissues such as the salivary glands and the fat body and provide nutrients for diploid cells of the imaginal discs; autophagy has been described in larvae only in the polyploid cells and only in response to starvation (37–39). Nonetheless, autophagy shares characteristics with apoptosis, including being detectable by AO staining and being dependent on caspases and the H99 locus (37, 40), and for this reason remains a formal possibility.
In conclusion, studies have shown that IR-induced apoptosis in two key models for apoptosis, C. elegans and Drosophila, depends on p53 (20–22, 26). We provide evidence that, contrary to the accepted view, Chk2 and p53 are not required for radiation-induced cell death in Drosophila. Furthermore, normal timing of apoptosis that depends on Chk2 and p53 is also not required for ensuring survival after irradiation. Radiation-induced cell death that is independent of Chk2 and p53 depends on radiation dose, has characteristics of apoptosis and is likely to rely on a novel mechanism(s) because no other members of the p53-family are known in Drosophila. This work is the first to establish a genetically tractable organism as a model for p53-independent apoptosis. Identification of genes required for Chk2-/p53-independent cell death in Drosophila is of potential therapeutic value because protein products of their human homologs may represent novel targets that can be activated clinically to eliminate p53-deficient cancer cells.
Materials and Methods
Fly Stocks.
WT flies were of Sevelin stock. All fly mutants used here have been described before. The p535A-1-4-null allele was generated by targeted deletion (41). The mnkp6 (chk2)-null allele was created by mobilization of a p element in a nearby gene. The lesion includes a small deletion that removes the start codon (20). The H99 deletion has been described before (31). mei-4129D was generated by imprecise excision of a p element in the coding region and is predicted to produce a 39-aa truncated protein (42). Homozygous mei-41 mutants were identified by the lack of GFP encoded by the balancer. The mnkp6, p535A-1-4, mei-4129D, p53, and mnkp6, H99/TM6-Tb double mutants were generated by standard fly crosses. The EnGal4, UAS-GFPS65T T2 and P[UAS-p35.H] BH3 stocks were obtained from L. Johnston (Columbia University, New York) and the Bloomington Stock Center (Indiana University, Bloomington), respectively.
Irradiation.
Embryos were collected for up to 24 hr and aged for 96 hr at 25°C to obtain third-instar larvae. Feeding larvae in food were irradiated by using a TORREX x-ray generator (Astrophysics Research, Long Beach, CA), set at 115 kV and 5 mA (producing 3.18 rad/s).
AO Staining.
Larvae were dissected in PBS, and imaginal discs were incubated for 5 min in PBS plus 0.5 mM AO (Sigma) at room temperature, washed twice with PBS, mounted in PBS, and imaged immediately.
Antibody Staining.
To detect activated caspase 3, larvae were dissected in PBS, fixed for 30 min at room temperature in PBS plus 4% formaldehyde, and washed twice in PBS for 5 min each and once in PBS plus 0.5% Triton X-100 for 10 min. The discs were washed in PBS and blocked for at least 1 hr at room temperature in PBS containing 0.1% Triton X-100 and 5% normal goat serum. Purified rabbit antibody to activated caspase 3 (catalog no. 9661; Cell Signaling, Beverly, MA) was applied at 1:100 in block for 2 hr at room temperature followed by a period overnight at 4°C. Larval discs were rinsed three times in block and incubated with secondary antibody (FITC-conjugated anti-rabbit; The Jackson Laboratory) diluted at 1:500 in block for 2 hr at room temperature. Larval discs were stained with 10 μg/ml Hoechst 33258 in PBS with 0.1% Triton X-100 for 2 min, rinsed five times in PBS with 0.1% Triton X-100, and mounted in Fluoromount G.
Image Acquisition and Quantification.
Images were taken by using a DMR fluorescence compound microscope (Leica, Deerfield, IL), a Sensicam CCD camera, and slidebook software (Intelligent Imaging Innovations, Santa Monica, CA). The AO signal was quantified by using the image j software from the National Institutes of Health (http://rsb.info.nih.gov/ij/).
RT-PCR.
Total RNA was isolated from larval imaginal discs by using the RNeasy kit (Qiagen, Valencia, CA). cDNA was prepared by using the iScript cDNA synthesis kit (Bio-Rad). Primers were designed against the indicated mRNAs by using primer express 2.0 (Applied Biosystems; sequences available on request). PCRs containing 1× SYBR Green Mix (Applied Biosystems), 2.5 ng of cDNA (rpr, hid, grim, and skl), or 0.1 ng of cDNA (actin and α-tubulin), and 500 nM primers were set up and read in a 7900HT RT-PCR instrument (Applied Biosystems). Relative levels in unirradiated and irradiated samples were determined by using a standard curve for each set of primers. The average of actin and α-tubulin controls was used to normalize for differences in cDNA amounts in each experiment.
Viability Assays.
Larvae were collected for 4–6 hr in bottles and aged for 4 days before irradiation. Irradiated or control larvae were allowed to develop into pupae at 25°C. The number of adults that eclosed and the number of empty pupae cases were scored for up to 10 days after irradiation. Percent eclosion is the number of empty pupae cases expressed as a percentage of total pupae formed. All irradiated larvae pupariated in these experiments.
Acknowledgments
We thank Laura Johnston, Joaquin Espinosa, and members of the Su laboratory for critical comments and protocols and Laura Johnston and the Bloomington Stock Center for Drosophila stocks. This work was supported by a National Institutes of Health grant (to T.T.S.).
Abbreviations
- IR
ionizing radiation
- AO
acridine orange
- RT-PCR
real-time PCR.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Vogelstein B., Lane D., Levine A. J. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Bartek J., Lukas J. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 3.Malkin D., Li F. P., Strong L. C., Fraumeni J. F., Jr., Nelson C. E., Kim D. H., Kassel J., Gryka M. A., Bischoff F. Z., Tainsky M. A., et al. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava S., Zou Z. Q., Pirollo K., Blattner W., Chang E. H. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- 5.Bell D. W., Varley J. M., Szydlo T. E., Kang D. H., Wahrer D. C., Shannon K. E., Lubratovich M., Verselis S. J., Isselbacher K. J., Fraumeni J. F., et al. Science. 1999;286:2528–2531. doi: 10.1126/science.286.5449.2528. [DOI] [PubMed] [Google Scholar]
- 6.Takai H., Naka K., Okada Y., Watanabe M., Harada N., Saito S., Anderson C. W., Appella E., Nakanishi M., Suzuki H., et al. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe S. W., Schmitt E. M., Smith S. W., Osborne B. A., Jacks T. Nature. 1993;362:847–849. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 8.Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr., Butel J. S., Bradley A. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 9.Ahn J. Y., Schwarz J. K., Piwnica-Worms H., Canman C. E. Cancer Res. 2000;60:5934–5936. [PubMed] [Google Scholar]
- 10.Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S. J. Proc. Natl. Acad. Sci. USA. 2000;97:10389–10394. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melchionna R., Chen X. B., Blasina A., McGowan C. H. Nat. Cell Biol. 2000;2:762–765. doi: 10.1038/35036406. [DOI] [PubMed] [Google Scholar]
- 12.Chehab N. H., Malikzay A., Appel M., Halazonetis T. D. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- 13.Shieh S. Y., Ahn J., Tamai K., Taya Y., Prives C. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 14.Khanna K. K., Keating K. E., Kozlov S., Scott S., Gatei M., Hobson K., Taya Y., Gabrielli B., Chan D., Lees-Miller S. P., Lavin M. F. Nat. Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 15.Canman C. E., Lim D. S., Cimprich K. A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M. B., Siliciano J. D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 16.Turenne G. A., Paul P., Laflair L., Price B. D. Oncogene. 2001;20:5100–5110. doi: 10.1038/sj.onc.1204665. [DOI] [PubMed] [Google Scholar]
- 17.Schuler M., Green D. R. Biochem. Soc. Trans. 2001;29:684–688. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 18.Urist M., Tanaka T., Poyurovsky M. V., Prives C. Genes Dev. 2004;18:3041–3054. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu J., Xin S., Du W. FEBS Lett. 2001;508:394–398. doi: 10.1016/s0014-5793(01)03103-9. [DOI] [PubMed] [Google Scholar]
- 20.Brodsky M. H., Weinert B. T., Tsang G., Rong Y. S., McGinnis N. M., Golic K. G., Rio D. C., Rubin G. M. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sogame N., Kim M., Abrams J. M. Proc. Natl. Acad. Sci. USA. 2003;100:4696–4701. doi: 10.1073/pnas.0736384100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J. H., Lee E., Park J., Kim E., Kim J., Chung J. FEBS Lett. 2003;550:5–10. doi: 10.1016/s0014-5793(03)00771-3. [DOI] [PubMed] [Google Scholar]
- 23.Peters M., DeLuca C., Hirao A., Stambolic V., Potter J., Zhou L., Liepa J., Snow B., Arya S., Wong J., et al. Proc. Natl. Acad. Sci. USA. 2002;99:11305–11310. doi: 10.1073/pnas.172382899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodsky M. H., Nordstrom W., Tsang G., Kwan E., Rubin G. M., Abrams J. M. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 25.Irwin M., Marin M. C., Phillips A. C., Seelan R. S., Smith D. I., Liu W., Flores E. R., Tsai K. Y., Jacks T., Vousden K. H., Kaelin W. G., Jr. Nature. 2000;407:645–648. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 26.Schumacher B., Hofmann K., Boulton S., Gartner A. Curr. Biol. 2001;11:1722–1727. doi: 10.1016/s0960-9822(01)00534-6. [DOI] [PubMed] [Google Scholar]
- 27.Abrams J. M., White K., Fessler L. I., Steller H. Development (Cambridge, U.K.) 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- 28.Yu S. Y., Yoo S. J., Yang L., Zapata C., Srinivasan A., Hay B. A., Baker N. E. Development (Cambridge, U.K.) 2002;129:3269–3278. doi: 10.1242/dev.129.13.3269. [DOI] [PubMed] [Google Scholar]
- 29.Yoo S. J., Huh J. R., Muro I., Yu H., Wang L., Wang S. L., Feldman R. M., Clem R. J., Muller H. A., Hay B. A. Nat. Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 30.Goyal L., McCall K., Agapite J., Hartwieg E., Steller H. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White K., Grether M. E., Abrams J. M., Young L., Farrell K., Steller H. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 32.Jaklevic B. R., Su T. T. Curr. Biol. 2004;14:23–32. doi: 10.1016/j.cub.2003.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jost C. A., Marin M. C., Kaelin W. G., Jr. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 34.Igaki T., Kanda H., Yamamoto-Goto Y., Kanuka H., Kuranaga E., Aigaki T., Miura M. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno E., Yan M., Basler K. Curr. Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 36.Trudel M., Lanoix J., Barisoni L., Blouin M. J., Desforges M., L’Italien C., D’Agati V. J. Exp. Med. 1997;186:1873–1884. doi: 10.1084/jem.186.11.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee C. Y., Baehrecke E. H. Development (Cambridge, U.K.) 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 38.Rusten T. E., Lindmo K., Juhasz G., Sass M., Seglen P. O., Brech A., Stenmark H. Dev. Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Scott R. C., Schuldiner O., Neufeld T. P. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Martin D. N., Baehrecke E. H. Development (Cambridge, U.K.) 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 41.Rong Y. S., Titen S. W., Xie H. B., Golic M. M., Bastiani M., Bandyopadhyay P., Olivera B. M., Brodsky M., Rubin G. M., Golic K. G. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurencon A., Purdy A., Sekelsky J., Hawley R. S., Su T. T. Genetics. 2003;164:589–601. doi: 10.1093/genetics/164.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]