Abstract
The mechanisms of malignant cell transformation mediated by the oncogenic, chimeric nucleophosmin/anaplastic lymphoma kinase (NPM/ALK) tyrosine kinase remain only partially understood. Here we report that the NPM/ALK-carrying T cell lymphoma (ALK+TCL) cells secrete IL-10 and TGF-β and express FoxP3, indicating their T regulatory (Treg) cell phenotype. The secreted IL-10 suppresses proliferation of normal immune, CD3/CD28-stimulated peripheral blood mononuclear cells and enhances viability of the ALK+TCL cells. The Treg phenotype of the affected cells is strictly dependent on NPM/ALK expression and function as demonstrated by transfection of the kinase into BaF3 cells and inhibition of its enzymatic activity and expression in ALK+TCL cells. NPM/ALK, in turn, induces the phenotype through activation of its key signal transmitter, signal transducer and activator of transcription 3 (STAT3). These findings identify a mechanism of NPM/ALK-mediated oncogenesis based on induction of the Treg phenotype of the transformed CD4+ T cells. These results also provide an additional rationale to therapeutically target the chimeric kinase and/or STAT3 in ALK+TCL.
Keywords: cell signaling, immune response evasion, oncogenesis
T cell lymphomas (TCL) that express anaplastic lymphoma kinase (ALK) represent a distinct lymphoma category. Ectopic expression of ALK in the affected CD4+ T lymphocytes results from various chromosomal translocations involving ALK gene and several different partners, most frequently the nucleophosmin (NPM) gene (1). The NPM/ALK chimeric protein is constitutively active (2, 3) and displays cell-transforming properties as demonstrated both in vitro (4, 5) and in vivo (6, 7). NPM/ALK mediates its oncogenic function by activating a number of key cell-signaling proteins including signal transducer and activator of transcription 3 (STAT3) (8–10).
Interleukin 10 (IL-10) is a well-characterized cytokine with multiple, primarily immunosuppressive activities (11). IL-10 seems to exert its immunosuppressive effect on several levels, mainly by inhibiting function of antigen-presenting cells (12) and T lymphocytes including CD4+ T helper cells from the type 1 (Th1) subcategory (13). IL-10 does so, among other mechanisms, by interfering with expression of MHC class II and costimulatory proteins, as well as synthesis and activity of several cytokines and chemokines. IL-10-mediated inhibition of immune response toward malignant cells seems to play the key role in carcinogenesis (14).
T regulatory (Treg) lymphocytes capable of down-regulating immune response have gained considerable attention in the recent years (15, 16). This functional subset of CD4+ T cells strongly expresses α chain (CD25) of the receptor for IL-2. Whereas some of the Treg cells develop during maturation process in the thymus, the other acquire the Treg phenotype as postthymic, fully mature cells because of an antigenic stimulation (15, 16). Besides secreting IL-10, a hallmark feature of the antigen-induced Treg lymphocytes (17), these immunoregulatory T cells secrete TGF-β (18) and, in the case of the preselected “natural” Treg cells, characteristically express FoxP3 protein (15, 16). The FoxP3 expression may, however, be induced also in the mature T cells by either TGF-β (19) or CD3/CD28 (20) stimulation. The wealth of experimental evidence indicates that Treg lymphocytes play a role in cancer by suppressing immune response toward the malignant cells. An increased frequency of Treg cells in the peripheral blood, draining lymph nodes, and tumor tissues, as well as the enhanced functional activity of Treg cells, has been observed in tumors as diverse as colorectal carcinoma, melanoma, ovarian carcinoma, and Hodgkin lymphoma (21–25). Malignant CD4+ T lymphocytes can also display features of Treg cells, either constitutively, as is the case in the human T cell lymphotropic virus (HTLV)-related adult-type lymphoma/leukemia (ATLL) (26, 27), or after in vitro cell stimulation, as demonstrated in cutaneous T cell lymphoma (CTCL) (28).
Here we report that ALK+TCL cells secrete IL-10 and TGF-β and express FoxP3 mRNA. The expression of IL-10, TGF-β, and FoxP3 is induced by the chimeric NPM/ALK tyrosine kinase itself through activation of STAT3. These findings identify a function of NPM/ALK and STAT3. They also provide a rationale to target therapeutically both these molecules in ALK+TCL.
Results
ALK+TCL Cells Express IL-10.
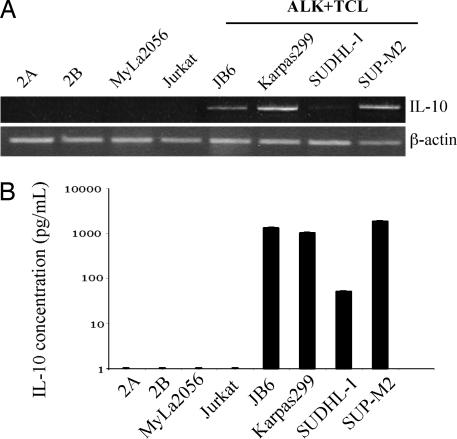
IL-10 has been suggested to play a key role in inhibition of immune response against malignant cells (13, 14). To determine whether ALK+TCL cells produce this cytokine, we examined by RT-PCR expression of IL-10 mRNA in four ALK+TCL-derived cell lines. As shown in Fig. 1A, all of the cell lines expressed the IL-10 message, albeit the signal generated by one of the cell lines, SUDHL-1, was reproducibly weaker than in the remaining cell lines, suggesting lower concentration of the mRNA. No IL-10 mRNA could be detected in four other ALK−TCL cell lines, indicating correlation between the expression of ALK and IL-10.
Fig. 1.
IL-10 expression by ALK+TCL cells. IL-10 expression in ALK+TCL cell lines (JB6, Karpas299, SUDHL-1, and SUP-M2), T cell lymphoma cell lines derived from lymphomas involving skin (2A, 2B, and MyLa2056), and T cell lymphoblastic lymphoma (Jurkat) detected at the mRNA level by RT-PCR (A) and the protein level by EIA (B).
To determine whether the ALK+TCL cells synthesize and secrete the IL-10 protein, we examined by enzyme immunoassay (EIA) the cell line supernatants. As depicted in Fig. 1B, the supernatants from three ALK+TCL cell lines contained as much as 1,000 pg/ml of IL-10; SUDHL-1 also secreted IL-10, although in the >10-fold lower amount. No IL-10 protein was detected in the supernatant from any of the four ALK−TCL cell lines examined.
ALK+TCL-Derived IL-10 Displays Immunosuppressive Properties and Enhances Viability of the ALK+TCL Cells.
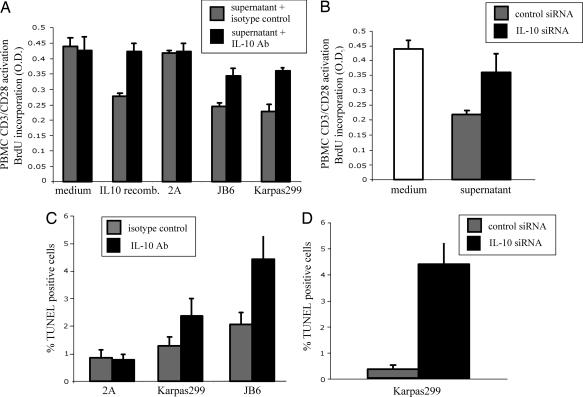
To determine whether the IL-10 secreted by the ALK+TCL cells is immunosuppressive, we examined the effect of the IL-10-containing supernatants on proliferation of the normal, T cell rich peripheral blood mononuclear cells (PBMCs) activated by the combination of bead-immobilized anti-CD3 and anti-CD28 antibodies that potently activate T lymphocytes. As shown in Fig. 2A, recombinant IL-10, as well as the supernatants from two ALK+TCL cell lines that secreted high amounts of IL-10, inhibited by >45% proliferation of the CD3/CD28 antibody-stimulated PBMCs. In contrast, the IL-10-free supernatant from the control 2A CTCL cell line had no inhibitory effect. Pretreatment of the supernatants with a blocking anti-IL-10 antibody completely inhibited the immunosuppressive activity of the recombinant IL-10 and markedly, but not completely, inhibited the immunosuppressive activity of the ALK+TCL cell supernatants. These findings indicate that IL-10 is the main, but not the only immunosuppressive, factor present in the supernatants.
Fig. 2.
ALK+TCL-derived IL-10 is immunosuppressive and enhances survival of the ALK+TCL cells. (A) Anti-IL-10 antibody inhibits immunosuppressive activity of the ALK+TCL cell supernatants. Inhibitory effect of the ALK+TCL cell line (JB6 and Karpas299) supernatants pretreated with the anti-IL-10 antibody or the isotype-matched antibody on proliferation of the T cell rich PBMCs stimulated by the anti-CD3/anti-CD28 antibody tandem. Medium alone, medium with added recombinant IL-10, and a supernatant from the IL-10 nonsecreting 2A ALK−TCL cell line served as additional controls. (B) siRNA-mediated depletion of IL-10 inhibits immunosuppressive activity of the ALK+TCL cell supernatants. Inhibitory effect of the supernatant from the IL-10 siRNA- vs. nonsense siRNA-treated Karpas299 cells on proliferation of the anti-CD3/anti-CD28 antibody-stimulated PBMCs. (C) Proapoptotic effect of the anti-IL-10 antibody on the ALK+TCL cells. Viability of the Karpas299 and JB6 cell lines cultured in the presence of the anti-IL-10 or isotype-matched antibody was determined by the DNA fragmentation (TUNEL) assay. ALK−TCL 2A cell line served as an additional control. (D) Proapoptotic effect of the IL-10 siRNA on the ALK+TCL cells. Viability of the Karpas299 cell line treated with IL-10-specific or nonsense siRNA as determined by the TUNEL assay.
To provide additional evidence that IL-10 is the key immunosuppressive component of the ALK+TCL-derived supernatants, we inhibited IL-10 expression in one of the high IL-10 secreting cell lines, Karpas299, by using IL-10-specific small interfering RNA (siRNA). The treatment decreased the amount of the secreted IL-10 by ≈80% as determined by measuring concentration of the cytokine in the supernatants as compared with untreated cells or cells treated with a nonspecific siRNA (data not presented). As shown in Fig. 2B Right, the supernatants of the IL-10 siRNA-treated cells were also much less effective as compared with the nonspecific siRNA-treated cells in inhibiting proliferation of the normal, CD3/CD28 antibody-stimulated PBMCs.
Apart from its well-established immunosuppressive role, IL-10 displays a plethora of other biological functions (11). One of them is the ability to protect T lymphocytes from apoptotic cell death (29). We decided, therefore, to examine whether the secreted IL-10 has a direct beneficial impact on the ALK+TCL cells, given their T cell derivation. We addressed this question by determining first whether the ALK+TCL cells express on their surface IL-10 receptor. All seven TCL cell lines examined, both ALK+ and ALK−, expressed IL-10 receptor (data not presented), suggesting that IL-10 may impact on malignant T cells and, in the case of ALK+TCL, may do so by creating an autocrine loop. Furthermore, addition of the anti-IL-10 antibody to the ALK+TCL cell cultures roughly doubled the spontaneous apoptotic cell rate as compared with the isotype-matched control antibody (Fig. 2C). A similar, if not even more pronounced, effect was observed in the ALK+TCL Karpas299 cells upon their transfection with IL-10 siRNA (Fig. 2D), supporting the conclusion that the endogeneous IL-10 enhances survival of the ALK+TCL cells. In contrast to the cell viability, no clear impact of IL-10 suppression on proliferation of the ALK+TCL cells could be appreciated (data not presented).
ALK+TCL Cells Display Treg Phenotype.
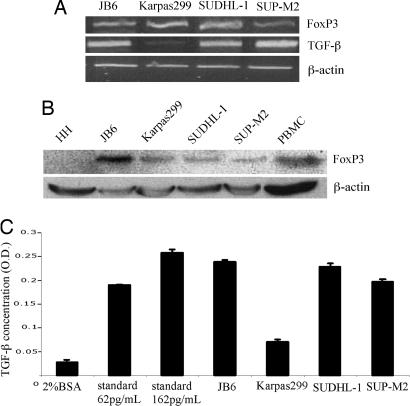
Because expression of IL-10 is characteristic for Treg lymphocytes, we examined the ALK+TCL cells for other hallmarks of this T cell subset. We began with flow cytometry analysis of CD4 and CD25 expression. Whereas we could detect CD4 in only two of the four ALK+TCL cell lines (Karpas299 and SUP-M2), all expressed CD25 with two expressing the protein very strongly (SUDHL-1 and SUP-M2; data not presented). As shown in Fig. 3A, all four ALK+TCL cell lines expressed mRNA coding for FoxP3 and TGF-β with Karpas299 expressing the latter message rather weakly. We also confirmed expression of FoxP3 and TGF-β on the protein level by Western blotting (Fig. 3B) and EIA (Fig. 3C), respectively.
Fig. 3.
ALK+TCL cells display Treg cell phenotype. Expression of mRNA coding for FoxP3 and TGF-β (A) and FoxP3 protein (B) and the secreted TGF-β protein (C) by the ALK+TCL lines.
Expression of IL-10, TGF-β, and FoxP3 Is Induced by NPM/ALK.
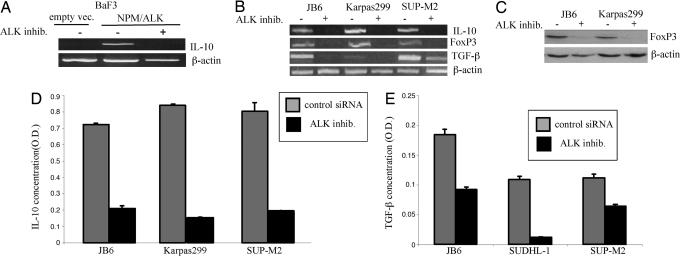
One could argue that ALK+TCL cells gained the ability to express IL-10 and the other features of the Treg phenotype prior or, alternatively, as a result of transformation by the NPM/ALK oncoprotein. To address the potential role of NPM/ALK in induction of the Treg phenotype, we examined lymphoid BaF3 cells transfected with a vector containing the NPM/ALK gene or the control, an empty vector (8). Although the parental BaF3 cells expressed TGF-β, they were negative for IL-10 (data not presented). As shown in Fig. 4A, transfection with NPM/ALK, but not vector alone, induced expression of the IL-10 mRNA. Notably, treatment of the NPM/ALK-expressing BaF3 cells with the small-molecule kinase inhibitor WHI-P154, recently identified by us as being able to suppress enzymatic activity of ALK (10), abrogated the expression of the IL-10 mRNA confirming the key role of NPM/ALK in promoting activation of the IL-10 gene.
Fig. 4.
Expression of IL-10, FoxP3, and TFG-β are induced by NPM/ALK expression and enzymatic activity. (A) Expression of IL-10 mRNA in BaF3 cells transfected with empty vector and NPM/ALK before and after treatment with an ALK inhibitor WHI-P154 (10). (B) Expression of IL-10, FoxP3, and TGF-β mRNA in the depicted ALK+TCL cell lines before and after treatment with the ALK inhibitor. (C) Expression of FoxP3 protein in the ALK+TCL cell lines before and after treatment with the ALK inhibitor. (D) Concentration of IL-10 in the ALK+TCL cell line supernatant before and after cell treatment with the ALK inhibitor. (E) Concentration of TGF-β in the ALK+TCL cell line supernatant before and after cell treatment with the ALK inhibitor.
To determine whether blocking NPM/ALK activity suppresses the Treg phenotype also in the ALK+TCL cells, we used the ALK inhibitor in three different ALK+TCL cell lines. As shown in Fig. 4B, inhibition of the NPM/ALK function suppressed expression of not only the IL-10 gene but also of the FoxP3 and TGF-β genes. In addition, we demonstrated the ALK inhibitor-mediated suppression of the FoxP3, IL-10, and TGF-β expression also on the protein level (Fig. 4 C–E, respectively).
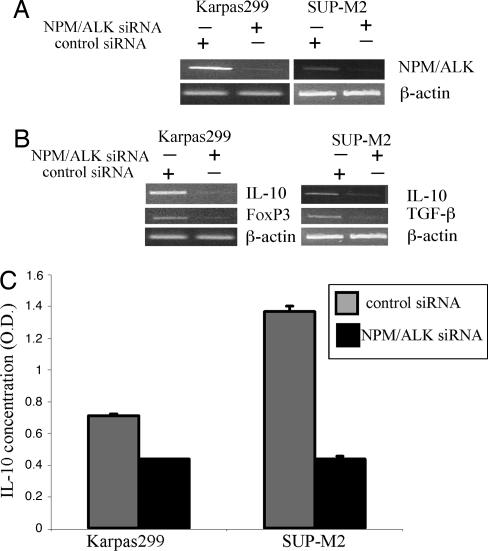
To provide the third independent piece of evidence that the Treg phenotype is induced by NPM/ALK, we performed siRNA-mediated depletion of NPM/ALK in two ALK+TCL cell lines, Karpas299 and SUP-M2. As shown in Fig. 5A, we achieved marked decrease in the NPM/ALK mRNA expression by this approach. The NPM/ALK depletion was associated with loss of IL-10, FoxP3, and TGF-β mRNA (Fig. 5B), as well as the IL-10 protein (Fig. 5C).
Fig. 5.
siRNA-mediated NPM/ALK depletion results in loss of the Treg phenotype. Two ALK+TCL cell lines, Karpas299 and JB6, were treated with the NPM/ALK-specific or control, nonsense siRNA and evaluated for expression of NPM/ALK (A), IL-10, FoxP3, and TGF-β (B), and the secreted IL-10 protein (C).
Induction of IL-10 Expression by NPM/ALK Is Mediated by STAT3.
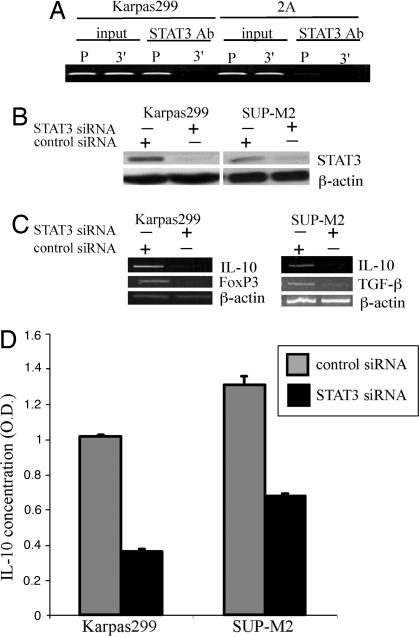
Because the NPM/ALK oncogenic kinase activates several signaling pathways (1, 30), we decided to examine which of these pathways is directly responsible for activation of transcription of the Treg phenotype-related genes. It has been shown by other investigators that STAT3 binds to a specific motif in the IL-10 gene promoter and is critical for activation of the gene by LPS (31) and IFN-α (32). Because NPM/ALK activates STAT3 (8, 10), we examined whether STAT3 is involved in activation of IL-10 gene in the ALK+TCL cells. Using a chromatin immunoprecipitation (ChIP) assay, we demonstrated that STAT3 binds very strongly to the IL-10 gene promoter in the IL-10 expressing ALK+TCL-derived Karpas299 cells but very weakly in the IL-10 nonexpressing CTCL-derived 2A cells (Fig. 6A). To demonstrate that STAT3 is functionally critical for the IL-10 expression, we depleted STAT3 (Fig. 6B) with the specific siRNA. STAT3 depletion resulted in the profoundly diminished expression of not only IL-10 mRNA (Fig. 6C) and protein (Fig. 6D) but also inhibited expression of the genes coding for FoxP3 and TGF-β (Fig. 6C). These findings identify STAT3 as the key effector of the NPM/ALK-mediated induction of the Treg phenotype in the T cells transformed by the kinase.
Fig. 6.
NPM/ALK induces the Treg phenotype through STAT3. (A) Binding of STAT3 to the IL-10 gene promoter. Protein cell lysates from the IL-10-expressing, ALK+TCL cell line Karpas299 and ALK−TCL cell line 2A were analyzed in the chromatin immunoprecipitation (ChIP) assay by using an anti-STAT3 antibody and primer pairs specific for promoter (P) and 3′ end (3′) of the IL-10 gene. Nonimmunoprecipitated lysates (input) served as positive controls. (B) Effect of the STAT3 siRNA on STAT3 expression. Two ALK+TCL cell lines, Karpas299 and SUP-M2, were treated with STAT3-specific and control, nonspecific siRNA and evaluated for expression of the STAT3 protein. (C) Effect of the siRNA-mediated STAT3 depletion on the Treg phenotype. Karpas299 (Left) and SUP-M2 (Right) were treated with STAT3-specific and nonspecific siRNA and evaluated for expression of the IL-10, FoxP3, and TGF-β mRNA. (D) Effect of the siRNA-mediated STAT3 depletion on expression of the secreted IL-10. Supernatants form the Karpas299 and SUP-M2 pretreated with STAT3 and nonspecific siRNA were analyzed for IL-10 concentration in an EIA assay.
Discussion
Here we report that ALK+TCL cells secrete IL-10 and TGF-β and express FoxP3. These features combined with derivation of the ALK+TCL cells from CD4+ T lymphocytes and expression of CD25, are hallmarks of Treg cells. Further, multifaceted analysis revealed that the Treg phenotype is conferred on the malignant cells by the chimeric NPM/ALK tyrosine kinase, whose expression resulting from the chromosomal translocation represents the critical event in the lymphoma pathogenesis (4–7, 10). Finally, we showed that NPM/ALK induces the Treg phenotype by activating its key effector, STAT3. These findings identify a function for NPM/ALK as an inducer of evasion of the immune response, document the central role of STAT3 in conferring on the ALK+TCL cells the Treg phenotype, and provide a rationale to therapeutically target NPM/ALK and STAT3.
Expression of the Treg phenotype has been recently postulated for two other malignancies derived from the mature CD4+ T cells: human T cell lymphotropic virus (HTLV)-related ATLL and CTCL. Accordingly, ATLL cells are nonresponsive to a mitogenic (ConA) stimulation and suppress proliferation of the normal T cells (26). They also universally and strongly express CD25 and frequently and relatively weakly express FoxP3 (33). HTLV-1 is likely responsible for induction of the Treg phenotype in the ATLL cells because prolonged exposure to the virus induced in the normal CD4+ T cells expression of CD25, FoxP3, and TGF-β (34). In turn, induction of the Treg phenotype in CTCL cells requires in vitro stimulation by the autologous dendritic cells loaded with apoptotic CTCL cells in the presence of four different cytokines GM-CSF, IL-2, IL-7, and IL-4 (28). This requirement indicates that the Treg phenotype is acquired rather than intrinsic to the CTCL cells. Furthermore, the exact mechanisms of the Treg phenotype induction in the CTCL and ATLL cells remain undefined.
Our findings indicate that ALK+TCL cells also display the Treg phenotype. Importantly, the phenotype is induced by the cell-transforming NPM/ALK tyrosine kinase. It seems remarkable that NPM/ALK is able to induce the Treg phenotype because, under physiological conditions, expression of ALK is expressed only by neural cells (1). This finding suggests that in the affected T cells, the aberrant NPM/ALK hybrid protein is capable of activating the intracellular cell signaling pathways that normally are involved in induction of the Treg differentiation. Considering that STAT3 which, as we show here, is instrumental for the Treg phenotype can be activated by a variety of quite diverse tyrosine kinases (35), it is perhaps less surprising that NPM/ALK is able to promote the Treg skewing. One might also argue that by the virtue of secreting IL-10, the ALK+TCL cells resemble more the induced, postthymic CD4+ T cell-derived Tregs rather than the thymus-programmed natural Tregs (15, 16, 17). Their ability to express FoxP3 does not contradict this conclusion because FoxP3 can be induced in the postthymic, fully mature CD4+ T cells by various stimuli (19, 20). It is also interesting that IL-10 secreted by the ALK+TCL cells simultaneously displays immunosuppressive (Fig. 2 A and B) and anti-apoptotic (Fig. 2 D and E) effects. This double activity is in agreement with the pleiotropic properties of the cytokine, including the ability to protect T lymphocytes from apoptotic cell death by up-regulating bcl-2 (29). Our data indicate that IL-10 synthesis benefits ALK+TCL cells both directly by enhancing their viability and indirectly by suppressing the immune response.
Our findings that the constitutively active NPM/ALK tyrosine kinase induces the Treg phenotype provide a rationale for therapeutically targeting the kinase (10, 36). As we show, depletion or inactivation of NPM/ALK inhibits expression of IL-10, TGF-β, and FoxP3, indicating the reversal of the Treg phenotype. It is interesting in this regard, that some degree of anti-lymphoma immune response, both humoral (37) and cellular (38), does develop in ALK+TCL patients with the NPM/ALK protein representing the stimulating antigen. One can only speculate that pharmacologic inhibition of NPM/ALK activity and the resulting abrogation of the Treg phenotype may boost the immune response and, possibly, open the door for an effective immunotherapy in ALK+TCL.
Our findings also define a role of STAT3 in the pathogenesis of ALK+TCL and, possibly, other malignancies. Persistently activated STAT3 has been identified in a large spectrum of cancers (35). The critical role of STAT3 in oncogenesis has recently been recently documented in vivo in the mouse models of cancers of skin (39) and breast (40) and, notably in the context of this report, ALK+TCL (41). STAT3 mediates its tumor-promoting function by regulating expression of key proteins involved in cancer cell survival, proliferation, and induction of angiogenesis. Interestingly, STAT3 has also been implicated in down-regulation of immune response in solid tumors by indirectly inhibiting activation of tumor-infiltrating antigen-presenting cells (42) and directly inducting anergy in such cells (43). However, the exact molecular mechanisms of this down-regulation remain undefined. Here we show that STAT3 is capable of inducing the Treg phenotype in the malignant CD4+ T lymphocytes by activating expression of genes that encode such Treg effector proteins as IL-10 and TGF-β. Whether similar immunosuppressive factors are produced in the other types of tumors in which STAT3 is persistently activated and oncogenic remains to be determined.
Finally, one could argue that STAT3 also represents in ALK+TCL cells an attractive therapeutic target (44). Although progress in development of inhibitors that interfere with protein-protein interactions has been rather slow, peptidomimetic small-molecule STAT3 inhibitors that impair STAT3 dimerization have been synthesized (45), suggesting that clinically suitable anti-STAT3 compound(s) might also be developed.
Materials and Methods
Malignant and Normal Cell Populations.
Most cell lines used in this study were described (8, 10, 29). In brief, PB-1, 2A, and 2B cell lines were established from a patient with a progressive CTCL. MyLa2056 was derived from advanced CTCL. SUDHL-1, JB6, Karpas299, and SUP-M2 lines were derived from ALK+TCL. Jurkat was developed from lymphoblastic T cell lymphoma. PBMCs from healthy individuals were obtained by centrifugation on Ficoll/Paque gradient. HH cell line was established form normal PBMCs by infection with an Epstein–Barr virus. BaF3 is an IL-3-dependent murine pro-B-cell lymphoblastic cell line.
ALK Inhibitor Treatment.
Cells were treated with 14 μM of WHI-P154 (Calbiochem) for the time periods indicated in Results.
RT-PCR.
Total RNA was isolated by using RNeasy Mini kit (Qiagen), treated with DNase I (Invitrogen), and reverse-transcribed by using Thermoscript RT-PCR system (Invitrogen) with random hexamers as cDNA synthesis primers. The following primer pairs were used for the cDNA amplification: β-actin, 5′ACCATTGGCAATGAGCGGT and 5′GTCTTTGCGGATGTCCACGT; NPM/ALK, 5′GACAGGCCCAACTTTGCCAT-CATT and 5′TGTTTCTGGATCCGTGGACCTTG; human IL-10, 5′GACAACTT-GTTGTTAAAGGAG and 5′CTAATTTATGTCCTAGAGTCT; human TGF-β, 5′GACCAGTGGGGAACACTACTG and 5′GTAGCTGGGACCACAGGTGTA; human FoxP3, 5′TCTTCAGAAACCATCCTGCCACCT and 5′AGGCAAGACAGT-GGAAACCTCACT; mouse IL-10, 5′CATGCTCCTAGAGCTGCGGAC and 5′CAGACTCAATACAGACTGCAG; mouse TGF-β, 5′TGATACGCCTGAGTGGC-TGTCTTT and 5′TGTACTGTGTGTCCAGGCTCCAAA; and mouse FOXP3, 5′TCAAAGAGCCCTC-ACAACCAGCTA and 5′TTGTGAAGGTTCCAGTGCT-GTTGC. PCR was performed by using Platium TaqDNA polymerase (Invitrogen) for 25 (β-actin) and 30 cycles (NPM/ALK, IL-10, TGF-β, and FoxP3) comprised of the denaturation step for 20 s at 94°C, annealing for 30 s at 58°C and elongation for 30 s at 72°C. The PCR products were visualized by ethium bromide staining in 2% agarose gel.
Human IL-10 EIA.
Concentration of IL-10 in culture supernatants was evaluated by using Quantikine Human IL-10 kit (R & D Systems). In brief, 200 μl per well of sample or standard was added to a plate precoated with IL-10 antibody. After 2-h incubation and washing, 200 μl of the IL-10 conjugate was added to each well. After 1-h incubation and washing, the concentration of IL-10 was determined by colorimetric conversion of the substrate and optical density (OD) determination using the EIA plate reader.
Human TGF-β EIA.
Cells were cultured in an RPMI medium 1640 supplemented with 2% of BSA for 4 h. Concentration of TGF-β in the harvested supernatants was evaluated by using Quantikine human TGF-β kit (R & D Systems). In brief, the latent TGF-β was activated to immunoreactive form by incubating 0.5 ml of supernatant with 0.1 ml of 1 M HCl. After 10 s, the samples were neutralized with 0.1 ml of 1.2 M NaOH/0.5 M Hepes. Next, 200 μl per well of the samples or standard were added to a plate precoated with the TGF-β antibody. After 3-h incubation and washing, 200 μl per well of TGF-β conjugate were to each well for 1.5 h. The concentration of TGF-β was determined by the OD determination.
Immunofluorescence Staining.
Cell surface protein expression was performed by using the conjugated antibodies CD25-FITC, CD4-PE, and IL-10R-PE (BD PharMingen) and flow cytometry (FACSort; BD Biosciences) by using cellquest pro software.
Western Blot.
These experiments were performed as described (8, 10). In brief, the cells were lysed in a buffer supplemented with phenylmethylsulfonyl fluoride, phosphatase inhibitor cocktails I and II (Sigma), and protease inhibitor mixture (Roche). For normalization of the gel loading, the protein extracts were assayed with Lowry method (Bio-Rad Dc protein assay). Cell lysates were separated on a 10% polyacrylamide/SDS gel, and transferred to poly(vinylidene difluoride) (Amersham Pharmacia) membranes. Proteins were detected with antibodies against ALK, β-actin, Stat3 (Santa Cruz Biotechnology), FoxP3 (eBioscience, San Diego, CA) and the secondary, peroxidase-conjugated antibodies (Santa Cruz Biotechnology). Blots were developed by using the SuperSignal West Dura nylon membranes (Pierce) and exposed to x-ray film (Kodak).
siRNA.
SUP-M2 and Karpas299 cell lines were transfected with 100 nM siRNA specific for STAT3 or nontargeting control twice at 24-h intervals and with NPM/ALK siRNA (all siRNA were from Dharmacon) either twice (SUP-M2) or three times (Karpas299) using lipofectamine (DMRIE-C; Invitrogen). Efficiency of the NPM/ALK and STAT3 suppression was assessed by Western blotting or RT-PCR (NPM/ALK).
In Vitro Proliferation Assay.
Bromodeoxyuridine (BrdUrd) incorporation was determined by using the Cell Proliferation ELISA kit (Roche) based on the colorimetric method as described (10). In the case of the CD3/CD28 stimulation, PBMCs from healthy blood donors were stimulated with beads coated with anti-CD3 and -CD28 antibodies.
Apoptotic Cell Death Assay.
Tunel staining was performed by using the ApoAlert DNA fragmentation assay kit from BD Biosciences (10). In brief, 3–5 × 106 cells were fixed with 1% formaldehyde/PBS, permeabilized with 70% ethanol, and incubated in the TdT buffer for 1 h at 37°C. After stopping the reaction with 20 mM EDTA, the cells were analyzed by flow cytometry.
Chromatin Immunoprecipitation (ChIP) Assay.
Soluble chromatin-containing lysates obtained from formaldehyde-fixed and sonicated cells were incubated with STAT3 antibody (Santa Cruz Biotechnology). DNA-protein immunocomplexes were precipitated with protein A-agarose beads and treated with RNase A and proteinase K; the DNA samples were extracted with phenol/chloroform, precipitated with ethanol, and PCR amplified using primers specific for IL-10 gene promoter (5′TGCTTACGATGCAAAAATTGA and 5′CTCAGGGAGGCCTCTTCAT) and, as a control, the 3′ end of the IL-10 gene (5′CCTCAGCCTCCCAAGTAGCTG and 5′ATCACTTGAGGCCAGGAGTTC3′).
Acknowledgments
This work was supported in part by National Cancer Institute Grants R01-CA89194 and R01-CA96856.
Abbreviations
- ALK
anaplastic lymphoma kinase
- ATLL
adult-type lymphoma/leukemia
- CTCL
cutaneous T cell lymphoma
- EIA
enzyme immunoassay
- IL-10
interleukin 10
- NPM
nucleophosmin
- PBMCs
peripheral blood mononuclear cells
- siRNA
small interfering RNA
- STAT3
signal transducer and activator of transcription 3
- TCL
T cell lymphoma
- Treg
T regulatory.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Wasik M. A. Am. J. Clin. Pathol. 2002;118:S81–S92. doi: 10.1309/YCRV-7H6U-6E8B-95GU. [DOI] [PubMed] [Google Scholar]
- 2.Shiota M., Fujimoto J., Semba T., Satoh H., Yamamoto T., Mori S. Oncogene. 1994;9:1567–1574. [PubMed] [Google Scholar]
- 3.Morris S. W., Naeve C., Mathew P., James P. L., Kirstein M. N., Cui X., Witte D. P. Oncogene. 1997;14:2175–2188. doi: 10.1038/sj.onc.1201062. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto J., Shiota M., Iwahara T., Seki N., Satoh H., Mori S., Yamamoto T. Proc. Natl. Acad. Sci. USA. 1996;93:4181–4186. doi: 10.1073/pnas.93.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischof D., Pulford K., Mason D. Y., Morris S. W. Mol. Cell. Biol. 1997;17:2312–2325. doi: 10.1128/mcb.17.4.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuefer M. U., Look A. T., Pulford K., Behm F. G., Pattengale P. K., Mason D. Y., Morris S. W. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- 7.Chiarle R., Gong J. Z., Guasparri I., Pesci A., Cai J., Liu J., Simmons W. J., Dhall G., Howes J., Piva R., et al. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q., Raghunath P. N., Xue L., Majewski M., Carpentieri D. F., Odum N., Morris S., Skorski T., Wasik M. A. J. Immunol. 2002;168:466–474. doi: 10.4049/jimmunol.168.1.466. [DOI] [PubMed] [Google Scholar]
- 9.Bacchiocchi R., Baldanzi G., Carbonari D., Capomagi C., Colombo E., van Blitterswijk W. J., Graziani A., Fazioli F. Blood. 2005;15:2175–2182. doi: 10.1182/blood-2005-01-0316. [DOI] [PubMed] [Google Scholar]
- 10.Marzec M., Kasprzycka M., Ptasznik A., Wlodarski P., Zhang Q., Odum N., Wasik M. A. Lab. Invest. 2005;85:1544–1554. doi: 10.1038/labinvest.3700348. [DOI] [PubMed] [Google Scholar]
- 11.Pestka S., Krause C. D., Sarkar D., Walter M. R., Shi Y., Fisher P. B. Annu. Rev. Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- 12.Wakkach A., Fournier N., Brun V., Breittmayer J. P., Cottrez F., Groux H. Immunity. 2003;18:605–617. doi: 10.1016/s1074-7613(03)00113-4. [DOI] [PubMed] [Google Scholar]
- 13.Taga K., Mostowski H., Tosato G. Blood. 1993;81:2964–2971. [PubMed] [Google Scholar]
- 14.Gamero A. M., Young H. A., Wiltrout R. H. Cancer Cell. 2004;5:111–112. doi: 10.1016/s1535-6108(04)00028-5. [DOI] [PubMed] [Google Scholar]
- 15.O’Garra A., Vieira P. Nat. Med. 2004;10:801–805. doi: 10.1038/nm0804-801. [DOI] [PubMed] [Google Scholar]
- 16.Chattopadhyay S., Chakraborty N. G., Mukherji B. Cancer Immunol. Immunother. 2005;54:1153–1161. doi: 10.1007/s00262-005-0699-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vieira P. L., Christensen J. R., Minaee S., O’Neill E. J., Barrat F. J., Boonstra A., Barthlott T., Stockinger B., Wraith D. C., O’Garra A. J. Immunol. 2004;172:5986–5993. doi: 10.4049/jimmunol.172.10.5986. [DOI] [PubMed] [Google Scholar]
- 18.Randolph D. A., Fathman C. G. Annu. Rev. Med. 2006;57:381–402. doi: 10.1146/annurev.med.57.121304.131337. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., Wahl S. M. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker M. R., Kasprowicz D. J., Gersuk V. H., Benard A., Van Landeghen M., Buckner J. H., Ziegler S. F. J. Clin. Invest. 2003;112:1437–1443. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somasundaram R., Jacob L., Swoboda R., Caputo L., Song H., Basak S., Monos D., Peritt D., Marincola F., Cai D., et al. Cancer Res. 2002;62:5267–5272. [PubMed] [Google Scholar]
- 22.Javia L. R., Rosenberg S. A. J. Immunother. 2003;26:85–93. doi: 10.1097/00002371-200301000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curiel T. J., Coukos G., Zou L., Alvarez X., Cheng P., Mottram P., Evdemon-Hogan M., Conejo-Garcia J. R., Zhang L., Burow M., et al. Nat. Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 24.Marshall N. A., Christie L. E., Munro L. R., Culligan D. J., Johnston P. W., Barker R. N., Vickers M. A. Blood. 2004;103:1755–1762. doi: 10.1182/blood-2003-07-2594. [DOI] [PubMed] [Google Scholar]
- 25.Viguier M., Lemaitre F., Verola O., Cho M. S., Gorochov G., Dubertret L., Bachelez H., Kourilsky P., Ferradini L. J. Immunol. 2004;173:1444–1453. doi: 10.4049/jimmunol.173.2.1444. [DOI] [PubMed] [Google Scholar]
- 26.Matsubara Y., Hori T., Morita R., Sakaguchi S., Uchiyama T. Leukemia. 2005;19:482–483. doi: 10.1038/sj.leu.2403628. [DOI] [PubMed] [Google Scholar]
- 27.Kohno T., Yamada Y., Akamatsu N., Kamihira S., Imaizumi Y., Tomonaga M., Matsuyama T. Cancer Sci. 2005;96:527–533. doi: 10.1111/j.1349-7006.2005.00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger C. L., Tigelaar R., Cohen J., Mariwalla K., Trinh J., Wang N., Edelson R. L. Blood. 2005;105:1640–1647. doi: 10.1182/blood-2004-06-2181. [DOI] [PubMed] [Google Scholar]
- 29.Cohen S. B., Crawley J. B., Kahan M. C., Feldmann M., Foxwell B. M. Immunology. 1997;92:1–5. doi: 10.1046/j.1365-2567.1997.00348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crockett D., Lin K. Z., Elenitoba-Johnson K. S., Lim M. S. Oncogene. 2004;23:2617–2629. doi: 10.1038/sj.onc.1207398. [DOI] [PubMed] [Google Scholar]
- 31.Benkhart E. M., Siedlar M., Wedel A., Werner T., Ziegler-Heitbrock H. W. J. Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 32.Ziegler-Heitbrock L., Lotzerich M., Schaefer A., Werner T., Frankenberger M., Benkhart E. J. Immunol. 2003;171:285–290. doi: 10.4049/jimmunol.171.1.285. [DOI] [PubMed] [Google Scholar]
- 33.Karube K., Ohshima K., Tsuchiya T., Yamaguchi T., Kawano R., Suzumiya J., Utsunomiya A., Harada M., Kikuchi M. Br. J. Haematol. 2004;126:81–84. doi: 10.1111/j.1365-2141.2004.04999.x. [DOI] [PubMed] [Google Scholar]
- 34.Walsh P. T., Benoit B. M., Wysocka M., Dalton N. M., Turka L. A., Rook A. H. J. Invest. Dermatol. 2006;126:690–692. doi: 10.1038/sj.jid.5700121. [DOI] [PubMed] [Google Scholar]
- 35.Yu H., Jove R. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 36.Wan W., Albom M. S., Lu L., Quail M. R., Becknell N. C., Weinberg L. R., Reddy D. R., Holskin B. P., Angeles T. S., Underiner T. L., et al. Blood. 2006;107:1617–1623. doi: 10.1182/blood-2005-08-3254. [DOI] [PubMed] [Google Scholar]
- 37.Pulford K., Falini B., Banham A. H., Codrington D., Roberton H., Hatton C., Mason D. Y. Blood. 2000;96:1605–1607. [PubMed] [Google Scholar]
- 38.Passoni L., Scardino A., Bertazzoli C., Gallo B., Coluccia A. M., Lemonnier F. A., Kosmatopoulos K., Gambacorti-Passerini C. Blood. 2002;99:2100–2106. doi: 10.1182/blood.v99.6.2100. [DOI] [PubMed] [Google Scholar]
- 39.Chan K. S., Sano S., Kiguchi K., Anders J., Komazawa N., Takeda J., DiGiovanni J. J. Clin. Invest. 2004;114:720–728. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling X., Arlinghaus R. B. Cancer Res. 2005;65:2532–2536. doi: 10.1158/0008-5472.CAN-04-2425. [DOI] [PubMed] [Google Scholar]
- 41.Chiarle R., Simmons W. J., Cai H., Dhall G., Zamo A., Raz R., Karras J. G., Levy D. E., Inghirami G. Nat. Med. 2005;11:623–629. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 42.Wang T., Niu G., Kortylewski M., Burdelya L., Shain K., Zhang S., Bhattacharya R., Gabrilovich D., Heller R., Coppola D., et al. Nat. Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 43.Cheng F., Wang H. W., Cuenca A., Huang M., Ghansah T., Brayer J., Kerr W. G., Takeda K., Akira S., Schoenberger S. P., et al. Immunity. 2003;19:425–436. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 44.Buettner R., Mora L. B., Jove R. Clin. Cancer Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 45.Turkson J., Kim J. S., Zhang S., Yuan J., Huang M., Glenn M., Haura E., Sebti S., Hamilton A. D., Jove R. Mol. Cancer Ther. 2004;3:261–269. [PubMed] [Google Scholar]