Abstract
The severity of epidemic and pandemic influenza outbreaks is dictated in part by the efficiency with which the causative strain transmits between human hosts. The mechanisms underlying influenza virus spread are poorly understood, in part because of the lack of a convenient animal model to study this phenomenon. Indeed, despite extremely efficient transmission among humans and virulence in the mouse model, we have shown that even the 1918 pandemic influenza virus does not transmit between mice. We therefore evaluated the guinea pig as a model mammalian host for influenza virus. Using the recent human isolate A/Panama/2007/99 (Pan/99) (H3N2) virus, we found that guinea pigs were highly susceptible to infection with the unadapted virus (ID50 = 5 plaque-forming units). Pan/99 virus grew to high titers in the upper respiratory tract and was shed in nasal washings of infected animals. Moreover, influenza virus was transmitted from infected guinea pigs to noninfected guinea pigs housed in the same cage, an adjacent cage, and a cage placed 91 cm away. Our results demonstrate that influenza virus can pass between guinea pigs by means of droplet spread and thereby establish the suitability of the guinea pig as a model host for influenza virus transmission studies.
Keywords: avian influenza virus, contact spread, droplet spread, pandemic, sentinel
Every 1–3 years, regional influenza epidemics lead to significant morbidity, mortality, and social disruption. In the United States alone, the influenza season is associated with an estimated 95,000–172,000 hospitalizations (1, 2) and 21,000–41,000 deaths (3, 4) due to influenza virus infection, costing billions of dollars in health care expenses and lost productivity (1, 5) annually. Furthermore, the three influenza pandemics of the last century, occurring in 1918, 1957, and 1968, caused an estimated 50 million (6), >2 million, and 1 million (7) deaths worldwide, respectively. Historical evidence suggests that influenza pandemics recur every 10–40 years, leading to speculation that the next pandemic is imminent. For a strain of influenza virus to cause a pandemic, it must be (i) novel to the human immune system, (ii) virulent in the human host, and (iii) transmissible from person to person. It is the failure of avian H5N1 influenza viruses to meet the last criterion that, to date, has prevented widespread human infection. The efficiency of transmission is also a key factor in determining the severity of influenza epidemics. An understanding of the mechanisms underlying influenza virus transmission would allow rational steps to be taken toward limiting viral spread in both pandemic and interpandemic years.
Unfortunately, the viral traits governing transmission efficiency have not been well characterized. Evidence from epidemiological studies and experimental infection of human volunteers suggests that influenza virus infection of humans can occur by inhalation of aerosols (8, 9). However, evidence in support of large droplet or contact transmission has also been reported (10, 11), and the relative contribution of each mode to the spread between humans is unknown. The paucity of information regarding influenza virus transmission can be attributed in part to the lack of a convenient animal model for the investigation of this topic. The most common mammalian model used for influenza research, the mouse, does not consistently transmit infection from one animal to another (see ref. 12 and data herein), making this system less than ideal for transmission studies. Ferrets, by contrast, are susceptible to infection with unadapted human influenza viruses, and such viruses have been shown to pass from an infected ferret to an uninfected contact (13, 14). Nevertheless, the ferret model presents several practical disadvantages (15): Ferrets are relatively large, and, as a result, the facilities required to house these animals are not widely available; ferrets are expensive; ferrets are not available from most laboratory animal suppliers; and it is difficult to obtain ferrets that have not had previous exposure to influenza viruses. For these reasons, we sought to develop an alternative animal model for influenza virus transmission studies.
In an article published in 1919 that details the progression of the 1918 influenza epidemic at Camp Cody, New Mexico, the authors describe a parallel outbreak of pneumonial disease among their laboratory guinea pigs (16). Although anecdotal, the account suggested that the infection was passed from guinea pig to guinea pig, and it prompted us to test the guinea pig as a model host for influenza virus. On occasion, the guinea pig has been used for influenza virus research in the past (17–19), but these reports did not examine the question of transmissibility. After determining that even the highly pathogenic 1918 influenza virus does not spread from infected mice to uninfected cagemates, we tested the propensity for transmission of a recent human influenza isolate, A/Panama/2007/99 (H3N2) (Pan/99), among guinea pigs. Unadapted Pan/99 virus was shown to replicate in the upper and lower respiratory tract of guinea pigs and shed at high titers in nasal secretions, and it was transmitted from infected guinea pigs to sentinel animals housed in the same cage or nearby cages. Our results indicate that droplet transmission of influenza virus occurs between guinea pigs, opening the door to mechanistic characterization of influenza virus transmission by using this animal model.
Results
Influenza Virus Does Not Transmit Readily Between Mice.
We evaluated the ability of the following to undergo transmission between mice: the 1918 influenza virus, the mouse-adapted WSN virus, a contemporary human influenza virus, a prototypical human influenza virus isolated in 1968, and a highly pathogenic H5N1 virus isolated from a human in 2004. Seven mice were inoculated intranasally, and naïve mice were subsequently placed in the same cage with the inoculated animals. Mice infected with 106 plaque-forming units (pfu) of 1918 or WSN viruses exhibited severe illness, and none of the mice survived past day 8 postinoculation (p.i.) (experiment 1 in Table 1). Although infectious virus was detected in the upper and lower respiratory tract of all H1N1 virus-inoculated animals, no virus was detected in any of the tissues collected from the contact mice on day 4 postcontact (p.c.), and convalescent sera collected from the contact animals were negative for the presence of H1N1 hemagglutination-inhibiting antibodies. Mice infected with a nonlethal dose (103 pfu) of 1918 virus exhibited less severe illness and survived the infection but still presented mean lung titers of 5.6 log10 50% egg infectious dose (eID50)/ml on day 4 p.i. (experiment 2 in Table 1). The highly pathogenic human influenza A/Vietnam/1203/04 (H5N1) isolate, previously shown to be lethal in mice (20), replicated to high titers in both the upper and lower respiratory tract and induced severe clinical illness, such as ruffled fur and listlessness during the first week of infection, and caused death by day 8 p.i. The human influenza A/Hong Kong/8/1968 (H3N2) virus induced only minimal clinical illness and weight loss and replicated to modest titers (4.2 log10 eID50/ml) in the lung tissue. Examination of contact animals housed with each virus-infected group again revealed a lack of transmission among mice: Virus was not recovered from any of the contact animals, nor did any of the contacts display weight loss (not shown) or seroconvert (Table 1).
Table 1.
Transmissibility of influenza viruses in BALB/c mice
| Experiment no. | Virus | Subtype | Inoculated mice |
Contact mice* |
||||
|---|---|---|---|---|---|---|---|---|
| Weight loss, %† | Nose titers‡ | Lung titers‡ | Seroconversion (HI antibody titer range)§ | Virus detected in nose or lung‡ | Seroconversion | |||
| 1 | Tx/91 | H1N1 | 2.7 | 2.4 ± 0.9 (2) | 3.6 ± 0.3 (3) | 4/4 (40–160) | (0/3) | 0/3 |
| 1 | WSN | H1N1 | 24.4 | 3.4 ± 0.5 (3) | 7.0 ± 0.4 (3) | ND¶ | (0/3) | 0/3 |
| 1 | 1918 | H1N1 | 22.3 | 2.1 ± 0.8 (2) | 7.5 ± 0.3 (3) | ND¶ | (0/3) | 0/3 |
| 2 | HK/8/68 | H3N2 | 12.4 | 2.2 ± 0.7 (2) | 4.2 ± 0.4 (3) | 4/4 (40–80) | (0/3) | 0/3 |
| 2 | VN/1203/04 | H5N1 | 21.5 | 5.8 ± 0.4 (3) | 6.8 ± 0.4 (3) | ND¶ | (0/3) | 0/3 |
| 2 | 1918 | H1N1 | 18.9 | 1.7 ± 0.4 (1) | 5.6 ± 0.3 (3) | 4/4 (80–160) | (0/3) | 0/3 |
In experiment 1, groups of BALB/c mice (n = 7) were inoculated intranasally with 106 pfu of the indicated virus. In experiment 2, groups of BALB/c mice (n = 7) were inoculated intranasally with 103 pfu of the indicated virus. HI, hemagglutination inhibition; ND, not determined.
*Age-matched BALB/c mice (n = 3) were added to the same cage at 24 h p.i. No weight loss was observed in contact mice.
†The percentage of mean maximum weight loss is shown (four mice per group).
‡Average tissue titers of three mice on day 4 p.i. expressed as 50% egg infectious dose/ml ± SD. The no. of positives is indicated in parentheses.
§Serum was collected on day 21 p.i., and homologous virus was used with horse RBCs for H5 virus or turkey RBCs for all other viruses in HI assays. The range of antibody titers obtained is indicated in parentheses.
¶Not determined because the four inoculated mice did not survive beyond 8 days p.i.
Guinea Pigs Are Highly Susceptible to Pan/99 Influenza Virus Infection.
To test the susceptibility of guinea pigs to infection with an unadapted human influenza virus, the ID50 of Pan/99 virus was determined. Groups of four animals were inoculated intranasally with 1, 10, 100, or 1,000 pfu [as determined by plaque assay on Madin–Darby canine kidney (MDCK) cells] of Pan/99 virus. The infection status of each guinea pig was then determined by performing plaque assays on nasal wash samples collected at 2 days p.i. The results obtained (shown in Table 2) were used to calculate the ID50 by the method of Reed and Muench (21). The ID50 was found to be 5 pfu, indicating that Hartley strain guinea pigs are highly susceptible to influenza virus infection.
Table 2.
Determination of ID50 of Pan/99 influenza virus in guinea pigs
| Dose, pfu | No. infected | No. uninfected |
|---|---|---|
| 1 | 1 | 3 |
| 10 | 3 | 1 |
| 100 | 4 | 0 |
| 1,000 | 4 | 0 |
Animals were scored as infected if >1,000 pfu was detected in nasal wash samples collected at 2 days p.i.
Kinetics of Growth of Pan/99 Virus in the Respiratory Tract of Guinea Pigs.
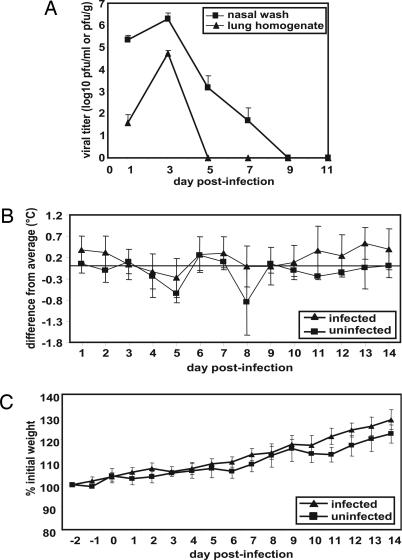
Viral replication in the upper and lower respiratory tract of guinea pigs, as well as the effects of infection on body temperature and weight, was assessed over a 14-day time course. Fourteen animals were inoculated intranasally with 103 pfu of Pan/99 virus, and two animals were mock-infected. On days 1, 3, 5, 7, and 9 p.i., two infected guinea pigs were killed, and their lungs were removed, homogenized in PBS, and subjected to plaque assay for the determination of viral titers. Viral growth in the upper respiratory tract was assessed by collecting nasal washes from all animals on days 1, 3, 5, 7, 9, and 11 p.i. and titrating them by plaque assay. The results indicate that viral growth peaked on day 3 p.i. in both the lungs and the nasal passages and that replication was more productive in the nasal passages (Fig. 1A). Furthermore, virus was cleared from the lungs by 5 days p.i. but persisted in the nose up to 9 days p.i.
Fig. 1.
Characteristics of Pan/99 virus infection in guinea pigs. Fourteen guinea pigs were infected intranasally with 103 pfu of Pan/99 virus. The number of infected animals evaluated on each day varied as follows: day 1, n = 14; days 2 and 3, n = 12; days 4 and 5, n = 10; days 6 and 7, n = 8; days 8 and 9, n = 6; days 10–14, n = 4. At all time points, two mock-infected animals were assessed. (A) Viral titers in nasal washings and lung homogenates. At the indicated time points, nasal washings were collected from all guinea pigs, and two animals were killed and their lungs were removed. Infectious content of nasal wash samples and lung homogenates were then determined by plaque assay (limit of detection was 10 pfu/ml or 30 pfu/g). Nasal wash titers are expressed in pfu/ml, and lung titers are expressed in pfu/g of lung. (B) Change in body temperature over the course of infection. Temperatures were measured twice daily from 2 days before infection to 14 days p.i. The average preinfection temperature for each animal was subtracted from each p.i. temperature to obtain the change in body temperature for each guinea pig; the average change in temperature for all guinea pigs was plotted. (C) Change in body weight over the course of infection. Body weight was assessed daily and is expressed as the average percentage change in body weight for each animal.
Body temperature was assessed twice daily and found to fluctuate between the morning and the evening. However, no effect of influenza virus infection on body temperature was observed at either time of day (Fig. 1B; only morning data are shown). Similarly, the weight of each animal was assessed once per day and found to steadily increase but was not influenced by influenza viral infection (Fig. 1C).
Transmission of Influenza Virus Between Cocaged Guinea Pigs.
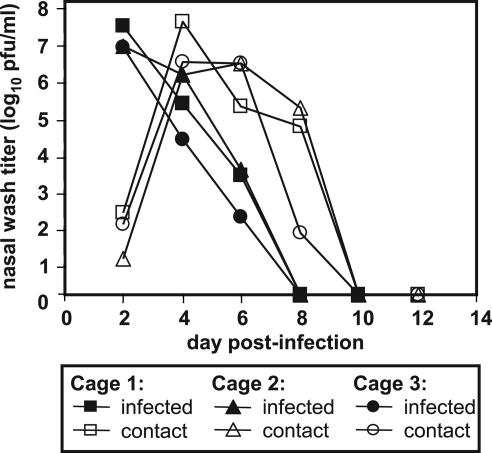
Our hypothesis that influenza virus would be transmitted from infected to uninfected guinea pigs was initially tested under conditions where direct contact between the two animals occurred. Specifically, three guinea pigs were infected intranasally with 102 pfu of Pan/99 virus and, at 24 h p.i., one uninfected animal was added to the cage of each infected animal. Nasal wash samples were collected every 2 days, beginning from day 2 p.i., until shedding ceased. Titers in the nasal passages of inoculated animals reached ≥5 × 106 pfu/ml on day 2 p.i. and dropped to undetectable levels by day 8 p.i. (Fig. 2). At day 1 p.c. (day 2 p.i.), low titers of virus were already detected in the nasal washings of contact guinea pigs. Levels of virus shed by these animals increased up to ≥106 pfu/ml on day 3 p.c. (Fig. 2), indicating that productive replication was occurring. Thus, the contact guinea pigs acquired influenza virus infections from their cagemates, demonstrating that guinea pig-to-guinea pig transmission of influenza virus had occurred.
Fig. 2.
Transmission of Pan/99 virus from intranasally infected guinea pigs to uninfected contacts. Three animals were inoculated with 102 pfu of Pan/99 virus. At 24 h p.i., each of these animals was placed in a cage (designated 1, 2, or 3) with one uninfected guinea pig. Nasal washes were performed at 48-h intervals, starting from 48 h p.i. for the inoculated animals (24 h p.c.).
Transmission of Influenza Virus by Means of Droplet Spread.
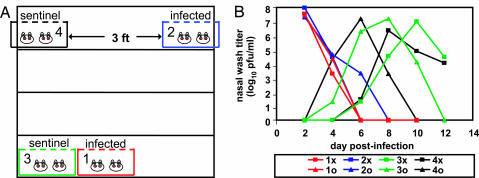
We next tested whether contact between guinea pigs was required for influenza virus transmission to occur or whether droplet and/or aerosol transmission was also possible. We assessed transmission between immediately adjacent cages and between cages separated by a 91-cm space. Two pairs of guinea pigs were inoculated intranasally with 103 pfu of Pan/99 virus, and, at 24 h p.i., a cage containing infected animals was placed either next to or 91 cm away from a cage containing two uninfected guinea pigs, as illustrated in Fig. 3A. The infection status of each animal was then determined by nasal washing at 48-h intervals. The results, shown in Fig. 3B, indicate that one sentinel animal in each cage acquired influenza virus infection between days 2 and 4 p.i. (days 1 and 3 p.c.). The second sentinel in each cage became infected between days 4 and 6 p.i., most likely from their respective cagemates. The results shown are representative of two similar experiments; however, transmission was not observed when the relative positions of the infected and uninfected animals were reversed, suggesting that spread depended on the direction of airflow in the room.
Fig. 3.
Droplet transmission of Pan/99 virus among guinea pigs. Representative results from two independent experiments are shown. Inoculated guinea pigs were given 103 pfu of Pan/99 virus intranasally and kept distant from sentinel animals until 24 h p.i. Nasal washes were performed at 48-h intervals, starting from 48 h p.i. for the inoculated animals. Cage layouts are shown in A; open, wire-top cages were placed on a shelf as shown; animals in cages 1 and 2 were infected. Viral titers in nasal wash samples are plotted in B; each curve corresponds to a single animal. “x” and “o” are used to distinguish two guinea pigs housed in the same cage; thus, 1x and 1o were housed in cage 1.
Discussion
In most cases, influenza A viruses must be adapted through serial passage in mice before they replicate to high titers or cause disease in this model host (22, 23). Previous work has indicated that exceptions to this rule include the reconstructed 1918 influenza virus (24) and some recent H5N1 isolates (20, 25). We have confirmed these findings and, in addition, determined that, despite the productive infection of mice by 1918 influenza or A/Vietnam/1203/04 viruses, these pathogens are not transmitted from infected mice to naïve contacts. Furthermore, even the highly mouse-adapted WSN virus was not transmitted from mouse to mouse. These results are contrary to those reported in a series of studies spanning from 1962 to 1968, which describe influenza virus transmission among experimentally infected mice (12, 26–29). The fact that, in these early studies, CFW or NCS mice were inoculated by exposure to aerosol may account for the discrepancy. However, even in these experiments, the efficiency of transmission was low (5–62.5%) (27, 29), and success with a given strain appeared to depend on the mouse rather than any property of the infection (12). Based on these data and our own findings, it is clear that mice are not an ideal model to mimic the transmission pattern of influenza viruses among humans.
In the absence of a feasible animal model, the question of which viral and host factors influence influenza virus spread has been largely neglected. However, recent fears of a pandemic have brought this issue to the forefront of the influenza field; indeed, the containment of epidemic and pandemic influenza depends on understanding what enables an influenza virus to transmit from person to person at a molecular level. To facilitate work toward such understanding, we propose the guinea pig as a mammalian model system for influenza virus transmission studies. We have shown that an unadapted human influenza virus replicates to high titers in the respiratory tract of guinea pigs and that the infection is passed from infected to sentinel guinea pigs. Furthermore, the presence of a physical barrier, or even 91 cm of intervening space, between infected and uninfected animals did not preclude transmission, indicating that not only contact but also large droplet and/or aerosol transmission occurred. Further studies assessing transmission over larger distances are required to determine whether true aerosol spread of influenza virus occurs in the guinea pig model. Although symptoms were not observed in the infected Hartley strain guinea pigs, this apparent lack of disease is also observed in a proportion of human influenza cases (30). Furthermore, influenza virus infection of inbred strain 13 guinea pigs (obtained from the U.S. Army Medical Research Institute of Infectious Diseases, Fort Detrick, MD) by means of droplet spread resulted in severe disease in these animals, including weight loss, lethargy, hair loss, and hypothermia (data not shown). It will be interesting to compare the proficiency of these two guinea pig strains in transmitting influenza viruses.
The ferret is well established as an animal model in influenza virus research because of its natural susceptibility to infection and the manifestation of influenza symptoms similar to those seen in human hosts (15, 31). In addition, transmission of influenza virus infection between ferrets has been reported to occur efficiently (13, 14, 31). Nevertheless, reasons of practicality limit the use of ferrets in academic research, with the result that the number of influenza virus studies performed in mice far outweighs that in ferrets, despite the superiority of the ferret model. Occasionally, hamsters have been used as an experimental model for influenza virus infection (32–34), and, in one report, they were shown to be competent for transmission. Hamster-to-hamster spread was not observed between adjacent cages, and the consistency with which transmission occurred varied with the virus strain that was analyzed (34).
It is not yet clear why influenza virus transmits efficiently from guinea pig to guinea pig but not from mouse to mouse. The lower susceptibility of mice to infection with most influenza virus strains likely contributes to this difference but does not account for the lack of transmission of mouse-adapted viruses, such as WSN. Perhaps the process of adaptation to the murine host concomitantly reduces the intrinsic transmissibility of an influenza virus. Our data indicated that, for all strains tested, influenza viruses grew to higher titers in the lungs of mice than in the nose, whereas the opposite was true for Pan/99 virus in the guinea pig; thus, the predominant site of viral replication in a given host may dictate whether transmission occurs. Transmission experiments combining mice and guinea pigs as either donors or receivers of influenza virus should shed some light on the reason that mice fail to propagate infection; furthermore, the physiological and/or virological basis for this observation may in turn provide clues as to why human hosts do not transmit avian influenza viruses.
Indeed, a guinea pig model of influenza virus transmission holds great potential for the elucidation of both host and viral factors contributing to influenza virus spread. Once the determinants of transmission have been mapped, effective strategies for the containment of influenza outbreaks at their inception might be developed.
Materials and Methods
Cells and Viruses.
MDCK cells were maintained in Eagle’s minimum essential medium supplemented with 10% FBS. Stocks of the influenza viruses Pan/99 (H3N2), A/Hong Kong/8/68 (H3N2), A/WSN/33 (H1N1), A/Texas/36/91 (H1N1), avian influenza A/Vietnam/1203/04 (H5N1), and the 1918 virus (24) were prepared by inoculation of 10-day-old embryonated hens’ eggs or MDCK cells followed by incubation at 37°C for 48 h for human viruses or at 37°C for 24 h for the avian virus. All experiments involving the 1918 and A/Vietnam/1203/04 viruses were performed under strict biosafety level 3 conditions (35). All guinea pigs were seronegative to Pan/99 virus on arrival, and those inoculated or secondarily infected were found to seroconvert.
Animals.
Eight-week-old female BALB/c mice and female Hartley strain guinea pigs weighing 300–350 g were obtained from Charles River Laboratories. Animals were allowed free access to food and water and kept on a 12-h light/dark cycle. During guinea pig transmission experiments, strict measures were followed to prevent aberrant cross-contamination between cages. Sentinel animals were handled before inoculated animals, gloves were changed between animal handlings, and work surfaces were sanitized after each use.
Mouse Transmission Experiments.
Mice were anesthetized with Avertin (tribromoethanol; Sigma) before infections. Fifty microliters of infectious virus diluted in PBS was given intranasally. Twenty-four hours later, three naïve mice were placed in the same cage with seven inoculated mice. Duplicate cages were set up for each virus to monitor for (i) virus replication in the respiratory tract and (ii) clinical signs and seroconversion at 21 days p.i. For virus determination, three inoculated or contact mice were killed on day 4 p.i.; lung and nose tissues were collected, immediately frozen on dry ice, and stored at −70°C until processing. Frozen tissues were later thawed, homogenized in 1 ml of cold PBS, and clarified by centrifugation (at 2,200 × g) at 4°C. Clarified tissue homogenates were titrated for virus infectivity in eggs. Weight loss was determined on alternate days for all mice for 21 days p.i.
Infection and Monitoring of Guinea Pigs.
Before infection, guinea pigs were anesthetized with a mixture of ketamine (30 mg/kg) and xylazine (2 mg/kg) administered intramuscularly. For intranasal infections, Pan/99 virus stock was diluted in PBS containing 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.3% BSA (PBS-PS-BA). A 300-μl volume of inoculum containing 102 or 103 pfu was instilled into the nostrils (150 μl on each side). Body temperature was measured twice daily from 2 days before infection up to 14 days p.i. by using s.c. implanted telemetric transponders from BioMedic Data Systems (Maywood, NJ). Body weight was measured once daily.
Collection of Guinea Pig Nasal Wash Samples and Lung Tissue.
Nasal washing was performed by instilling a total of 1 ml of PBS-PS-BA into the nostrils and allowing it to drain onto a sterile Petri dish. Samples were collected in 1.5-ml tubes and centrifuged for 5 min at 2,000 × g and 4°C; supernatants were stored at −80°C before analysis by plaque assay. For the collection of lungs, animals were killed through i.p. injection of 100 mg/kg sodium pentobarbital (Nembutal; Ovation Pharmaceuticals, Deerfield, IL). Extracted lung tissue was weighed, homogenized in PBS, and subjected to centrifugation at 12,000 × g and 4°C for 10 min to pellet debris. Supernatants were stored at −80°C before analysis by plaque assay.
Quantification of Viral Titers.
Virus titers were determined by plaque assay of 10-fold serial dilutions on MDCK cells or by limiting dilution in embryonated chicken eggs, starting from an initial dilution of 1:10 in PBS. The limit of virus detection was 101.2 50% egg infectious dose/ml or 10 pfu/ml.
Acknowledgments
This work was supported by National Institutes of Health Grants P01 AI58113, U54 AI057158, and UC19AI-062623 (to A.G.-S. and P.P.) and R01 AI18998-25 (to P.P.). P.P. is a senior fellow of the Ellison Medical Foundation. S.M. was supported by Sunnybrook Health Sciences Centre, University of Toronto (Toronto).
Abbreviations
- p.i.
postinoculation
- p.c.
postcontact
- pfu
plaque-forming units
- Pan/99
A/Panama/2007/99.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Barker W. H. Am. J. Public Health. 1986;76:761–765. doi: 10.2105/ajph.76.7.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson W. W., Shay D. K., Weintraub E., Brammer L., Bridges C. B., Cox N. J., Fukuda K. J. Am. Med. Assoc. 2004;292:1333–1340. doi: 10.1001/jama.292.11.1333. [DOI] [PubMed] [Google Scholar]
- 3.Simonsen L., Clarke M. J., Williamson G. D., Stroup D. F., Arden N. H., Schonberger L. B. Am. J. Public Health. 1997;87:1944–1950. doi: 10.2105/ajph.87.12.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dushoff J., Plotkin J. B., Viboud C., Earn D. J. D., Simonsen L. Am. J. Epidemiol. 2006;163:181–187. doi: 10.1093/aje/kwj024. [DOI] [PubMed] [Google Scholar]
- 5.Nichol K. L. Arch. Intern. Med. 2001;161:749–759. doi: 10.1001/archinte.161.5.749. [DOI] [PubMed] [Google Scholar]
- 6.Johnson N. P. A. S., Mueller J. Bull. Hist. Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Avian Influenza: Assessing the Pandemic Threat. [Accessed May 10, 2006];2005 Available at www.who.int/csr/disease/influenza/WHO_CDS_2005_29/en/index.html. [Google Scholar]
- 8.Alford R. H., Kasel J. A., Gerone P. J., Knight V. Proc. Soc. Exp. Biol. Med.; 1966. pp. 800–804. [DOI] [PubMed] [Google Scholar]
- 9.Moser M. R., Bender T. R., Margolis H. S., Noble G. R., Kendal A. P., Ritter D. G. Am. J. Epidemiol. 1979;110:1–6. doi: 10.1093/oxfordjournals.aje.a112781. [DOI] [PubMed] [Google Scholar]
- 10.Bean B., Moore B. M., Sterner B., Peterson L. R., Gerding D. N., Balfour H. H. J. J. Infect. Dis. 1982;146:47–51. doi: 10.1093/infdis/146.1.47. [DOI] [PubMed] [Google Scholar]
- 11.Blumenfeld H. L., Kilbourne E. D., Louria D. B., Rogers D. E. J. Clin. Invest. 1959;38:199–212. doi: 10.1172/JCI103789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulman J. L., Kilbourne E. D. J. Exp. Med. 1963;125:267–275. doi: 10.1084/jem.118.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herlocher M. L., Elias S., Truscon R., Harrison S., Mindell D., Simon C., Monto A. S. J. Infect. Dis. 2001;184:542–546. doi: 10.1086/322801. [DOI] [PubMed] [Google Scholar]
- 14.Herlocher M. L., Truscon R., Elias S., Yen H.-L., Roberts N. A., Ohmit S. E., Monto A. S. J. Infect. Dis. 2004;190:1627–1630. doi: 10.1086/424572. [DOI] [PubMed] [Google Scholar]
- 15.Maher J. A., DeStefano J. Lab. Anim. 2004;33:50–53. doi: 10.1038/laban1004-50. [DOI] [PubMed] [Google Scholar]
- 16.Lamb F. H., Brannin E. B. J. Am. Med. Assoc. 1919;72:1056–1062. [Google Scholar]
- 17.Azoulay-Dupuis E., Lambre C. R., Soler P., Moreau J., Thibon M. J. Comp. Pathol. 1984;94:273–283. doi: 10.1016/0021-9975(84)90046-x. [DOI] [PubMed] [Google Scholar]
- 18.Phair J. P., Kauffman C. A., Jennings R., Potter C. W. Med. Microbiol. Immunol. 1979;165:241–254. doi: 10.1007/BF02152923. [DOI] [PubMed] [Google Scholar]
- 19.Wetherbee R. E. J. Immunol. 1973;111:157–163. [PubMed] [Google Scholar]
- 20.Maines T. R., Lu X. H., Erb S. M., Edwards L., Guarner J., Greer P. W., Nguyen D. C., Szretter K. J., Chen L.-M., Thawatsupha P., et al. J. Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed L. J., Muench H. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- 22.Hoyle L. In: The Influenza Viruses. Gard S., Hallauer C., Meyer K. F., editors. New York: Springer; 1968. pp. 170–171. [Google Scholar]
- 23.Wright P. F., Webster R. G. In: Fields Virology. Knipe D. M., Howley P. M., editors. Vol. 1. New York: Lippincott Williams & Wilkins; 2001. pp. 1533–1579. [Google Scholar]
- 24.Tumpey T. M., Basler C. F., Aguilar P. V., Zeng H., Solorzano A., Swayne D. E., Cox N. J., Katz J. M., Taubenberger J. K., Palese P., Garcia-Sastre A. Science. 2005;310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 25.Salomon R., Franks J., Govorkova E. A., Ilyushina N. A., Yen H.-L., Hulse-Post D. J., Humberd J., Trichet M., Rehg J. E., Webby R. J., et al. J. Exp. Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulman J. L., Kilbourne E. D. Nature. 1962;195:1129–1130. doi: 10.1038/1951129a0. [DOI] [PubMed] [Google Scholar]
- 27.Schulman J. L., Kilbourne E. D. J. Exp. Med. 1963;118:257–266. doi: 10.1084/jem.118.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulman J. L. J. Exp. Med. 1967;125:479–488. doi: 10.1084/jem.125.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulman J. L. Am. J. Public Health. 1968;58:2092–2096. doi: 10.2105/ajph.58.11.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treanor J. J. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Mandell G. L., Bennett J. E., Dolin R., editors. Vol. 2. Philadelphia: Churchill Livingstone; 2005. pp. 2062–2078. [Google Scholar]
- 31.Reuman P. D., Keely S., Schiff G. M. J. Virol. Methods. 1989;24:27–34. doi: 10.1016/0166-0934(89)90004-9. [DOI] [PubMed] [Google Scholar]
- 32.Subbarao E. K., Kawaoka Y., Murphy B. R. J. Virol. 1993;67:7223–7228. doi: 10.1128/jvi.67.12.7223-7228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath A. W., Addison C., Ali M., Teale D., Potter C. W. Antiviral Res. 1983;3:241–252. doi: 10.1016/0166-3542(83)90003-7. [DOI] [PubMed] [Google Scholar]
- 34.Ali M. J., Teh C. Z., Jennings R., Potter C. W. Arch. Virol. 1982;72:187–197. doi: 10.1007/BF01348964. [DOI] [PubMed] [Google Scholar]
- 35.U.S. Department of Health and Human Services/Centers for Disease Control/National Institutes of Health. Biosafety in Microbiological and Biomedical Laboratories. 5th Ed. Washington, DC: U.S. Government Printing Office; 2006. [Google Scholar]