Abstract
Polycomb group (PcG) genes contribute to the maintenance of cell identity, cell cycle regulation, and oncogenesis. We describe the expression of five PcG genes (BMI-1, RING1, HPC1, HPC2, and EZH2) in normal breast tissues, invasive breast carcinomas, and their precursors. Members of the HPC-HPH/PRC1 PcG complex, including BMI-1, RING1, HPC1, and HPC2, were detected in normal resting and cycling breast cells. The EED-EZH/PRC2 PcG complex protein EZH2 was only found in rare cycling cells, whereas normal resting breast cells were negative for EZH2. PcG gene expression patterns in ductal hyperplasia (DH), well-differentiated ductal carcinoma in situ (DCIS), and well-differentiated invasive carcinomas closely resembled the pattern in healthy cells. However, poorly differentiated DCIS and invasive carcinomas frequently expressed EZH2 in combination with HPC-HPH/PRC1 proteins. Most BMI-1/EZH2 double-positive cells in poorly differentiated DCIS were resting. Poorly differentiated invasive carcinoma displayed an enhanced rate of cell division within BMI-1/EZH2 double-positive cells. We propose that the enhanced expression of EZH2 in BMI-1+ cells contributes to the loss of cell identity in poorly differentiated breast carcinomas, and that increased EZH2 expression precedes high frequencies of proliferation. These observations suggest that deregulated expression of EZH2 is associated with loss of differentiation and development of poorly differentiated breast cancer in humans.
Keywords: Polycomb; breast cancer; BMI-1, EZH2; tissue array
Introduction
Polycomb group (PcG) proteins play a key role in the maintenance of cell identity [1–3] and contribute to the regulation of various processes, including lymphocyte development [4–6] and the cell cycle [7,8]. PcG proteins function as large multimeric complexes [9], containing various enzymes involved in the modification of histone tails [10–14]. This suggests that stable gene silencing by PcG complexes is related to alteration of chromatin composition. Two of these complexes have been identified and were shown to be evolutionarily conserved. The HPC-HPH “maintenance complex” is the mammalian counterpart of polycomb repressive complex-1 (PRC1) in Drosophila [15,16] and consists of the BMI-1, RING1, HPC, and HPH PcG proteins [17–22]. The mammalian EED-EZH complex is similar to the Drosophila Esc-E(Z)/PRC2 “initiation complex” [23,24] and contains the EED, EZH, and YY1 PcG proteins [25–28]. PcG complexes exhibit cell type-specific composition, which is most likely related to their specificity for different target genes [9,29].
The role of PcG genes in the maintenance of cell identity is underscored by the fact that several PcG genes can be classified as oncogenes and tumor suppressor genes [30,31]. For instance, overexpression of HPC2, RING1, and BMI-1 in experimental model systems resulted in cellular transformation of cell lines, or produced lymphomas in mutant mice [22,32–35]. Whether PcG genes are capable of producing cancer in humans is unclear, but recent studies that addressed this question concluded that neoplastic cells are associated with altered expression of PcG genes. For instance, Reed-Sternberg cells of Hodgkin's lymphoma and neoplastic cells in B-NHL display altered expression of BMI-1 [36,37]. Recent studies of human solid tumors demonstrated that BMI-1 is differentially expressed in non small cell lung cancer [38], whereas disease progression in prostate cancer, which shows many similarities to breast cancer, appears related to upregulation of EZH2 [39,40].
In this paper, we describe the expression of the BMI-1, RING1, HPC1, HPC2, and EZH2 PcG proteins in normal breast tissues, invasive breast carcinomas, and their precursors by means of conventional tissue sections and tissue arrays. We found that poorly differentiated ductal carcinoma in situ (DCIS) and poorly differentiated invasive carcinoma displayed enhanced expression of EZH2 in cells using PcG proteins of the HPC-HPH/PRC1 complex. This pattern was associated with an increased frequency of cell proliferation in EZH2-positive cells of invasive carcinomas, and suggests that altered EZH2 expression is related to loss of differentiation in breast carcinomas.
Materials and Methods
Human Tissue
Four normal breast tissues, 20 preinvasive breast lesions, and 15 invasive breast lesions were collected from the archives of the Department of Pathology of the VU University Medical Center (Amsterdam, The Netherlands). Anonymous use of leftover tumor material is part of the standard treatment agreement with patients in our hospital [41]. The normal breast tissues, obtained from breast reduction surgery, had no indications for the presence of hyperplasia, (in situ) carcinoma, fibroadenoma, or papilloma. Only adjacent nonproliferative changes, such as apocrine metaplasia, duct ectasia, or sclerosing adenosis, could be present. Preinvasive lesions were classified according to the criteria of Page et al. [42,43] and included four usual ductal hyperplasias (DHs), six atypical DHs, five well-differentiated, and five poorly differentiated ductal carcinomas in situ (DCIS). All preinvasive lesions were “pure,” meaning that there were no more advanced stages present (no invasion in the case of DCIS; no DCIS or invasion in the case of hyperplasia). Invasive breast carcinomas were graded according to the Elston and Ellis Grading System [44] and included four grade I (well-differentiated), six grade II (intermediately differentiated), and five grade III (poorly differentiated) lesions. A tissue microarray containing 172 invasive breast cancer cases with long-term follow-up was constructed as described before [45].
PcG Protein Detection by Immunohistochemistry
Four-micrometer sections were cut from paraffin-embedded tissues and mounted on Superfrost Plus slides (Omnilabo, Breda, The Netherlands). Following deparaffinization, endogenous peroxidase was inhibited by incubation of the tissue sections for 30 minutes at room temperature in 0.3% H2O2, diluted in methanol. Antigens were retrieved by boiling for 10 minutes in citrate buffer (pH = 6), followed by successive rinses in phosphate-buffered saline (PBS) containing 0.5% Triton (1 x 5 minutes), and PBS only (3 x 5 minutes). Slides were then incubated for 10 minutes in 0.1 M glycine (diluted in PBS), and rinsed in PBS only (3 x 5 min). Expression of PcG expression was detected using monoclonol and polyclonal antisera directed against PcG proteins [20,22,25,32], as indicated in Table 1. Tissues of known PcG reactivity were included in each experiment as positive controls for antiserum reactivity, and negative controls were obtained by omission of primary antibodies. Before application of the primary antiserum or antibody, sections were incubated for 10 minutes in normal swine serum (diluted 1:10 in PBS + 1% BSA) or normal rabbit serum (diluted 1:50 in PBS + 1% BSA). Secondary antisera were biotinylated goat antimouse or biotinylated swine anti-rabbit. Immunostaining was performed with 3-amino-9-ethylcarbazole (AEC) using the streptavidin-biotin complex/horseradish peroxidase (sABC-HRP) method and tyramine intensification, or with diaminobenzidine (DAB)/peroxidase and imidazole intensification. Sections were briefly counterstained with hematoxylin. Photographs were taken with a Zeiss Axiophot microscope (Zeiss, Oberkochen, Germany) and digitized using an Agfa duoscan scanner (Agfa, Mortsel, Belgium). The tissue microarray was stained for EZH2 only.
Table 1.
Overview of the Different PcG Antibodies Used.
| Target | Antibody | Source | Dilution |
| BMI-1 | 6C9 | Mouse monoclonal | Undiluted |
| RING1 | K320 | Rabbit polyclonal | 1:100 |
| HPC1 | K350 | Rabbit polyclonal | 1:100 |
| HPC2 | K326 | Rabbit polyclonal | 1:100 |
| EZH2 | K358 | Rabbit polyclonal | 1:100 |
Immunofluorescence Double and Triple Staining
Double immunofluorescence stainings with antisera against BMI-1, EZH2, and MIB-1/Ki-67 were performed on 3-µm frozen sections. After fixation in 2% formaldehyde, endogenous peroxidase was inhibited with 1% H2O2, diluted in PBS. Sections were preincubated with 5% BSA, and primary antibodies against BMI-1 and EZH2 were applied in combination with antiserum against MIB-1/Ki-67. BMI-1 was detected by incubation with GaMIgG2bHRP using the streptavidin-biotin-avidin complex/HRP method and rhodamine/tyramine intensification (excitation 550, emission 570; red fluorescence). EZH2 was detected by incubation with GaR antiserum coupled to ALEXA (excitation 495, emission 519; green fluorescence). MIB-1/Ki-67 was detected by incubating the slides with GaMIgGBIO1, followed by incubation with StrepAPC [streptavidin coupled to allophycocyanin (excitation 650, emission 660); infrared interpreted as blue fluorescence by the computer]. Cross-reactivity of the antisera was excluded by appropriate controls, and PcG expression patterns were confirmed in at least three separate experiments on tissues derived from different individuals. Sections were analyzed with a Leica DMR Confocal LaserScan microscope (Leica, Rijswijk, The Netherlands). Images were stored digitally at 1024 dpi and processed using Corel Photo-Paint 8.
Results
PcG Expression in Normal Breast Tissue
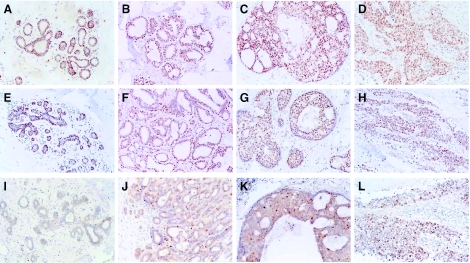
We first determined the nuclear expression of BMI-1, RING1, HPC1, HPC2, and EZH2 in normal breast ducts by immunohistochemistry. Normal breast ducts primarily expressed PcG proteins belonging to the HPC-HPH/PRC1 complex (Table 2, Figure 1). The majority of myoepithelial cells (outer layer) strongly expressed BMI-1, RING1, and HPC2, with HPC1 being detectable but at an apparently lower intensity (Figure 1, A and E). Most columnar epithelial cells (inner layer) weakly expressed BMI-1, RING1, and HPC1, with a minority of these cells being strongly positive for HPC2. Expression EED-EZH/PRC2 complex protein EZH2 was virtually undetectable in normal ducts (Figure 1I). Only very occasionally, a positive nucleus was seen in premenopausal breast tissues, including a positive mitotic figure.
Table 2.
PcG Expression in Normal Breast Tissues, Invasive Breast Carcinomas, and Precursors.
| Tissue | BMI-1 | RING1 | HPC1 | HPC2 | EZH2 |
| Healthy (n = 4) | 3 | 4 | 2 | 3 | 1 |
| DH, usual type (n = 4) | 4 | 4 | 1 | 4 | 1 |
| Atypical DH (n = 6) | 4 | 4 | 1 | 4 | 1 |
| DCIS, well differentiated (n = 5) | 4 | 4 | 2 | 4 | 1 |
| DCIS, poorly differentiated (n =5) | 4 | 4 | 1 | 4 | 2 |
| Invasive carcinoma, well differentiated (n = 4) | 4 | 3 | 1 | 4 | 1 |
| Invasive carcinoma, intermediately differentiated (n = 6) | 4 | 4 | 1 | 4 | 1 |
| Invasive carcinoma, poorly differentiated (n = 5) | 4 | 4 | 1 | 4 | 3 |
Nuclear PcG gene expression was investigated using immunohistochemistry and PcG-specific antibodies. Neoplastic cells were scored as follows: 1 = 0–25% positive cells; 2 = 26–50% positive cells; 3 = 51–75% positive cells; 4 = <75% positive cells.
Figure 1.
Expression of PcG genes in normal breast glands, invasive carcinomas, and their precursor lesions. Shown is the nuclear expression pattern of the BMI-1 (A–D), RING1 (E–H), and EZH2 (I–L) PcG genes in normal breast tissues (A, E, I), ductal hyperplasia (B, F, J), well-differentiated DCIS (C and G), poorly differentiated DCIS (K), and invasive carcinoma (D, H, L).
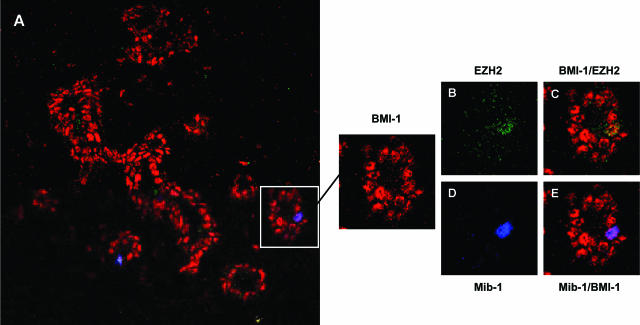
We then used immunofluorescence double and triple staining for BMI-1 and EZH2 to confirm the expression of the HPC-HPH/PRC1 and EED-EZH/PRC2 complexes in normal breast tissues. The majority of epithelial cells expressed the BMI-1 protein, confirming widespread expression of BMI-1 (Figure 2A, red fluorescence). EZH2 expression was rarely detected and always associated with cycling cells. This is illustrated by colocalization of the signal for EZH2 (Figure 2B, green fluorescence) and MIB-1/Ki-67 (Figure 2D, blue fluorescence) in Figure 2. Note that the EZH2/MIB-1 double-positive cycling cell has retained expression of BMI-1 (Figure 2, C and E).
Figure 2.
Expression of BMI-1, EZH2, and MIB-1 in normal breast glands. BMI-1, EZH2, and MIB-1 were detected by red, green, and blue fluorescence, respectively. The majority of cells in normal breast glands express BMI-1 (red signal) and are resting (as indicated by the relative absence of staining for MIB-1). Rare cycling MIB-1POS cells (blue signal) are occasionally visible. These cells retain expression of BMI-1 and are faintly stained for EZH2 (see insets and details, a BMI-1+/EZH2+/MIB-1+ cell is indicated by an arrow).
PcG Expression in Preinvasive Lesions
PcG expression was analyzed in lesions through which invasive breast cancer is thought to develop [46,47], including usual and atypical DH, and DCIS. Expression of genes encoding the HPC-HPH PcG complex resembled that of normal tissues or appeared slightly increased (Table 2, Figure 1, B, C, F, and G). In DH, most cells expressed HPC2 and were variably positive for BMI-1, RING1, and HPC1. In DCIS, neoplastic cells strongly expressed BMI-1, RING1, and HPC2, with HPC1 being detectable at variable levels. There were no obvious differences in the detection of PcG proteins belonging to the HPC-HPH/PRC1 complex between well differentiated and poorly differentiated DCIS.
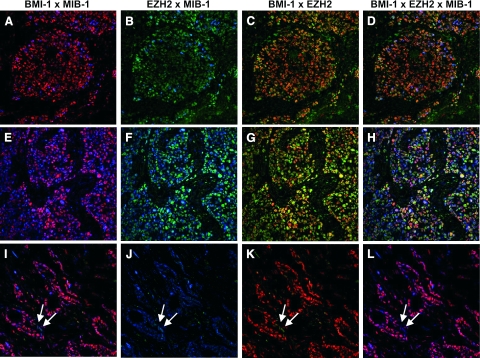
In contrast to normal tissues, expression of the EZH2 PcG protein was frequently detected in poorly differentiated DCIS, but only sporadically in DH or well differentiated DCIS (Table 2, Figure 2, J and K). Analysis of these tissues by immunofluorescence double and triple staining confirmed that EZH2 expression was widespread in neoplastic cells of poorly differentiated DCIS, and infrequent in well-differentiated DCIS and DH. In Figure 3, A–D, a case of poorly differentiated DCIS is shown, where combination of the BMI-1 signal (red fluorescence) and the EZH2 signal (green fluorescence) produces a yellow signal in the nucleus of BMI-1/EZH2 coexpressing cells (Figure 3C). Whereas expression of EZH2 in normal epithelial cells of breast glands was associated with cell division (Figure 2, C and E), BMI-1/EZH2 double-positive cells in DH and DCIS were resting in most instances. As shown in Figure 3, A–D, the majority of BMI-1/EZH2 double positive in poorly differentiated DCIS cells did not express MIB-1/Ki-67.
Figure 3.
Immunofluorescence detection of BMI-1, EZH2, and MIB-1 in breast carcinomas. Expression of BMI-1, EZH2, and MIB-1 was determined in poorly differentiated DCIS (A–D), poorly differentiated invasive carcinoma (E–H), and well-differentiated invasive carcinoma (I–L). Nuclear expression of BMI-1, EZH2, and MIB-1 was detected by red, green, and blue fluorescence, respectively. Shown are double fluorescence images of BMI-1 + MIB-1 (A, E, I), EZH2 + MIB-1 (B, F, J), BMI-1 + EZH2 (C, G, K), and triple immunofluorescence for BMI-1, EZH2, and MIB-1 (D, H, L). In DCIS of poor differentiation, widespread expression of BMI-1 (A) and EZH2 (B) resulted in frequent coexpression of BMI-1 and EZH2 in neoplastic cells (C). Only a limited number of these cells were MIB-1POS and cycling (A, B, D). Neoplastic cells in badly differentiated invasive carcinoma also displayed widespread expression of BMI-1 (E) and EZH2 (F), leading to frequent BMI-1/EZH2 coexpression (G). In contrast to badly differentiated DCIS, the majority of the BMI-1+/EZH2+ cells were MIB-1+ (blue signal in E, F, and H) and cycling. The majority of neoplastic cells in well-differentiated invasive carcinoma were BMI-1+ (I and K) and growing (as indicated by the blue signal in J and L). In contrast to poorly differentiated tumors, however, expression of EZH2 was infrequent, as indicated by the rare occurrence of neoplastic cells that faintly stained for EZH2 (indicated by the arrows in I–L).
PcG Expression in Invasive Carcinomas
Invasive breast carcinomas expressed HPC-HPH/PRC1 complex proteins in a pattern similar to that of preinvasive lesions. BMI-1, RING1, and HPC2 were strongly expressed in invasive neoplastic cells, with variable nuclear positivity for HPC1 (Table 2). This pattern of expression was similar in well-differentiated invasive carcinomas, and intermediately or poorly differentiated carcinomas. As noted for DCIS lesions, detection of the EED-EZH/PRC2 complex protein EZH2 depended on differentiation grade. EZH2 was most frequently detected in the majority of neoplastic cells of poorly differentiated invasive carcinomas (Table 2), whereas well-differentiated and intermediately differentiated lesions were mostly negative for EZH2. This positive correlation between EZH2 expression and histologic grade was confirmed on the tissue microarray (P < .004, chi-square test). EZH2 expression was analyzed in 172 tumors, graded G1 (n = 24), G2 (n = 96), and G3 (n = 52). Of these tumors, 4.2%, 11.5%, and 53.8% stained for EZH2, respectively. EZH2 did, however, not have prognostic values in Kaplan-Mayer survival analysis (log rank test).
We next confirmed the expression pattern of EZH2 in relation with BMI-1 and MIB-1/Ki-67 in invasive carcinomas by imunofluorescence double and triple staining. An example of a poorly differentiated invasive carcinoma is shown in Figure 3, E–H, and of a well-differentiated invasive carcinoma in Figure 3, I–L. Neoplastic cells in well-differentiated invasive carcinomas were all BMI-1+ and expressed MIB-1/Ki-67 (Figure 3, I and J). Low-level EZH2 expression was detected in a minority of these cells (Figure 3K, arrows). Detection of BMI-1, EZH2, and MIB-1/Ki-67 in poorly differentiated lesions (Figure 3, E–H) revealed a highly diverse pattern in neoplastic cells. The vast majority of these cells were MIB-1+ and expressed BMI-1 (Figure 3E). EZH2 expression was more frequent than in well-differentiated invasive carcinomas and was detected at varying levels of intensity. This is illustrated by the “autumn leaf expression pattern” of BMI-1 and EZH2 in Figure 3G. Combination of the three immunofluorescent signals showed that poorly differentiated invasive carcinomas contained highly diverse populations of neoplastic cells with extensive fluctuation in expression of BMI-1, EZH2, and MIB-1 (Figure 3H).
Discussion
PcG genes form a “cellular memory system” that is essential for the maintenance of cell identity by preserving gene silencing patterns after cell division [1–3]. Various processes, including regulation of the cell cycle and hematopoiesis, were recently shown to be controlled by PcG genes [4–8]. The essential role of PcG genes in these processes is underscored by the relationship between inordinate PcG gene expression and malignant transformation. The best known example is induction of lymphomas in BMI-1 transgenic mice, where upregulation of BMI-1 results in downregulation of the cell cycle regulators p16INK4a/p19ARF [35,48]. Investigation of BMI-1 in human malignancies showed altered expression patterns in Hodgkin's lymphoma [36], B-cell non-Hodgkin's lymphoma [37,49], and lung carcinoma [38], suggesting that human BMI-1 may also contribute to oncogenic transformation. Yet, despite these observations, our knowledge of a role for PcG genes in human tumors remains limited.
In the current paper, we describe the expression pattern of individual PcG genes in healthy breast gland cells and tumors derived from these cells. We showed that healthy resting breast gland cells primarily express PcG genes encoding the HPC-HPH/PRC1 PcG complex, including BMI-1, RING1, HPC1, and HPC2. In contrast, rare dividing cells were associated with expression of the EZH2 gene belonging to the EED-EZH/PRC2 PcG complex. This expression pattern is reminiscent of the PcG expression profile of healthy mature lymphoid cells, where expression of BMI-1 is associated with resting lymphocytes whereas EZH2 is found in dividing cells [50,51]. Of note is, however, that BMI-1 remains detectable in healthy cycling breast gland cells, whereas BMI-1 expression is lost in cycling lymphocytes. The most likely explanation for this disparity is that breast gland cells and lymphocytes have different cellular identities. They are, therefore, expected to possess distinct patterns of PcG gene expression, which is in line with the observation that the HPC1 PcG protein is detectable in breast gland cells, whereas mature lymphocytes do not express HPC1 (van Galen and Raaphorst, in preparation).
Neoplastic cells in DH, DCIS, and invasive carcinoma displayed frequent and strong staining for core proteins of the HPC-HPH/PRC1 complex, including BMI-1, RING1, HPC1, and HPC2. Because several of these proteins have been associated with oncogenic properties, the question arises as to whether they contribute to the development of breast cancer. Earlier studies of the BMI-1 binding partner Mel-18 demonstrated that its expression levels are decreased in human breast cancer cell lines, and that haploinsufficiency for this gene predisposes mice to the development of mammary tumors [52]. In addition, overexpression of BMI-1 in human mammary epithelial cells resulted in downregulation of the cell cycle regulators p16INK4a/p19ARF, upregulation of telomerase, and immortalization [34,35,48]. These results suggest that PcG proteins of the HPC-HPH/PRC1 complex are likely contributing factors in the development of breast tumors. We observed that normal breast gland cells stain for BMI-1 and its binding partners with variable intensity, whereas staining of neoplastic cells was frequent and strong. This might suggest that HPC-HPH/PRC1 genes are overexpressed in DH, DCIS, and ductal carcinoma, but we believe that this interpretation should be made with caution because rare cycling healthy breast cells also express HPC-HPH/PRC1 complex genes. Future molecular studies should determine whether frequent detection of BMI-1 and its binding partners in breast carcinoma is reflective of actual overexpression of these genes.
In contrast to HPC-HPH/PRC1 complex proteins, the expression profile of the EED-EZH/PRC2 complex gene EZH2 was clearly related to loss of differentiation and development of breast carcinoma. Similar to healthy breast gland cells, EZH2 was infrequently detected in DH, lowgrade DCIS, and well-differentiated invasive carcinoma. However, EZH2+ cells were abundant in DCIS and invasive carcinomas of high grade, as confirmed using a tissue array of a separate group of 172 breast carcinomas. Interestingly, increased expression of EZH2 in breast carcinomas is in line with the recent observation that Myc-derived experimental breast tumors in mice overexpress the EZH2 homologue Enx1 [53]. In the current study, we show that increased expression of EZH2 in poorly differentiated DCIS results in coexpression of EZH2 with BMI-1 and other PcG proteins of the HPC-HPH complex. A minority of these BMI-1/EZH2 double-positive cells expressed MIB-1/Ki-67, suggesting that only a fraction of them was cycling. The high frequency of EZH2 expression in high-grade invasive carcinomas also resulted in frequent BMI-1/EZH2 coexpression, but in this case, virtually all BMI-1/EZH2 double-positives were MIB-1/Ki-67+ and dividing. This pattern suggests an increased rate of cell division within this population as opposed to BMI-1+/EZH2+ cells in high-grade DCIS. We were unable to determine whether increased EZH2 expression in BMI-1+ cells contributed to enhanced cell division or malignant transformation because our study is cross-sectional. The limited expression of MIB-1 in BMI-1+/EZH2+ neoplastic cells of poorly differentiated DCIS, in addition to the fact that cycling neoplastic cells of low-grade invasive carcinomas rarely express EZH2, indicates that EZH2 expression in breast carcinoma is not necessarily related to proliferation. However, transfection experiments of EZH2 to the Ramos cell line recently demonstrated that increased expression of EZH2 results in increased proliferation [49], whereas treatment of EZH2+ prostate cancer cells with small interfering RNA targeted against EZH2 produced inhibition of cell proliferation [39]. These experimental data suggest that a contribution of EZH2 to altered proliferative behavior of high-grade breast cancer cells is at least theoretically possible. Increasing data suggest that low-grade and high-grade breast cancers evolve through quite different genetic pathways [54]. Therefore, some of the differences in PcG expression level may reflect these different backgrounds, rather than being causal events. This will be the subject of further investigations. To which extent the described changes in expression of PcG proteins relate to the putative cell of origin within the glandular/myoepithelial cell concept of breast carcinogenesis [55] is difficult to say. It is possible that EZH2 is expressed in cells with a more stem cell phenotype, especially when they are dividing. To investigate this, further double stainings for EZH2 and cytokeratins are necessary.
An estimated one-third to half of DCIS are predicted to progress to invasive carcinoma if left untreated. We propose that enhanced expression of EZH2 in BMI-1+ cells of DCIS contributes to loss of cell identity in high-grade breast carcinomas, and that increased EZH2 expression precedes high proliferation. The recent observation that progression of prostate cancer to an invasive phenotype is also associated with enhanced expression of EZH2 [39,56] strongly suggests that altered expression of PcG genes is an important contributing factor in the development of neoplasms in humans.
Abbreviations
- DCIS
ductal carcinoma in situ
- DH
ductal hyperplasia
- PcG
polycomb group
- PRC
polycomb repressive complex
References
- 1.Pirrotta V. Polycomb silencing and the maintenance of stable chromatin states. Results Probl Cell Differ. 1999;25:205–228. doi: 10.1007/978-3-540-69111-2_10. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- 3.Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- 4.Raaphorst FM, Otte AP, Meijer CJ. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001;22:682–690. doi: 10.1016/s1471-4906(01)02082-8. [DOI] [PubMed] [Google Scholar]
- 5.van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takihara Y, Hara J. Polycomb-group genes and hematopoiesis. Int J Hematol. 2000;72:165–172. [PubMed] [Google Scholar]
- 7.Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the rb and polycomb pathways. Mol Cell. 2001;8:557–569. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- 8.Brock HW, van Lohuizen M. The Polycomb group—no longer an exclusive club? Curr Opin Genet Dev. 2001;11:175–181. doi: 10.1016/s0959-437x(00)00176-3. [DOI] [PubMed] [Google Scholar]
- 9.Satijn DP, Otte AP. Polycomb group protein complexes: do different complexes regulate distinct target genes? Biochim Biophys Acta. 1999;1447:1–16. doi: 10.1016/s0167-4781(99)00130-x. [DOI] [PubMed] [Google Scholar]
- 10.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 11.Tie F, Furuyama T, Prasad-Sinha J, Jane E, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 12.Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller MB, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell. 2002;11:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 13.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 15.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8:545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 16.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duncan IM. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics. 1982;102:49–70. doi: 10.1093/genetics/102.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Satijn DP, Gunster MJ, van der Vlag J, Hamer KM, Schul W, Alkema AJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunster MJ, Satijn DP, Hamer KM, den Blaauwen JL, de Bruijn D, Alkema MJ, van Lohuizen M, van Driel R, Otte AP. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alkema MJ, Bronk M, Verhoeven E, Otte A, van't Veer LJ, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto N, Brock HW, Nomura M, Kyba M, Hodgson J, Fujita Y, Takihara Y, Shimada K, Higashinakagawa T. RAE28, BMI1, and M33 are members of heterogeneous multimeric mammalian Polycomb group complexes. Biochem Biophys Res Commun. 1998;245:356–365. doi: 10.1006/bbrc.1998.8438. [DOI] [PubMed] [Google Scholar]
- 22.Satijn DP, Otte AP. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng J, Hart CM, Morgan K, Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furuyama T, Tie F, Harte PJ. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis. 2003;35:114–124. doi: 10.1002/gene.10173. [DOI] [PubMed] [Google Scholar]
- 25.Sewalt RG, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DP, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones CA, Ng J, Peterson AJ, Morgan K, Simon J, Jones RS. The Drosophila esc and E(z) proteins are direct partners in polycomb group-mediated repression. Mol Cell Biol. 1998;18:2825–2834. doi: 10.1128/mcb.18.5.2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gunster MJ, Raaphorst FM, Hamer KM, den Blaauwen JL, Fieret E, Meijer CJ, Otte AP. Differential expression of human Polycomb group proteins in various tissues and cell types. J Cell Biochem Suppl. 2001;36:129–143. doi: 10.1002/jcb.1093. [DOI] [PubMed] [Google Scholar]
- 30.Muyrers-Chen I, Paro R. Epigenetics: unforeseen regulators in cancer. Biochim Biophys Acta. 2001;1552:15–26. doi: 10.1016/s0304-419x(01)00032-4. [DOI] [PubMed] [Google Scholar]
- 31.Caldas C, Aparicio S. Cell memory and cancer—the story of the trithorax and Polycomb group genes. Cancer Metastasis Rev. 1999;18:313–329. doi: 10.1023/a:1006333610078. [DOI] [PubMed] [Google Scholar]
- 32.Satijn DP, Olson DJ, van der Vlag J, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goebl MG. The bmi-1 and mel-18 gene products define a new family of DNA-binding proteins involved in cell proliferation and tumorigenesis. Cell. 1991;66:623. doi: 10.1016/0092-8674(91)90106-9. [DOI] [PubMed] [Google Scholar]
- 34.Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, van Lohuizen J, Campisi J, Wazer DE, Band V. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002;62:4736–4745. [PubMed] [Google Scholar]
- 35.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raaphorst FM, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin's disease. Am J Pathol. 2000;157:709–715. doi: 10.1016/S0002-9440(10)64583-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- 38.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner M, Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha MG, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 40.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 41.van Diest PJ. No consent should be needed for using leftover body material for scientific purposes. BMJ. 2002;325:648–651. [PubMed] [Google Scholar]
- 42.Page DL, Anderson TJ, Rogers LW. Carcinoma in situ. In: Page DL, Anderson TJ, editors. Diagnostic Histopathology of the Breast. Edinburgh, UK: Livingstone; 1987. pp. 157–192. [Google Scholar]
- 43.Page DL, Anderson TJ, Rogers LW. Epithelial hyperplasia. In: Page DL, Anderson TJ, editors. Diagnostic Histopathology of the Breast. Edinburgh, UK: Livingstone; 1987. pp. 120–156. [Google Scholar]
- 44.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer: I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 45.Korsching E, Packeisen J, Agelopoulos K, Eisenacher M, Voss R, Isola PJ, van Diest PJ, Brandt B, Boecker W, Buerger H. Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest. 2002;82:1525–1533. doi: 10.1097/01.lab.0000038508.86221.b3. [DOI] [PubMed] [Google Scholar]
- 46.van Diest PJ. Ductal carcinoma in situ in breast carcinogenesis. J Pathol. 1999;187:383–384. doi: 10.1002/(SICI)1096-9896(199903)187:4<383::AID-PATH299>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 47.Bos R, Zhong H, Hanrahan CF, Mommers EC, Semenza GL, Pinedo MD, Abeloff MD, Simons JW, van Diest PJ, van der WE. Levels of hypoxia-inducible factor-1 alpha during breast carcinogenesis. J Natl Cancer Inst. 2001;93:309–314. doi: 10.1093/jnci/93.4.309. [DOI] [PubMed] [Google Scholar]
- 48.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 49.Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 50.Raaphorst FM, Otte AP, van Kemenade FJ, Blokzijl T, Fieret E, Hamer DP, Satijn DP, Meijer CJ. Distinct bmi-1 and ezh2 expression patterns in thymocytes and mature T cells suggest a role for polycomb genes in human T cell differentiation. J Immunol. 2001;166:5925–5934. doi: 10.4049/jimmunol.166.10.5925. [DOI] [PubMed] [Google Scholar]
- 51.Raaphorst FM, van Kemenade FJ, Fieret E, Hamer KM, Satijn DP, Otte CJ, Meijer CJ. Cutting edge: polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J Immunol. 2000;164:1–4. doi: 10.4049/jimmunol.164.1.1. [DOI] [PubMed] [Google Scholar]
- 52.Matsuo F, Yano K, Saito H, Morotomi K, Kato M, Yoshimoto M, Kasumi F, Akiyama F, Sakamoto G, Miki Y. Mutation analysis of the mel-18 gene that shows decreased expression in human breast cancer cell lines. Breast Cancer. 2002;9:33–38. doi: 10.1007/BF02967544. [DOI] [PubMed] [Google Scholar]
- 53.Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Gerald WL, Hunter K, Kucherlapati R, Simon R, Liu ET, Green JE. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buerger H, Mommers C, Littmann R, Simon R, Diallo R, Poremba C, Dockhorn-Dworniczak B, van Diest PJ, Boecker W. Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol. 2001;194:165–170. doi: 10.1002/path.875. [DOI] [PubMed] [Google Scholar]
- 55.Bocker W, Moll R, Poremba C, Holland R, van Diest PJ, Dervan P, Burger H, Wai D, Ina DR, Brandt B, Herbst H, Schmidt A, Lerch MM, Buchwallow IB. Common adult stem cells in the human breast give rise to glandular and myoepithelial cell lineages: a new cell biological concept. Lab Invest. 2002;82:737–746. doi: 10.1097/01.lab.0000017371.72714.c5. [DOI] [PubMed] [Google Scholar]
- 56.LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]