Abstract
Cancer cells invade by secreting degradative enzymes, which are sequestered in lysosomal vesicles. In this study, the impact of an acidic extracellular environment on lysosome size, number, and distance from the nucleus in human mammary epithelial cells (HMECs) and breast cancer cells of different degrees of malignancy was characterized because the physiological microenvironment of tumors is frequently characterized by extracellular acidity. An acidic extracellular pH (pHe) resulted in a distinct shift of lysosomes from the perinuclear region to the cell periphery irrespective of the HMECs' degree of malignancy. With decreasing pH, larger lysosomal vesicles were observed more frequently in highly metastatic breast cancer cells, whereas smaller lysosomes were observed in poorly metastatic breast cancer cells and HMECs. The number of lysosomes decreased with acidic pH values. The displacement of lysosomes to the cell periphery driven by extracellular acidosis may facilitate exocytosis of these lysosomes and increase secretion of degradative enzymes. Filopodia formations, which were observed more frequently in highly metastatic breast cancer cells maintained at acidic pHe, may also contribute to invasion.
Keywords: Breast cancer, metastasis, fluorescence microscopy, lysosome, trafficking
Introduction
Lysosomes are membranous organelles of ∼0.5 µm diameter [1] containing about 40 different types of hydrolytic enzymes, including proteases, that are optimally active at low pH. An intralysosomal pH between 4.6 and 5.0 is maintained by an H+ pump localized in the lysosomal membrane [2]. Lysosomes function as the terminal degradation compartment of the endocytic pathway [3], and are also involved in degradation of phagocytosed material [4], autophagy [5], crinophagy [6], and proteolysis of cytosolic proteins [7]. Lysosomal movement through the cell is achieved by the cooperation of actin filaments, myosin Iα, and microtubules [8]. Recent evidence also suggests a role for lysosomes in plasma membrane repair, which is mediated by calcium-regulated exocytosis [9,10].
Lysosomes may play a pivotal role in the degradation of extracellular matrix (ECM) proteins, cell invasion, and cell migration into ECM because several proteases that contribute to ECM degradation are directly or indirectly associated with lysosomes, endocytosis, or exocytosis. For instance, cathepsins L, B, and D, which are sequestered in lysosomal vesicles [11,12], exhibit proteolytic activity when activated by the acidic lysosomal environment [13]. During invasion of cancer cells, cathepsins participate in the digestion of matrix proteins in an extracellular [14], pericellular [15], or intracellular manner [16], or through activation of other zymogenic proteases, such as urokinase-type plasminogen activator (uPA) [17,18]. A second protease system associated with lysosomes is the uPA system itself. The intracellular urokinase-type plasminogen activator receptor (uPAR) is localized in the cell membrane, perinuclear region, and granule-like structures, some of which were identified as lysosomes in breast cancer cells [19,20]. First, uPAR, as part of a receptor complex, is internalized by receptormediated endocytosis [21] and then recycled back to the cell surface [22], whereas other parts of the initial complex are transferred to lysosomes for degradation. As a third protease system, two members of the matrix metalloprotease (MMP) family require lysosomal trafficking. Membrane-type MMP-1 was shown to be regulated by endocytosis and is probably routed to the lysosome for destruction [23]. Gelatinase A (MMP-2) was shown to be secreted by fibroblasts in close contact with breast cancer cells. These breast cancer cells then bind MMP-2 to their cell surface and internalize it by endocytosis [24]. Thus, endocytosis and lysosomal trafficking may play an important role in protease regulation associated with ECM degradation, invasion, and metastasis. Therefore, it is important to systematically investigate lysosomal trafficking in human breast cancer cells, as well as normal human mammary epithelial cells (HMECs).
Hypoxia [25] and acidic extracellular pH (pHe) [26,27] are frequently detected in the microenvironment of tumors. Hypoxia in solid tumors is a consequence of poor blood supply and poor perfusion [25]. This tumor hypoxia causes an elevated cellular acid production and, combined with poor tumor perfusion, leads to acidic pHe in tumors. Typically, tumor pHe is heterogeneous with a range of 6.44 < pHe < 6.79 [28]. The tumor microenvironment plays a critical role in cancer cell invasion and metastasis [29,30], as acidic pH induces invasive, migratory behavior in vitro [30] and metastasis in vivo [31], through the activation and release of proteases from cancer cells [32–35]. Because these proteases are sequestered in lysosomes, or regulated by endocytosis as discussed earlier, lysosomes may be a key mediator of this acidosis-mediated activation or release of proteases in cancer cell invasion.
We used in vitro confocal immunofluorescence imaging of two human lysosome-associated membrane proteins (hLAMP-1 and hLAMP-2), which are widely used as lysosomal markers [36]. LAMP-1/LAMP-2 are ubiquitously expressed, highly glycosylated proteins that are located in lysosomal membranes [37], where they are tightly packed. They represent more than 50% of the total membrane proteins of lysosomes, and protect the lysosomal membrane from degradation by lysosomal hydrolases [38]. LAMP-2 additionally functions as a receptor for the selective uptake and degradation of cytosolic proteins by lysosomes [7], and in chaperone-mediated autophagy [39]. We chose LAMPs for our study because they are ubiquitously expressed, highly abundant, and localized to lysosomes. Lysosometo-nucleus distance, lysosomal diameter, and the number of lysosomes were quantified in confocal images of HMECs and breast cancer cells cultured at different pH values. To study lysosomes under acidic pHe conditions in live cells, highly glycosylated lysosomal proteins were labeled with 5-amino-3,4,6-trihydroxy-tetrahydro-pyran-2-ylmethyl ester (6-O-dansyl-GlcNH2) and imaged using confocal laser scanning fluorescence microscopy, while cells were perfused in a microscopy-compatible cell perfusion chamber [40]. We consistently observed that acidic pHe led to a shift of lysosomes to the cell periphery, whereas the total number of lysosomes per cell decreased. Lysosomal diameters increased with extracellular acidification in highly metastatic breast cancer cells, but decreased in poorly metastatic breast cancer cells and HMECs.
Materials and Methods
Cell Culture
Four HMECs representing different stages of malignancy were used in this study. MCF-12A, a spontaneously immortalized HMEC line established from MCF-12M mortal cells [41], was obtained from the American Type Culture Collection (ATCC; Rockville, MD) and cultured in DMEM-Ham's F12 medium (Invitrogen Corporation, Carlsbad, CA) supplemented as described previously [41]. MCF-7, an estrogensensitive, poorly metastatic human breast cancer cell line was also obtained from ATCC and cultured in EMEM medium (Mediatech, Inc., Herndon, VA) supplemented with 10% fetal bovine serum and antibiotics [42]. MDA-MB-231 and MDA-MB-435, two estrogen-independent highly metastatic breast cancer cell lines, were kindly provided by Dr. R. J. Gillies (Arizona Health Sciences, Tuscon, AZ). These cell lines were maintained in RPMI-1640 medium (Invitrogen Corporation) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen Corporation). The three human breast cancer cell lines were originally isolated from pleural effusions of patients with breast cancer. All cells were kept in a humidified atmosphere of 5% CO2 in air at 37°C.
Immunofluorescence Staining of Breast Sections and Cell Cultures
HMECs were grown on Permanox chamber slides (Nalge Nunc, Naperville, IL) to 60% to 70% confluence. Cells were incubated with cell culture medium of either pH 7.4 (control), or pH 6.8, or pH 6.4 for 24 hours each. The cell culture medium contained 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid (HEPES; 10 mM) and 2-(N-morpholino)-ethanesulfonic acid (MES; 10–20 mM) as additional buffers to stabilize pH. After incubation with media at different pH values, cells were washed twice with ice-cold phosphatebuffered saline (PBS) and fixed with 4% paraformaldehyde in PBS for 20 minutes on ice. Cells were washed three times with ice-cold PBS and incubated with 5% normal goat serum in PBS for 30 minutes at room temperature. Formaldehydefixed, paraffin-embedded breast cancer sections (4–6 µm thickness), kindly donated by Dr. Saraswati Sukumar (Johns Hopkins University, Baltimore, MD), were treated with xylene for 30 minutes at room temperature to remove paraffin; rehydrated for 5 minutes each with 100%, 95%, and 70% ethanol; and finally treated with 0.05 M glycine-HCl, pH 3.5, for 15 minutes in a microwave oven for antigen retrieval. For histological grading of the breast cancers, neighboring sections of the same tumors were stained with hematoxylin and eosin. Cells and breast sections were incubated with a 1:50 dilution (dilution buffer consisting of 0.5% bovine serum albumin and 0.01% sodium azide in PBS) of the monoclonal antibodies H4A3 (hLAMP-1) or H4B4 (hLAMP-2) [36] (kindly provided by Dr. J. Thomas August, Johns Hopkins University) overnight at room temperature. This was followed by three washes with PBS. Subsequently, cells or sections were incubated with a 1:50 dilution of a Cy3-labeled goat anti-mouse antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 hour at room temperature and washed five times with PBS. Cell nuclei were counterstained with a 1:3000 dilution of Hoechst H-33342 (Molecular Probes, Eugene, OR) for 10 minutes at room temperature, cells were washed, and a cover glass was attached with Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL). Fluorescence microscopy was performed with a Zeiss LSM 410 laser scanning microscope (Zeiss, Goöttingen, Germany) using a 40 x /1.2 water immersion lens. Cy3 was excited with a 543-nm laser and fluorescence emission was detected with a photomultiplier using a 570-nm longpass filter. Simultaneously, H-33342 was excited with UV lasers at 351 and 364 nm, and the fluorescence emission was detected with a second photomultiplier by applying a 560-nm dichroic beam splitter and a 400-to 470-nm bandpass filter to achieve simultaneous detection of both fluorophores. Confocal z sections of 1 µm thickness containing the maximal number of labeled lysosomes were imaged. Cy3 fluorescence was assigned red, H-33342 fluorescence was assigned green, and both images were superimposed. Additional magnification was achieved electronically by application of different ZOOM factors. Differential interference contrast (DIC) images were acquired for each field of view (FOV) to analyze the number and lengths of filopodia per cell.
Western Blot Analysis
HMEC and breast cancer cell lines were cultured in T-75 tissue culture flasks until they reached 70% to 80% confluence. Cells were scraped and homogenized with lysis buffer containing 150 mM NaCl, 100 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5% Triton X-100, and a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). The amount of soluble proteins was determined using a modified Lowry assay (Bio-Rad, Richmond, CA). Twenty micrograms of total protein was loaded to each lane. Two lanes were loaded with prestained protein ladder (BenchMark; Life Technologies, Rockville, MD) as a molecular weight standard. Proteins were electrophoretically resolved by 9% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Immobilon PVDF; Millipore, Bedford, MA) by Western blotting. After blocking with 5% dry milk overnight at 4°C, PVDF membranes were incubated for 1 hour with 1:100 dilutions of hLAMP-1 (H4A3)- or hLAMP-2 (H4B4)-specific monoclonal antibody (mouse). Subsequently, membranes were washed three times with wash/dilution solution (PBS containing 1% dry milk and 0.1% Tween 20), and incubated for 1 hour with 1:3000 diluted sheep anti-mouse horseradish peroxidase-conjugated second step antibody (Amersham, Arlington Heights, IL). This was followed by five washes with wash solution. Protein bands were visualized by an enhanced chemiluminescence Western blotting detection system (Amersham) and recorded on X-ray film (Kodak Biomax; Kodak, Rochester, NY). Molecular weights of the protein bands were assigned using the prestained protein ladder (BenchMark).
Image Processing
To analyze confocal fluorescence images of HMECs and breast cancer cells, customized software was developed inhouse to detect boundaries of lysosomes and the nucleus using a boundary detection algorithm. The boundary is computed as a contour that tries to best fit the pool of pixels (pixel size is 0.15625 x 0.15625 µm) and at the same time maintains smoothness along the boundary to avoid rough edges as may arise due to imaging quality. By adjusting the smoothness factor, the user can choose the acceptable boundary by quick visual judgement. Based on this boundary, the software calculates the center point and effective diameter of each detected lysosome. Subsequently, it computes, for each lysosome, the shortest distance from the center of the lysosome to the pixel that represents the boundary of the nucleus. The software provides a histogram of lysosome-to-nucleus distance for all lysosomes in the cell, a histogram of the lysosomal diameter distribution per cell, and the total number of lysosomes per cell. Quantitation of filopodia from DIC images of cells was performed manually. One hundred cells from four independent experiments were analyzed per cell line and pH. The number of filopodia per cell was counted and the length of each filopodium was measured.
Quantitative Analysis, Statistical Analysis, and Graphical Representation of Data
The following analyses were conducted to determine if there was a significant displacement of lysosomes relative to the nucleus, as well as changes in lysosomal diameters or lysosomal number, at different pH values. Because the respective distributions were unknown, the median lysosome distance, lysosomal diameter, or number of lysosomes was determined for every cell of a given cell line at a given pH. To assess the effect of varying pH (from 7.4 to 6.8, and from 7.4 to 6.4) on lysosomal displacement, lysosomal diameter, or lysosomal number, the nonparametric equivalent of the unpaired t-test (i.e., the Mann-Whitney U-test) with P = .05 as the significance threshold was used. The Mann-Whitney U-test for difference in medians (pH 7.4 vs 6.8 and pH 7.4 vs 6.8) was first performed to test if this difference (D) was equal to zero. If D ≠ 0, the test was performed two more times to determine if D > 0 or if D < 0 (i.e., to determine whether the median lysosomal displacements, lysosomal diameters, or number of lysosomes increased or decreased with a change in pH). The P values reported in the Results section were the ones obtained for the latter test (D > 0, D < 0) wherever applicable.
Box-and-whisker plots were created to graphically display the relationship between the lysosomal displacement (as estimated from the lysosome-to-nuclear distance), lysosomal diameter, and the number of lysosomes at different pH values for each cell line. In these plots, a box is drawn with the top at the third quartile and the bottom at the first quartile of the respective distribution. The length of the box is a visual representation of the interquartile range (i.e., the box represents 50% of the data). The location of the midpoint or median of the distribution is indicated by a horizontal line in the box. In addition, straight lines or whiskers extend 1.5 times the interquartile range above and below the 75th and 25th percentiles to help define outliers, if any.
The lysosomal burden per cell was calculated from the median of the lysosomal diameter distributions, assuming a spherical volume 4/3Πr3, multiplied by the median of the number of lysosomes per cell.
Biosynthetic Labeling of Lysosomes and Fluorescence Detection in Living Cells
Labeling of lysosomes in living HMECs was performed using a dansylated glucosamine that specifically labels lysosomal proteins as previously described [40]. Briefly, HMECs or breast cancer cells were grown on sterilized 22 x 22mm2 cover glasses in the presence of 500 µM 6-O-dansyl-GlcNH2 for 4 days. This cover glass was then sealed within a microscopy-compatible perfusion chamber (designed by Drs. Montrose and Maouyo; Ross Confocal Microscope Facility, Johns Hopkins University) and continuously perfused with cell culture medium at 37°C throughout the duration of the experiment, as previously described [40]. Living cells were analyzed by fluorescence microscopy using a Zeiss LSM 410 laser scanning microscope and a 40 x /1.2 water immersion lens. The dansyl group was excited with 351-and 364-nm lasers and the emission above 515 nm was detected with a photomultiplier using a 515-nm long-pass filter. DIC images were acquired for each FOV to analyze the localization of the lysosomes in relation to the cell borders.
Results
LAMP-1 and LAMP-2 Expression in HMECs
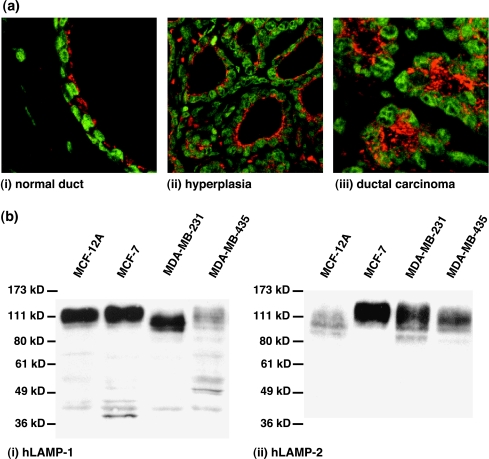
Because we studied HMECs and human breast cancer cell lines representing different stages of malignancy, we sought to verify the ubiquitous expression of the lysosomal proteins of choice, hLAMP-1 and hLAMP-2, in breast tissue specimens and the cell lines used in our study. This was necessary because the expression of hLAMP-1/hLAMP-2 has not been tested in either HMECs or breast cancer cells before, and for our method of studying lysosomal trafficking, it was crucial that hLAMP-1/hLAMP-2 were not differentially expressed in malignant cells. We therefore performed confocal immunofluorescence analysis of hLAMP-1 and hLAMP-2 in tumor sections from six breast carcinoma patients to verify the ubiquitous expression of these lysosomal proteins in breast tissue specimens. Microscopic evaluation of adjacent hematoxylin and eosin-stained sections of breast tissue revealed that normal (Figure 1a(i)), hyperplastic (Figure 1a(ii)), and malignant (Figure 1a(iii)) HMECs express both hLAMP-1 and hLAMP-2, as shown in Figure 1a. Western blot analyses revealed that the spontaneously immortalized HMEC line, MCF-12A, as well as the estrogen-sensitive, poorly metastatic breast cancer cell line, MCF-7, and two estrogen-independent highly metastatic breast cancer cell lines, MDA-MB-231 and MDA-MB-435, expressed both hLAMP-1 (Figure 1b(i)) and hLAMP-2 (Figure 1b(ii)), as shown in Figure 1b. Immunoreative bands were detected between 100 and 140 kDa, which is typical of mature highly glycosylated hLAMP-1 and hLAMP-2. Thus, hLAMP-1 and hLAMP-2 are ubiquitously expressed at different stages of breast carcinogenesis and, in particular, in the cell lines used in this study, which makes them suitable marker proteins for imaging lysosomes in all of these cell lines.
Figure 1.
(a) Representative immunofluorescence staining of hLAMPs in tumor sections of breast carcinoma patients. The LAMP fluorescence is displayed in red; the fluorescence of the cell nuclei is displayed in green. The image on the left shows a (i) normal duct (FOV 80 x 80 m). The central image shows (ii) hyperplasia (160 x 160 m). The image on the right shows a (iii) ductal carcinoma (80 x 80 m). Histological grading of the tumor sections was verified by hematoxylin and eosin staining of neighboring sections of the same tumor. (b) Western blots probing for (i) hLAMP-1 or (ii) hLAMP-2 in cell lysates from HMECs and human breast cancer cell lines used in this study. Left to right lane: MCF-12A, MCF-7, MDA-MB-231, and MDA-MB-435 cells. hLAMP-1 and hLAMP-2 antibodies revealed immunoreactive bands between 100 and 140 kDa that are typical of mature, highly glycosylated LAMP proteins. Immunofluorescence staining of hLAMP-1/hLAMP-2 in tumor sections as well as Western blots for hLAMP-1/hLAMP-2 prove that this protein is ubiquitously expressed in breast tissues and human breast cancer cell lines with different degrees of malignancy.
Automated Image Analysis to Quantitate Lysosome-to- Nucleus Distance, Lysosomal Diameters, and Number of Lysosomes
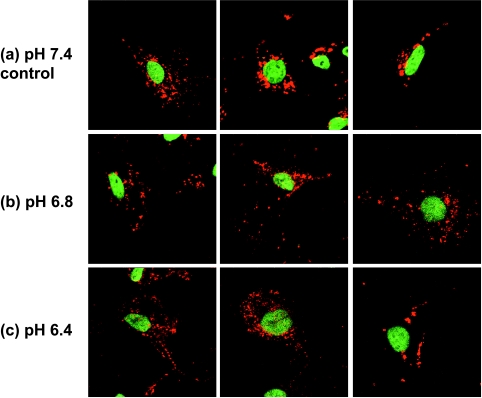
Because hLAMP-1 and hLAMP-2 were ubiquitously expressed in HMECs and the different human breast cancer cell lines studied, confocal immunofluorescence microscopy of either of these LAMPs allowed us to study lysosomal trafficking under different steady-state conditions of pHe. Figure 2 shows three representative immunofluorescence images of MDA-MB-231 cells at a physiological pH of 7.4 (Figure 2a) and two acidic pHe values of 6.8 (Figure 2b) and 6.4 (Figure 2c) as an example (raw data of the other cell lines studied are not shown). Cells maintained at an acidic extracelluar pH value of 6.4 or 6.8 for 24 hours exhibited altered lysosome-to-nucleus distance (lysosomal displacement) as evident in the images in the (b) central and (c) lower panels of Figure 2. Also, lysosomes were more scattered and shifted toward the cell periphery in cells maintained at acidic pHe. In control cells (pH 7.4; Figure 2a), lysosomes were mainly observed in the perinuclear region.
Figure 2.
Representative immunofluorescence staining of invasive/metastatic MDA-MB-231 breast cancer cells exposed to (a) pH 7.4 (control), (b) pH 6.8, or (c) pH 6.4. Examples from three images (80 x 80 m) are shown for each pH to demonstrate that in control cells (pH 7.4), lysosomes were mainly perinuclear, whereas in cells at acidic extracellular pH (pH 6.8 and pH 6.4), lysosomes were more scattered and shifted toward the cell periphery.
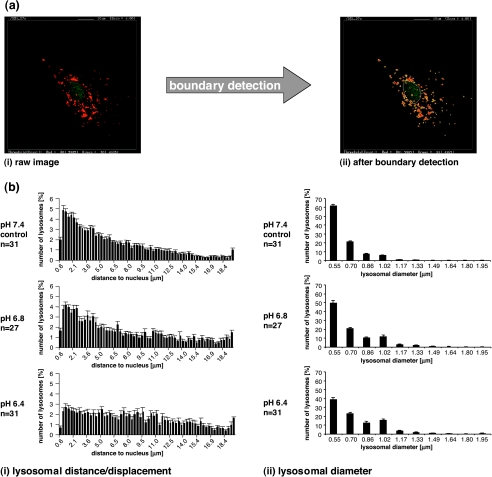
Customized software was developed in-house to analyze these fluorescence images. Figure 3a displays snapshots of the image analysis window of this software. A representative raw image is shown in Figure 3a(i), in which lysosomes are displayed in red and the cell nucleus is displayed in green. The software detects the boundaries of the lysosomes (displayed in yellow) as well as the boundary of the nucleus (displayed in white), as shown in Figure 3a(ii), and computes the shortest distance from the center point of each lysosome to the boundary of the nucleus, as described in detail in the Materials and Methods section. The software also computes the distribution of lysosomal diameters and lysosomal number per cell. Figure 3b displays typical histograms of the lysosomal distance distribution (Figure 3b(i)) and lysosomal diameter distribution (Figure 3b(ii)) generated by quantitative analyses of fluorescence images using the in-house software, reflecting the changes observed in the raw images (Figure 2). As evident from the example of MDA-MB-231 cells, the software computed the frequency distribution of the number of lysosomes in cells under different pH conditions versus distance from the nucleus, as shown in Figure 3b(i), and versus lysosomal diameter, as shown in Figure 3b(ii). At acidic pH values, a higher number of lysosomes at a larger distance from the nucleus were detected when compared with cells at pH 7.4 (Figure 3b(i)). Also, there were fewer small-diameter lysosomes and more largediameter lysosomes at acidic pH values compared to pH 7.4 in this example of MDA-MB-231 breast cancer cells.
Figure 3.
(a) Snapshots displaying the image analysis window of our in-house software to demonstrate the boundary detection of the nucleus (white line) and the lysosomes (yellow lines) performed by the software. The image on the left is a (i) raw fluorescence image of a single MDA-MB-231 cell (80 x 80 m) and the image on the right shows (ii) the same image after boundary detection by the software. (b) The frequency distribution of (i) lysosome-to-nucleus distance and (ii) lysosomal diameter in MDA-MB-231 breast cancer cells exposed to pH 7.4 (upper panel, control), pH 6.8 (central panel), or pH 6.4 (lower panel). Individual cells (typically from one to two cells per image) from four independent stainings per pH value were analyzed.
Acidic pHe Shifts Lysosomes to the Cell Periphery
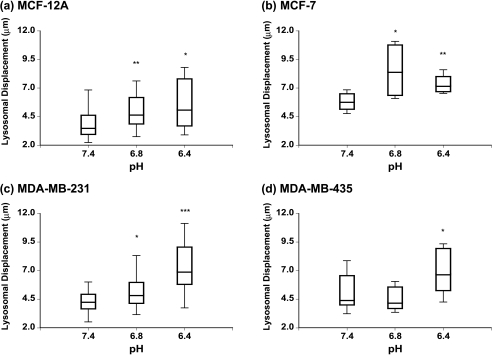
Lysosomes shifted toward the cell periphery at lower pH values, in all the cell lines, as shown in Figure 4. MCF-12A (Figure 4a) and MDA-MB-231 (Figure 4c) cells exhibited an increase in the median lysosomal displacement (as estimated from the lysosome-to-nuclear distance) with decreasing pH. As shown in Figure 4, the largest median distance was apparent at the lowest pH of 6.4 in all cell lines except MCF-7 cells (Figure 4b), for which the largest median distance occurred at pH 6.8. The Mann-Whitney U-test revealed a statistically significant (P < .05) increase in lysosomal displacement for pH 6.4 vs 7.4 in all four human breast epithelial cell lines independent of their degree of malignancy (Figure 4). However, the increase in lysosomal displacement at pH 6.8 vs 7.4 was significant (P < .05) only in MCF-12A, MCF-7, and MDA-MB-231 cells (Figure 4). To make sure that the observed changes in lysosomal displacement were not corrupted by changes in cell shape (e.g., cell swelling, cell shrinking, or changes in cell polarity or spreading), cell size was characterized at different pH values by determining the maximum lysosome-to-nucleus distance. This maximum distance, obtained from lysosomes localized at the plasma membrane, was used as an index for alterations in cell size at different pH values. The results of these measurements are summarized in Table 1. No significant differences of these values were detected in the HMECs or any of the breast cancer cells under different pH conditions, demonstrating that there were no changes in cell shape with extracellular acidosis.
Figure 4.
Box-and-whisker plots of changes in lysosomal displacement with extracellular pH in (a) MCF-12A, (b) MCF-7, (c) MDA-MB-231, and (d) MDA-MB-435 cells. The box represents the range between the third quartile and the first quartile of the respective distribution displaying the length of the interquartile range, which is 50% of the data. The median of the distribution is indicated by the horizontal line in the box. *Represents P < .05. **Represents P < .01, pH 6.8 vs 7.4 and pH 6.4 vs 7.4 (Mann-Whitney U-test; if D ≠ 0, P values for D > 0, D < 0).
Table 1.
Maximal Lysosome-to-Nucleus Distance in Microns for pH 7.4 (Control), pH 6.8, and pH 6.4 in an HMEC Line and Three Breast Cancer Cell Lines Representing Different Stages of Breast Carcinogenesis.
| Cell Line | Maximal Lysosome Distance [µm] (Number of Images) | ||
| pH 7.4 (n) | pH 6.8 (n) | pH 6.4 (n) | |
| MCF-12A | 15.8 ± 1.1 (34) | 16.7 ± 1.0 (23) | 16.6 ± 0.7 (10) |
| MCF-7 | 27.1 ± 2.8 (10) | 27.0 ± 4.3 (4) | 26.9 ± 1.0 (9) |
| MDA-MB-231 | 19.2 ± 1.1 (31) | 19.1 ± 1.9 (27) | 19.1 ± 0.8 (31) |
| MDA-MB-435 | 25.1 ± 2.6 (14) | 24.8 ± 3.0 (5) | 25.3 ± 2.2 (9) |
Data were obtained by analyzing n images from four different stainings per pH and cell line using our in-house software. Values are mean ± SE.
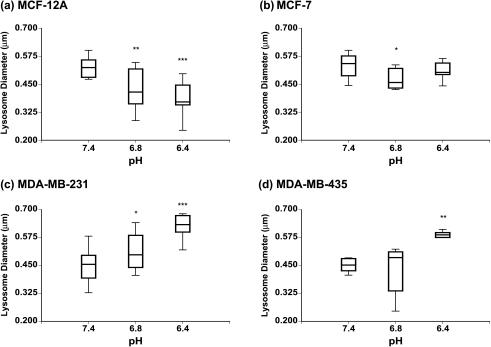
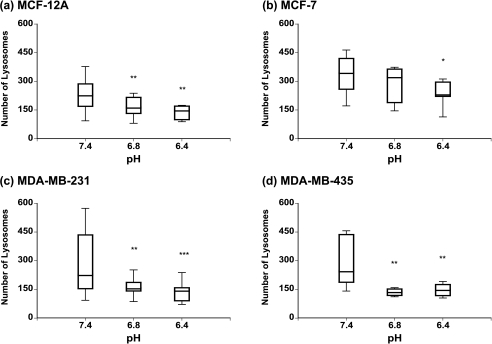
Acidic pHe Leads to Changes in Lysosomal Diameter and Lysosomal Number
Lysosomal diameters of the HMECs and the different breast cancer cell lines, under different conditions of pHe as measured by the in-house image analysis software, are displayed in Figure 5. A shift toward larger lysosomal vesicles was observed in the highly metastatic human breast cancer cell lines MDA-MB-231 (Figure 5c) and MDA-MB-435 (Figure 5d) with decreasing pH (Figure 5, lower panel). This increase in lysosomal diameter was statistically significant (P < .001) for pH 6.4 vs 7.4 in MDA-MB-231 and MDA-MB-435 (P < .01) cells, whereas for pH 6.8 vs 7.4, only MDA-MB-231 cells displayed a statistically significant (P < .05) increase in lysosomal diameter (Figure 5, lower panel). In contrast, exposure to decreasing pH values resulted in smaller lysosomes in the poorly metastatic breast cancer cell line MCF-7 (Figure 5b, upper panel) and the HMEC line MCF-12A (Figure 5a, upper panel). This decrease in lysosomal diameter was significant (P < .01) for pH 6.8 vs 7.4 in MCF-12A and MCF-7 (P < .05) cells; for pH 6.4 vs 7.4, only nonmalignant MCF-12A cells exhibited a significant (P < .001) decrease in lysosomal diameter (Figure 5, upper panel). The number of lysosomes decreased at acidic pH values as compared to physiological pH 7.4 in the HMEC line as well as all three breast cancer cell lines (Figure 6). There was a greater decrease in the two highly metastatic breast cancer cell lines (Figure 6, lower panel) compared to the poorly metastatic breast cancer cell line and the HMEC line (Figure 6, upper panel). Lysosomal numbers were significantly reduced (P < .05) at pH 6.4 vs 7.4 in all four cell lines (Figure 6). At pH 6.8 vs 7.4, all cell lines except MCF-7 cells displayed a significantly (P < .01) decreased number of lysosomes (Figure 6).
Figure 5.
Box-and-whisker plots of changes in lysosomal diameter with extracellular pH in (a) MCF-12A, (b) MCF-7, (c) MDA-MB-231, and (d) MDA-MB-435 cells. The box represents the range between the third quartile and the first quartile of the respective distribution displaying the length of the interquartile range, which is 50% of the data. The median of the distribution is indicated by the horizontal line in the box. *Represents P < .05. **Represents P < .01. ***Represents P < .001, pH 6.8 vs 7.4 and pH 6.4 vs 7.4 (Mann-Whitney U-test; if D ≠ 0, P values for D > 0, D < 0).
Figure 6.
Box-and-whisker plots of changes in lysosomal number with extracellular pH in (a) MCF-12A, (b) MCF-7, (c) MDA-MB-231, and (d) MDA-MB-435 cells. The box represents the range between the third quartile and the first quartile of the respective distribution displaying the length of the interquartile range, which is 50% of the data. The median of the distribution is indicated by the horizontal line in the box. *Represents P < .05. **Represents P < .01. ***Represents P < .001, pH 6.8 vs 7.4 and pH 6.4 vs 7.4 (Mann-Whitney U-test; if D ≠ 0, P values for D > 0, D < 0).
The lysosomal burden, given as the volume of all lysosomes per cell in Table 2, decreased with decreasing pH in the HMEC line MCF-12A and the poorly metastatic breast cancer cell line MCF-7. In contrast, lysosomal burden significantly increased in both highly metastatic breast cancer cell lines MDA-MB-231 and MDA-MB-435 (Table 2).
Table 2.
Lysosomal Burden Per Cell in Cubic Micrometer for pH 7.4 (Control), pH 6.8, and pH 6.4 in an HMEC Line and Three Breast Cancer Cell Lines Representing Different Stages of Breast Carcinogenesis.
| Cell Line | Lysosomal Burden [µm3/cell] (Number of Images) | ||
| pH 7.4 (n) | pH 6.8 (n) | pH 6.4 (n) | |
| MCF-12A | 16.8 (34) | 6.0 (23) | 3.7 (10) |
| MCF-7 | 28.9 (10) | 17.0 (4) | 15.4 (9) |
| MDA-MB-231 | 11.2 (31) | 9.9 (27) | 18.5 (31) |
| MDA-MB-435 | 11.6 (14) | 7.3 (5) | 14.5 (9) |
Data were obtained by analyzing n images from four different stainings per pH and cell line using our in-house software. Lysosomal burden was calculated from the median of the lysosomal diameter distributions (Figure 5) by assuming a spherical volume (4/3Πr3) and the median of the number of lysosomes per cell (Figure 6) to give the total volume of lysosomes per cell (µm3/cell).
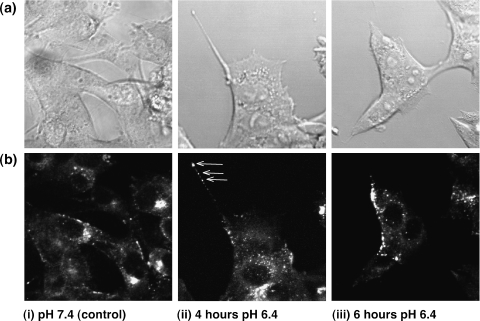
Acidic pHe Shifts Lysosomes to the Cell Periphery in Living Cells and Increases Cellular Filopodia Formation
To further validate the results obtained in fixed cells, the effects of extracellular acidification were studied in live cells. 6-O-dansyl-GlcNH2 was used to specifically label highly glycosylated lysosomal proteins and image lysosomes by laser scanning confocal fluorescence microscopy in living cells continuously perfused in a microscopy-compatible cell perfusion system, as previously described [40]. A comparison of DIC (Figure 7a) and fluorescence images (Figure 7b) obtained from the same FOV allowed us to observe changes in lysosomal trafficking with respect to the cell border and the nucleus, as shown in Figure 7. After 4 hours of switching cell perfusion to pH 6.4 medium, MDA-MB-435 breast cancer cells started displaying a more scattered and more peripheral lysosome distribution (Figure 7b(ii)) than control cells maintained with medium at pH 7.4 (Figure 7b(i)). Additionally, breast cancer cells displayed filopodia (Figures 7(ii) and 8, (ii) and (iii)) more frequently at acidic pH as compared to cells maintained at physiological pH (Figures 7(i) and 8(i)). These filopodia often contained lysosomes or lysosomal proteins at their distal ends as marked by arrows in Figure 7b(ii). Figure 7(iii) shows MDA-MB-435 breast cancer cells in which lysosomes were translocated to the cell periphery within 6 hours of switching the perfusion medium to pH 6.4. Comparable results were obtained with live MDA-MB-231 cells. Living MCF-7 and MCF-12A cells also exhibited a more scattered and peripheral lysosome distribution at pH 6.4 within 4 hours, but did not extend filopodia.
Figure 7.
Representative (a) differential interference contrast images (80 x 80 m) and (b) dansyl group fluorescence images (80 x 80 m) of living MDA-MB-435 human breast cancer cells perfused in the microscopy-compatible cell perfusion system using cell culture medium at (i) pH 7.4 (control), (ii) pH 6.4 for 4 hours, and (iii) pH 6.4 for 6 hours. Highly glycosylated lysosomal proteins of these cells were biosynthetically prestained prior to the experiment with the dansyl group carrying glucosamine 6-O-dansyl-GlcNH2, as previously described [40]. Some cells displayed filopodia that contained lysosomes as well (b(ii), arrows).
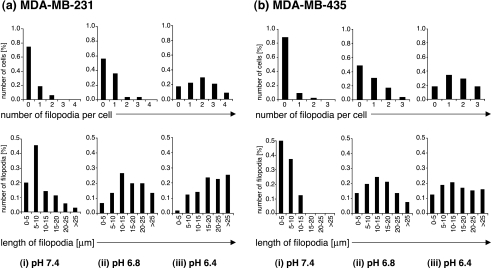
Figure 8.
Quantitation of the number of filopodia per cell (upper panel) and the length of filopodia (lower panel) in invasive/metastatic human breast cancer cells (a) MDA-MB-231 and (b) MDA-MB-435 at (i) pH 7.4 (control), (ii) pH 6.8, and (iii) pH 6.4. 100 cells from four independent experiments were measured per cell line and pH employing DIC images. In both metastatic breast cancer cell lines, the number of cells with a high number of filopodia as well as the number of long filopodia increased gradually with decreasing pH. No filopodia were observed in MCF-12A HMECs and poorly invasive human breast cancer cells MCF-7.
Figure 8 displays the number of filopodia per cell (Figure 8, upper panel) and the lengths of filopodia (Figure 8, lower panel) as quantified from 100 cells after 24 hours of incubation at pH 7.4 (control; Figure 8(i)), pH 6.8 (Figure 8(ii)), and pH 6.4 (Figure 8(iii)) in highly malignant MDA-MB-231 (Figure 8a) and MDA-MB-435 (Figure 8b) breast cancer cells. The frequency of filopodia formations gradually increased with acidic pH (Figure 8, (ii) and (iii)) only in the highly metastatic human breast cancer cell lines MDA-MB-231 (Figure 8a) and MDA-MB-231 (Figure 8b). The lengths of filopodia gradually increased with low pH (Figure 8, (ii) and (iii)) in the highly metastatic breast cancer cell lines as well, as shown in the lower panel of Figure 8. No filopodia formation whatsoever was observed in HMECs and poorly metastatic breast cancer cells.
Discussion
Maintaining HMECs and human breast cancer cells at an acidic pHe resulted in profound changes in lysosomal displacement, number, and diameter. Acidic pHe values of pH 6.8 and pH 6.4 caused a significant shift of lysosomes from the perinuclear region to the cell periphery in the HMEC line as well as in the breast cancer cell lines representing different stages of malignancy. Lysosomal displacement to the cell periphery occurred irrespective of whether the cells were malignant or nonmalignant. Cells are exposed to acidic pHe during hypoxic periods in normal tissues during wound healing [43], or in the pathophysiological microenvironment of tumors [26,27]. Lysosomes participate in the regulation and function of matrix metalloproteinases, serine proteases, and cathepsins, which are sequestered in lysosomal vesicles. Lysosomal displacement to the cell periphery, detected in response to an acidic pHe in this study, may be a mechanism to facilitate increased secretion of degradative enzymes. This would be the first step in the recruitment of lysosomes toward the cell surface before lysosomal exocytosis resulted in the actual secretion. A recent study has demonstrated that lysosomes are the vesicles responsible for exocytosis in nonsecretory cells [10]. The results presented in this paper are consistent with studies where cathepsin B redistribution toward the cell periphery was observed at acidic pHe in cancer cells [33]. The significant reduction in lysosomal numbers after chronic exposure to acidic pH values suggests that increased exocytosis, not compensated for by cellular biogenesis of lysosomes, may have occurred.
One interesting finding to emerge from this study was that lysosomal diameter and lysosomal burden decreased at low pHe in HMECs as well as poorly metastatic breast cancer cells, and increased at low pHe in highly metastatic breast cancer cells. Although lysosomal diameters increased with decreasing pH in highly metastatic breast cancer cell lines, we did not observe lysosomal vesicles larger than 2 µm as previously reported [16,44] in breast cancer cells in a Matrigel migration assay. Our data suggest that extracellular acidification alone is not sufficient to induce formation of large acidic vesicles (LAVs) with a diameter of 5 to 10 µm in breast cancer cells. Access to particles such as ECM, glass beads, or dextrans, which can be endocytosed by breast cancer cells, may be required for the formation of LAVs in these cells. Increased formation of cellular filopodia extension mediated by an acidic pericellular environment was observed in the highly metastatic breast cancer cells only. This filopodia formation may increase the cell's ability to perform directional cell migration. Localization of lysosomes or lysosomal proteins to filopodia indicates that lysosomes were recruited to these filopodia extensions to potentially facilitate directional invasion at these sites.
Previous observations in macrophages suggested an outward movement of lysosomes along microtubules, with decreasing cytoplasmic pH [45]. In cancer cells, however, chronic acidic pHe does not cause chronic acidification of cytoplasmic pH but adaptation of cellular pH regulation enabling these cells to maintain a normal intracellular pH while the pHe is acidic [46,47]. In vivo pH measurements have revealed regions of acidic pHe in tumors although the intracellular pH is maintained in the neutral range [48,49]. Therefore, cytoplasmic acidification as a consequence of extracellular acidification seems to be an unlikely explanation for the outward movement of lysosomes, observed in the HMECs and the breast cancer cells, in the present study. Previous studies have demonstrated that mammalian cells contain two populations of lysosomes based on their spatial localization [10]. Consistent with these results, most lysosomes are present in the perinuclear region under physiological pH conditions in HMECs and breast cancer cells, and only a small subpopulation is present close to the plasma membrane. Only lysosomes predocked at the plasma membrane undergo calcium-dependent exocytosis [10]. This indicates that recruitment of lysosomes to the cell surface during extracellular acidification is a process that differs from lysosomal movement during calcium-dependent exocytosis, as evident from the greater range of lysosomal displacement. Microtubules involved in the long-range movement of lysosomes, or actin filaments performing their local transport [8,50,51] may be the first target of extracellular acidosis-induced lysosomal displacement. Certain steps in the actin polymerization process and actin binding to membrane-anchoring proteins were shown to be associated with changes in cytoplasmic pH [52], which may be translated into alterations in lysosomal movement. pHe may thus play a role in signaling events that cause the lysosomal rearrangement. However, further studies are necessary to understand potential signaling pathways, which may mediate the observed outward shift of lysosomes during extracellular acidification.
Recent findings suggested that, in addition to their classical role as terminal digestive compartment of the cell [3,53,54], lysosomes have other functions, such as calcium-dependent exocytosis in nonsecretory cells [10,55], calcium-dependent plasma membrane repair in mammalian cells through exocytosis [9], as well as developmentally specialized lysosomal function during dendritic cell maturation [56]. In this study, we have demonstrated, for the first time, that extracelluar pH conditions affect the lysosometo- nucleus distance, diameter, and total numbers of lysosomes in HMECs and breast cancer cells. This is of critical importance because the microenvironment in breast tumors is usually characterized by acidic pHe values [28]. Filopodia formation was triggered by acidic pH in the highly metastatic breast cancer cells only, which may be related to their invasive behavior. Future studies should clarify the specific pathways involved in pH-mediated lysosomal trafficking and the effects of an acidic environment on directed migration, and address potential secondary effects of lysosomal trafficking on proteases.
Acknowledgements
We thank Djikolngar Maouyo for expert technical assistance in performing the experiments with the microscopy-compatible cell perfusion system. We would also like to thank B. Douglas Ward (Biophysics Research Institute at the Medical College of Wisconsin) for his helpful suggestions regarding statistical analyses. We thank Gary Cromwell for maintaining the cell lines and J. Thomas August for providing us with monoclonal antibodies against hLAMP-1 (H4A3) and hLAMP-2 (H4B4).
Abbreviations
- DIC
differential interference contrast
- ECM
extracellular matrix
- FOV
field of view
- HMEC
human mammary epithelial cell
- hLAMP
human lysosome-associated embrane protein
- LAV
large acidic vesicle
- LSM
laser scanning microscopy
- MMP
matrix metalloprotease
- pHe
extracellular pH
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- uPA
urokinase-type plasminogen activator
- uPAR
urokinase-type plasminogen activator receptor
Footnotes
This work was supported by P20 CA86346 and P50 CA103175.
Current address: Optimer Pharmaceuticals, Inc., San Diego, CA 92121, USA.
References
- 1.Holtzmann E. Lysosomes. New York: Plenum Press; 1989. [Google Scholar]
- 2.Mellman I, Fuchs R, Helenius A. Acidification of the endocytic and exocytic pathways. Annu Rev Biochem. 1986;55:663–700. doi: 10.1146/annurev.bi.55.070186.003311. [DOI] [PubMed] [Google Scholar]
- 3.Kornfeld S, Mellman I. The biogenesis of lysosomes. Annu Rev Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 4.Funato K, Beron W, Yang CZ, Mukhopadhyay A, Stahl PD. Reconstitution of phagosome-lysosome fusion in streptolysin O-permeabilized cells. J Biol Chem. 1997;272:16147–16151. doi: 10.1074/jbc.272.26.16147. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence BP, Brown WJ. Autophagic vacuoles rapidly fuse with pre-existing lysosomes in cultured hepatocytes. J Cell Sci. 1992;102:515–526. doi: 10.1242/jcs.102.3.515. [DOI] [PubMed] [Google Scholar]
- 6.Noda T, Farquhar MG. A non-autophagic pathway for diversion of ER secretory proteins to lysosomes. J Cell Biol. 1992;119:85–97. doi: 10.1083/jcb.119.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuervo AM, Dice JF. receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 8.Cordonnier MN, Dauzonne D, Louvard D, Coudrier E. Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol Biol Cell. 2001;12:4013–4029. doi: 10.1091/mbc.12.12.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy A, Caler EV, Andrews NW. Plasma membrane repair is mediated by Ca(2+)-regulated exocytosis of lysosomes. Cell. 2001;106:157–169. doi: 10.1016/s0092-8674(01)00421-4. [DOI] [PubMed] [Google Scholar]
- 10.Jaiswal JK, Andrews NW, Simon SM. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J Cell Biol. 2002;159:625–635. doi: 10.1083/jcb.200208154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rochefort H, Capony F, Garcia M, Cavailles V, Freiss G, Chambon M, Morisset M, Vignon F. Estrogen-induced lysosomal proteases secreted by breast cancer cells: a role in carcinogenesis? J Cell Biochem. 1987;35:17–29. doi: 10.1002/jcb.240350103. [DOI] [PubMed] [Google Scholar]
- 12.Sloane BF, Moin K, Krepela E, Rozhin J. Cathepsin B and its endogenous inhibitors: the role in tumor malignancy. Cancer Metastasis Rev. 1990;9:333–352. doi: 10.1007/BF00049523. [DOI] [PubMed] [Google Scholar]
- 13.McGrath ME. The lysosomal cysteine proteases. Annu Rev Biophys Biomol Struct. 1999;28:181–204. doi: 10.1146/annurev.biophys.28.1.181. [DOI] [PubMed] [Google Scholar]
- 14.Linebaugh BE, Sameni M, Day NA, Sloane BF, Keppler D. Exocytosis of active cathepsin B enzyme activity at pH 7.0, inhibition and molecular mass. Eur J Biochem. 1999;264:100–109. doi: 10.1046/j.1432-1327.1999.00582.x. [DOI] [PubMed] [Google Scholar]
- 15.Sloane BF, Moin K, Sameni M, Tait LR, Rozhin J, Ziegler G. Membrane association of cathepsin B can be induced by transfection of human breast epithelial cells with c-Ha-ras oncogene. J Cell Sci. 1994;107:373–384. doi: 10.1242/jcs.107.2.373. [DOI] [PubMed] [Google Scholar]
- 16.Montcourrier P, Mangeat PH, Valembois C, Salazar G, Sahuquet A, Duperray C, Rochefort H. Characterization of very acidic phagosomes in breast cancer cells and their association with invasion. J Cell Sci. 1994;107:2381–2391. doi: 10.1242/jcs.107.9.2381. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi H, Schmitt M, Goretzki L, Chucholowski N, Calvete J, Kramer M, Gunzler WA, Janicke F, Graeff H. Cathepsin B efficiently activates the soluble and the tumor cell receptor-bound form of the proenzyme urokinase-type plasminogen activator (Pro-uPA) J Biol Chem. 1991;266:5147–5152. [PubMed] [Google Scholar]
- 18.Kobayashi H, Ohi H, Sugimura M, Shinohara H, Fujii T, Terao T. Inhibition of in vitro ovarian cancer cell invasion by modulation of urokinase-type plasminogen activator and cathepsin B. Cancer Res. 1992;52:3610–3614. [PubMed] [Google Scholar]
- 19.Bastholm L, Nielsen MH, De Mey J, Dano K, Brunner N, Hoyer-Hansen E, Ronne E, Elling F. Confocal fluorescence microscopy of urokinase plasminogen activator receptor and cathepsin D in human MDA-MB-231 breast cancer cells migrating in reconstituted basement membrane. Biotech Histochem. 1994;69:61–67. doi: 10.3109/10520299409106263. [DOI] [PubMed] [Google Scholar]
- 20.Bastholm L, Elling F, Brunner N, Nielsen MH. Immunoelectron microscopy of the receptor for urokinase plasminogen activator and cathepsin D in the human breast cancer cell line MDA-MB-231. APMIS. 1994;102:279–286. doi: 10.1111/j.1699-0463.1994.tb04876.x. [DOI] [PubMed] [Google Scholar]
- 21.Heegaard CW, Simonsen AC, Oka K, Kjoller L, Christensen A, Madsen L, Ellgaard L, Chan L, Andreasen PA. Very low density lipoprotein receptor binds and mediates endocytosis of urokinase-type plasminogen activator-type-1 plasminogen activator inhibitor complex. J Biol Chem. 1995;270:20855–20861. doi: 10.1074/jbc.270.35.20855. [DOI] [PubMed] [Google Scholar]
- 22.Nykjaer A, Conese M, Christensen EI, Olson D, Cremona O, Gliemann F, Blasi F. Recycling of the urokinase receptor upon internalization of the uPA:serpin complexes. EMBO J. 1997;16:2610–2620. doi: 10.1093/emboj/16.10.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang A, Lehti K, Wang X, Weiss SJ, Keski-Oja J, Pei D. Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc Natl Acad Sci USA. 2001;98:13693–13698. doi: 10.1073/pnas.241293698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polette M, Gilbert N, Stas I, Nawrocki B, Noel A, Remacle A, Stetler-Stevenson WG, Birembaut P, Foidart M. Gelatinase A expression and localization in human breast cancers. An in situ hybridization study and immunohistochemical detection using confocal microscopy. Virchows Arch. 1994;424:641–645. doi: 10.1007/BF00195779. [DOI] [PubMed] [Google Scholar]
- 25.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. [PubMed] [Google Scholar]
- 26.Stubbs M, McSheehy PM, Griffiths CL. Causes and consequences of tumour acidity and implications for treatment. Mol Med Today. 2000;6:15–19. doi: 10.1016/s1357-4310(99)01615-9. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths JR, McIntyre DJ, Howe FA, Stubbs M. Why are cancers acidic? A carrier-mediated diffusion model for H+ transport in the interstitial fluid. Novartis Found Symp. 2001;240:46–62. doi: 10.1002/0470868716.ch4. discussion 62–67, 152–53. [DOI] [PubMed] [Google Scholar]
- 28.van Sluis R, Bhujwalla ZM, Raghunand N, Ballesteros P, Alvarez J, Cerdan S, Galons JP, Gillies RJ. In vivo imaging of extracellular pH using 1H MRSI. Magn Reson Med. 1999;41:743–750. doi: 10.1002/(sici)1522-2594(199904)41:4<743::aid-mrm13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 29.Bhujwalla ZM, Artemov D, Aboagye E, Ackerstaff E, Gillies RJ, Natarajan K, Solaiyappan M. The physiological environment in cancer vascularization, invasion and metastasis. Novartis Found Symp. 2001;240:23–38. doi: 10.1002/0470868716.ch3. discussion 38–45, 152–53. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Zaguilan R, Seftor EA, Seftor RE, Chu YW, Gillies RJ, Hendrix MJ. Acidic pH enhances the invasive behavior of human melanoma cells. Clin Exp Metastasis. 1996;14:176–186. doi: 10.1007/BF00121214. [DOI] [PubMed] [Google Scholar]
- 31.Schlappack OK, Zimmermann A, Hill RP. Glucose starvation and acidosis: effect on experimental metastatic potential, DNA content and MTX resistance of murine tumour cells. Br J Cancer. 1991;64:663–670. doi: 10.1038/bjc.1991.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montcourrier P, Silver I, Farnoud R, Bird I, Rochefort H. Breast cancer cells have a high capacity to acidify extracellular milieu by a dual mechanism. Clin Exp Metastasis. 1997;15:382–392. doi: 10.1023/a:1018446104071. [DOI] [PubMed] [Google Scholar]
- 33.Rozhin J, Sameni M, Ziegler G, Sloane BF. Pericellular pH affects distribution and secretion of cathepsin B in malignant cells. Cancer Res. 1994;54:6517–6525. [PubMed] [Google Scholar]
- 34.Cuvier C, Jang A, Hill RP. Exposure to hypoxia, glucose starvation and acidosis: effect on invasive capacity of murine tumor cells and correlation with cathepsin (L + B) secretion. Clin Exp Metastasis. 1997;15:19–25. doi: 10.1023/a:1018428105463. [DOI] [PubMed] [Google Scholar]
- 35.Webb SD, Sherratt JA, Fish RG. Modelling tumour acidity and invasion. Novartis Found Symp. 2001;240:169–181. doi: 10.1002/0470868716.ch12. discussion 181–85. [DOI] [PubMed] [Google Scholar]
- 36.Furuta K, Yang XL, Chen JS, Hamilton SR, August JT. Differential expression of the lysosome-associated membrane proteins in normal human tissues. Arch Biochem Biophys. 1999;365:75–82. doi: 10.1006/abbi.1999.1147. [DOI] [PubMed] [Google Scholar]
- 37.Fukuda M. Lysosomal membrane glycoproteins. Structure, biosynthesis, and intracellular trafficking. J Biol Chem. 1991;266:21327–21330. [PubMed] [Google Scholar]
- 38.Kundra R, Kornfeld S. Asparagine-linked oligosaccharides protect Lamp-1 and Lamp-2 from intracellular proteolysis. J Biol Chem. 1999;274:31039–31046. doi: 10.1074/jbc.274.43.31039. [DOI] [PubMed] [Google Scholar]
- 39.Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–2456. doi: 10.1074/jbc.M001394200. [DOI] [PubMed] [Google Scholar]
- 40.Glunde K, Guggino SE, Ichikawa Y, Bhujwalla ZM. A novel method of imaging, lysosomes in living human mammary epithelial cells. Mol Imaging. 2003;2:24–36. doi: 10.1162/15353500200302142. [DOI] [PubMed] [Google Scholar]
- 41.Paine TM, Soule HD, Pauley RJ, Dawson PJ. Characterization of epithelial phenotypes in mortal and immortal human breast cells. Int J Cancer. 1992;50:463–473. doi: 10.1002/ijc.2910500323. [DOI] [PubMed] [Google Scholar]
- 42.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 43.Crowther M, Brown NJ, Bishop ET, Lewis CE. Microenvironmental influence on macrophage regulation of angiogenesis in wounds and malignant tumors. J Leukoc Biol. 2001;70:478–490. [PubMed] [Google Scholar]
- 44.Montcourrier P, Mangeat PH, Salazar G, Morisset M, Sahuquet A, Rochefort H. Cathepsin D in breast cancer cells can digest extracellular matrix in large acidic vesicles. Cancer Res. 1990;50:6045–6054. [PubMed] [Google Scholar]
- 45.Heuser J. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J Cell Biol. 1989;108:855–864. doi: 10.1083/jcb.108.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook JA, Fox MH. Effects of chronic pH 6.6 on growth, intracellular pH, and response to 42.0 degrees C hyperthermia of Chinese hamster ovary cells. Cancer Res. 1988;48:2417–2420. [PubMed] [Google Scholar]
- 47.Owen CS, Pooler PM, Wahl ML, Coss RA, Leeper DB. Altered proton extrusion in cells adapted to growth at low extracellular pH. J Cell Physiol. 1997;173:397–405. doi: 10.1002/(SICI)1097-4652(199712)173:3<397::AID-JCP12>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 48.Griffiths JR. Are cancer cells acidic? Br J Cancer. 1991;64:425–427. doi: 10.1038/bjc.1991.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raghunand N, Gillies RJ. pH and chemotherapy. Novartis Found Symp. 2001;240:199–211. doi: 10.1002/0470868716.ch14. discussion 265–68. [DOI] [PubMed] [Google Scholar]
- 50.Matteoni R, Kreis TE. Translocation and clustering of endosomes and lysosomes depends on microtubules. J Cell Biol. 1987;105:1253–1265. doi: 10.1083/jcb.105.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson J, Bushnell A, Silverstein SC. Tubular lysosome morphology and distribution within macrophages depend on the integrity of cytoplasmic microtubules. Proc Natl Acad Sci USA. 1987;84:1921–1925. doi: 10.1073/pnas.84.7.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Tang J, Edmonds BT, Murray J, Levin S, Condeelis J. F-actin sequesters elongation factor 1alpha from interaction with aminoacyl-tRNA in a pH-dependent reaction. J Cell Biol. 1996;135:953–963. doi: 10.1083/jcb.135.4.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cohn ZA, Ehrenreich BA. The uptake, storage, and intracellular hydrolysis of carbohydrates by macrophages. J Exp Med. 1969;129:201–225. doi: 10.1084/jem.129.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehrenreich BA, Cohn ZA. The fate of peptides pinocytosed by macrophages in vitro. J Exp Med. 1969;129:227–245. doi: 10.1084/jem.129.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews NW. Lysosomes and the plasma membrane: trypanosomes reveal a secret relationship. J Cell Biol. 2002;158:389–394. doi: 10.1083/jcb.200205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]