Abstract
Background:
Rat Nb2-11C lymphoma cells are dependent on prolactin for proliferation and are widely used to study prolactin signaling pathways. To investigate the role of this hormone in the transcriptional mechanisms that underlie prolactin-stimulated mitogenesis, five different techniques were used to isolate differentially expressed transcripts: mRNA differential display, representational difference analysis (RDA), subtractive suppressive hybridization (SSH), analysis of weakly expressed candidate genes, and differential screening of an organized library.
Results:
About 70 transcripts were found to be modulated in Nb2 cells following prolactin treatment. Of these, approximately 20 represent unknown genes. All cDNAs were characterized by northern blot analysis and categorized on the basis of their expression profiles and the functions of the known genes. We compared our data with other cell-cycle-regulated transcripts and found several new potential signaling molecules that may be involved in Nb2 cell growth. In addition, abnormalities in the expression patterns of several transcripts were detected in Nb2 cells, including the constitutive expression of the immediate-early gene EGR-1. Finally, we compared the differential screening techniques in terms of sensitivity, efficiency and occurrence of false positives.
Conclusions:
Using these techniques to determine which genes are differentially expressed in Nb2 lymphoma cells, we have obtained valuable insight into the potential functions of some of these genes in the cell cycle. Although this information is preliminary, comparison with other eukaryotic models of cell-cycle progression enables identification of expression abnormalities and proteins potentially involved in signal transduction, which could indicate new directions for research.
Introduction
Prolactin is a pleiotropic hormone whose numerous actions are associated with reproduction, growth and development, water and electrolyte balance, metabolism, behavior and immunoregulation [1]. The prolactin-dependent rat Nb2 lymphoma (Nb2-11C) is widely used as a model in which to study signal transduction and transcriptional mechanisms that underlie prolactin-stimulated mitogenesis (reviewed in [1,2,3]). Deprivation of lactogen induces a block in the early G1 phase of the Nb2 cell cycle [4]. The addition of physiological concentrations of prolactin to synchronized G0/G1-arrested cultures reinitiates cell-cycle progression [5], which is characterized by the induction of growth-related genes such as those for c-Myc, β-actin and ornithine decarboxylase (ODC) and hsp70-like genes [6]. This list has recently been extended to include the genes for interferon regulatory factor-1 (IRF-1) [7], cyclins D2 and D3 [8], proto-oncogene Pim-1 [9], growth factor independence-1 (Gfi-1) [10], and rat nuclear distribution c (Rnudc) [11]. The identification of other prolactin-regulated genes in proliferating Nb2 cells would help to elucidate the relationships between prolactin-activated proteins and genes induced or repressed by prolactin, and lead to a better understanding of the role of prolactin in proliferation regulatory mechanisms. In this study, five differential screening techniques were applied at different stages of the Nb2 cell cycle. Differential display [12], representational difference analysis (RDA [13]) and suppressive subtractive hybridization (SSH [14]) consist of selective and/or suppressive cycles of PCR using cDNA prepared from the cell populations or tissues to be compared. The two other techniques used were the screening of an organized library [15] and the analysis of weakly expressed candidate genes. These two methods are based on the hybridization of DNA macro- or microarrays on nylon filters using complex probes generated from radiolabeled transcribed cDNA isolated from the cell populations to be compared.
We have characterized known and unknown transcripts identified by these five techniques, adding information relative to their expression peak or expression variations during Nb2 cell proliferation. Whenever possible, prolactin-induced transcripts were compared with those in other eukaryotic models of cell-cycle progression such as Saccharomyces cerevisiae and normal human fibroblasts. This comparison allowed us to establish non-exhaustive lists of cell-cycle-regulated transcripts. Regulated mRNAs were classified with respect to their functional characteristics and to their conservation from yeast to vertebrates. On the basis of this analysis, new signaling molecules presumably involved in Nb2 proliferation are proposed. Furthermore, we have detected expression profile abnormalities in Nb2 lymphoma cells, and we discuss the consequence of one, the constitutive expression of the immediate-early gene EGR-1.
Results
Application of the different screening techniques to Nb2 cells
When deprived of lactogen, 80-85% of an Nb2 cell culture is synchronized in growth arrest [5] (Figure 1). Addition of prolactin to the culture reinitiates cell-cycle progression and cell proliferation. Using differential display, we first compared RNAs from synchronized Nb2 cells stimulated for various times with prolactin. In addition, three different RDA and SSH subtractive libraries were prepared. One RDA library allowed the identification of transcripts expressed at a higher level during proliferation (12 hours prolactin-stimulated) compared with growth arrest, and two SSH subtractive libraries were used to compare expression profiles in growth arrest and G1 (mix of cells stimulated with prolactin for 2, 4, 6 and 8 hours) and vice versa. Messenger RNAs from Nb2 cells were used to differentially screen an organized library of rat brain cDNA. Finally, the expression of 91 weakly expressed candidate genes was also compared in growth-arrested, early (mix of 2, 4, 6 and 8 hours prolactin-stimulated Nb2 cells), intermediate (10, 12 and 14 hours) or late (20, 22 and 24 hours) proliferative phase and unsynchronized Nb2 cells.
Figure 1.
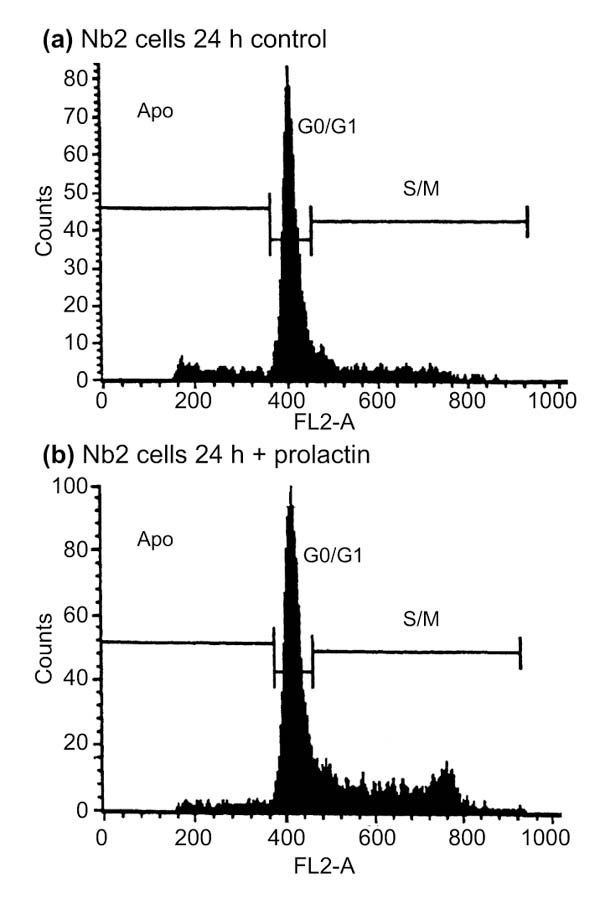
Cell-cycle analysis of synchronized Nb2 cells stimulated by prolactin. Nb2 cells were serum deprived for 24 hours, then incubated with no PRL or PRL at 20 ng/ml before cells were collected for analysis. DNA content (FL2-A) versus cell number is presented in each panel. (a) Profiles obtained with control cells; (b) profiles obtained with cells incubated with PRL. From left to right, profiles correspond to cells in apoptosis (Apo, area below 400 on the x axis), in G0/G1 (peak centered on 400 on the x axis), or in S/M phase (area above 400 on the x axis).
Most of the potential positive clones isolated by differential display, RDA, SSH and screening of rat brain organized library were analyzed by northern blot to eliminate false positives and to evaluate variations in the expression of each clone during the Nb2 proliferative response. The remaining cDNA clones were tested using reverse northern blot to rapidly eliminate false-positive cDNAs. Briefly, PCR products corresponding to potential positive clones were screened by hybridization with complex probes generated from the populations tested. This step was necessary because of the high rate of false-positive clones generated by the earlier protocols used for differential display. The method has since been improved and may now have a better readout. Sequencing of these clones enabled us to identify known transcripts and to determine which ones were of unknown genes.
Together, the different techniques enabled the isolation of about 70 known or unknown differentially expressed transcripts potentially involved in the resumption of cell proliferation by quiescent cells. A summary of the data is presented in Table 1. Examples of expression profiles obtained by northern blot using the isolated cDNAs as probes are shown in Figure 2. We did not isolate transcripts for known molecules such as histones or cyclins; it is, however, of interest to note that the expression of the rat homolog of Cdc21, the adenosine nucleotide translocator Ant-2, the nuclear export factor CRM-1 (exportin), and unknown transcripts DD3, 4-16 and 4-15 (Figure 2) are induced during Nb2 cell proliferation. In contrast, expression of the unknown transcript 6-4 is decreased during G1 phase, but this transcript is much more abundant in unsynchronized Nb2 cells. Northern blots indicate that some of these cDNA probes identify several distinct transcripts, probably generated by alternative splicing (ANT-2, CRM-1, CD45 (leukocyte common antigen), DD3, 4-15), which are not necessarily all induced in the same manner. Interestingly, two opposite expression profiles are observed for the two transcripts recognized by the cDNA probe identified as CD45 (clone from the SSH library G1 phase growth arrest). Indeed, the longer transcript is progressively repressed during Nb2 cell-cycle progression, whereas the shorter form is induced. These examples emphasize that northern blot analysis provided new information that could not be obtained using other methods (such as reverse northern blot, arrays or DNA chips). As shown in Table 1, about 20 of the differentially expressed cDNAs that were isolated correspond to unknown transcripts whose expression is modulated during Nb2 proliferation. Most of these unknown cDNAs share significant homology with several mouse and human expressed sequence tags (ESTs) isolated from various libraries, suggesting that the corresponding transcripts are ubiquitously expressed and have a role in cell proliferation in one of the different functional categories described below.
Table 1.
Differentially expressed transcripts found in Nb2 cells during cell-cycle progression using five different screening techniques
| Identity | Accession number | Expression variations | |
| Differential display | |||
| Unknown DD3 | No EST | Three transcripts (2, 2.8, 3.7 kb) | |
| expressed in G1 and G1/S, | |||
| (>4-fold induction) | |||
| Representational difference analysis: G1/S-GA | |||
| ATP synthase β subunit | M19044 | Induced in G1/S, G2 (2-fold induction) | |
| Aldehyde dehydrogenase | U79118 | Induced in late G1 (2-fold induction) | |
| Enolase α1 | NM012554 | Induced in late G1 (2-fold induction) | |
| Dynein heavy chain | D13896 | Induced in G1, G1/S (3-fold induction) | |
| TCP-1 ε | D43950(h) | Induced in G1/S, G2 (3-fold induction) | |
| αCOP (COPA) | U24105 | Induced in late G1 (2-fold induction) | |
| Cdc21 homolog | D26089 (m) | Peaks in G1/S (>8-fold induction) | |
| Ribosomal protein L26 | A1716351 | Induced in late G1 (2-fold induction) | |
| Itm1 | A1956728 (h) | Induced in G1/S (2-fold induction) | |
| Unknown T22 | A1053031 (h) | 6 kb, 2-fold induction in G1/S | |
| Unknown T34 | A1235326 | 2 kb, 2-fold induction in G1/S | |
| Subtractive suppressive hybridization: G1-GA | |||
| Galectin-8 | U09824 | Induced in G1 (2-fold induction) | |
| Hsp86 | X16857 | Induced in late G1 (>3-fold induction) | |
| TCP-1 η | AA900460 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L13A | X68282 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L12 | AA900142 | Induced in late G1 (2-fold induction) | |
| Ribosomal protein L3 | A1687295 | Induced in G1 (2-fold induction) | |
| Y00441 | Induced in G1 (2-fold induction) | ||
| β2-microglobulin | D12771 | Induced from late G1 to G2 (>5 fold induction) | |
| Adenine nucleotide translocator ANT-2 | NM011342 (m) | Induced in G1 (2-fold induction) | |
| Sec-22 | NM012562 | Induced in G1 (2-fold induction) | |
| L-fucosidase | U91538 | Induced in G1/S (>3-fold induction) | |
| CRM-1 homolog/exportin 1 | X81839 | Induced in G1/S (>3-fold induction) | |
| Ubiquitin/ribosomal protein S27a | A47416 | Induced in G1/S (>3-fold induction) | |
| Ubiquitin/ribosomal protein S30 (FAU) | AF195142 (m) | 3-fold induction in G1 (1.5 kb) | |
| Unknown 4-2 (mouse selenoprotein R mRNA) | H35219 | 2-fold induction in G1/S (2 and 4 kb) | |
| Unknown 4-4 (human KIAA0081) | No EST | 2-fold induction in G1 (1 kb) | |
| Unknown 4-11 | AU035826 | 4-fold induction in G1 (1 and 1.5 kb) | |
| Unknown 4-15 | AF046001 | 4-fold induction in G1 (2.5 kb) | |
| Unknown 4-16 (human ZNF207 or mouse Zep) | No EST | 3-fold induction in G1 (1.5 kb) | |
| Unknown 4-20 | AW435432 | 2-fold induction in G1 | |
| Unknown 4-27 (new ribosomal protein L15 type) | A1121996 (m) | 2-fold induction in G1 | |
| Unknown 4-49 | AW246248 (h) | 2-fold induction in G1 | |
| Unknown 4-58 (SH3, Rab GAP, TBC domain) | No EST | 2-fold induction in G1 | |
| Unknown 4-59 | |||
| Subtractive suppressive hybridization: GA-G1 | |||
| Spermidine/spermine N-acetyl transferase (SSAT) | AA955996 | Repressed transiently in G1 | |
| Leukocyte common antigen (alternative splicing) CD45 | Y00065 | Switch between two transcripts (one | |
| repressed, the other induced in G1) | |||
| ZFX | X75171 | Repressed transiently in G1 | |
| Ribosomal protein S8 | AA874997 | Repressed transiently in G1 | |
| Ribosomal protein S13 | L01123 | Repressed transiently in G1 | |
| Unknown 6-2 | No EST | Repressed transiently in G1 | |
| Unknown 6-3 (hypothetical protein expressed in thymocytes) | AJ237585 (m) | Repressed transiently in G1 | |
| Unknown 6-4 | No EST | Repressed in G1 | |
| Unknown 6-9 | No EST | Repressed transiently in G1 | |
| Unknown 6-10 | No EST | Repressed transiently in G1 | |
| Unknown 6-12 (human protein KIAA0710) | AB014610 (h) | Repressed transiently in G1 | |
| Unknown 6-45 (homolog to mouse PARP-2) | NM009632 (m) | Repressed transiently in G1 | |
| Differential screening of a rat organized library: G2-GA | |||
| Prothymosin α | M86564 | Induced in G2 | |
| Cyclophilin | M19533 | Induced in G2 | |
| ATP synthase β subunit | M19044 | Induced in G1/S and G2 | |
| Tubulin α2 | AA686718 | Induced in G1/S and G2 | |
| GaPDH | X02231 | Induced in G2 | |
| Phosphoglycerate kinase | M31788 | Induced in G1/S and G2 | |
| MRG1 related protein | U65093 | Induced in G1/S and G2 | |
| Unknown BO1 | No EST | Induced in G2 | |
| Analysis of weakly expressed candidate genes: UN, A, GI, G1/S, G2 | |||
| Name | Accession number | Fold induction | Kinetics |
| Ganglioside synthase (GD3) | D84068 | > 4 |  |
| P13 kinase | D64045 | > 3 | |
| Phospholipase Cγ1 | M34667 | > 3 | |
| Bax | S76511 | > 2 |  |
| P53 | X13058 | > 2 | |
| FAKp125 | AF020777 | > 3 | |
| 14-3-3 ε | M84416 | > 2 | |
| 14-3-3 η | D17445 | > 2 |  |
| Vitamin D3 receptor | J09838 | 3 | |
| Glycine transporter | M88595 | 3 |  |
| Thromboxane A2 receptor | D32080 | 2 |  |
| Phosphatidylinositol transfer protein | D17445 | 2 |  |
| RexB/NSP | U17604 | > 2 |  |
| Glucocorticoid receptor | M14053 | 2 |  |
| Ga3PDH | J04147 | 2 |  |
| Zif268 = EGR1 | U75398 | 2 |  |
Nb2 cells: UN, unsynchronized; GA, growth arrested; G1, G1 phase; G1/S, G1/S transition; G2, G2 phase. PC12, rat pheochromocytoma PC12 cells. 18S and 28 S, ribosomal RNA. Whenever possible, rat accession numbers (GenBank) are written in the second column. When this sequence is not in known for the rat, (h) or (m) indicates that the accession number corresponds, respectively, to a human and a mouse cDNA homologous to those of the rat. `No EST' means that our rat sequence does not correspond to any previously described EST in mammals. For some of the unknown cDNAs, an estimation of the size of the corresponding transcript(s) as well as the fold induction is given in the right column `Expression variations', and for weakly expressed candidate genes, the expression kinetics are shown.
Figure 2.
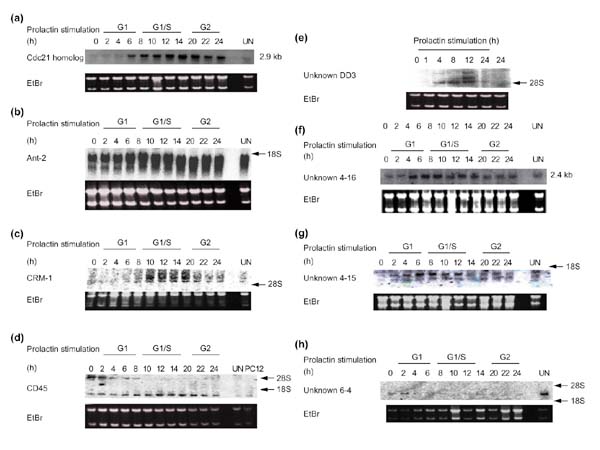
Expression profiles of various known and unknown transcripts during Nb2 cell cycle progression. Samples of total RNA (10 μg) were loaded per lane and blots were hybridized with the indicated cDNA probes. Ethidium bromide staining (EtBr) of the gels is shown as a control (18S and 28S rRNA). (a) Cdc21 homolog; (b) Ant-2; (c) CRM-1; (d) CD45; (e) unknown DD3; (f) unknown 4-16; (g) unknown 4-15; (h) unknown 6-4.
Putative signaling molecules potentially involved in Nb2 cell survival and/or cell-cycle progression
The list of differentially expressed transcripts has been completed with Nb2 cell transcripts described in previous reports (Table 2). These differentially expressed genes can be found in almost all the subclasses listed in Table 2, including, for example, those for receptors (such as the prolactin receptor [16], the T-cell receptor γ chain [17], the vitamin D3 receptor, the thromboxane A2 and the prostaglandin F2 receptors (our present results)), transcription factors (IRF-1 [7], c-Myc [18], Zfx (our present results)), and T-cell survival and apoptosis molecules (Bag-1 [19], Bcl-2, Bax [20], Ant-2 (our present results)). Although all the signaling molecules are not necessarily regulated at the transcriptional level, it can be hypothesized that those translated from cell-cycle-modulated transcripts are involved in Nb2 cell-cycle progression. This hypothesis seems to be confirmed, as we have found that several molecules previously described as transducers in Nb2 cells are encoded by cell-cycle-modulated transcripts (for example, phosphatidylinositide 3-kinase (PI 3-kinase), phospholipase Cγ1 (PLCγ1), and focal adhesion kinase (FAK) p125). Consequently, it is tempting to speculate that at least some of the cell-cycle-modulated transcripts may encode transducers of Nb2 cell proliferation. For example, the stress kinase p38 mitogen-activated protein kinase (p38 MAP kinase), whose transcript is induced in Nb2 cells upon prolactin stimulation [21], maybe involved in prolactin-induced signaling pathways. This hypothesis is reinforced by the fact that p38 MAP kinase seems to be required for the optimal activation of T cells by interleukin (IL)-12 and IL-2 and for the regulation of serine phosphorylation of STAT transcription factors [22]. The GD3 ganglioside synthase, which mediates the propagation of CD95-generated apoptotic signals in hematopoietic cells [23], may also be involved in the regulation of survival and apoptosis in Nb2 cells. It is also possible that receptors, such as the prostaglandin F2, thromboxane A2 and vitamin D3 receptors, galectin-8 and CD45 and their ligands, may be involved in the signaling pathways required for Nb2 cell survival and proliferation.
Table 2.
Functional classification of cell-cycle-regulated transcripts found in Nb2 cells
| Category | Transcript |
| Cell cycle: cyclins and cell-cycle regulators | |
| Transcripts that have a cell-cycle-modulated homolog in yeast | Cyclin E1 [78], peaks in G1 |
| EGR-1, immediate-early gene (constitutive expression in Nb2 cells) | |
| Cdc5-like protein [79], peaks in M | |
| Cdk2, Cdk5 [78], peak in G1 | |
| Cell-cycle-modulated transcripts with a yeast homolog (not modulated) | Cyclin B1, peaks in G2/M |
| Cell-cycle-modulated transcripts with no yeast homolog | Cyclin B2, peaks in G2 |
| Cyclin D2 [8], peaks in G1 | |
| Cyclin D3 [8], peaks in G1 | |
| Nucleotide metabolism, DNA replication and repair | Cdc21 homolog |
| Spermidine/spermine N-acetyl transferase (SSAT) | |
| Ornithine decarboxylase (ODC) [80] | |
| S-adenosylmethionine decarboxylase [80] | |
| Prothymosin α | |
| PARP-2 (Unknown 6-45) | |
| Chromatin structure | Histones H2A, H2B, peak in S in mammals and in yeast |
| Cytoskeleton | Myosin heavy chain |
| Tubulin α, β | |
| β-actin [6] | |
| Clone 15 = rNUDC [81] | |
| FAK p125 | |
| Cell-surface antigens, adhesion molecules and signaling molecules | |
| (involved in apoptosis, survival and/or proliferation) | |
| Growth factor | FGF-2 [82] |
| Surface molecules | Prolactin receptor Nb2 form [16] |
| T-cell receptor γ chain [17] | |
| T-cell receptor α chain [17] | |
| GnRH receptor [83] | |
| Glucocorticoid receptor | |
| Galectin-8 | |
| Leukocyte membrane glycoprotein, CD45 | |
| Vitamin D3 receptor | |
| Thromboxane A2 receptor | |
| β2-microglobulin | |
| Cytoplasmic and/or nuclear signaling molecules | p38 Map kinase [21] |
| Pim-1 [9] | |
| Gfi-1 [10] | |
| Stathmin [84] | |
| P13 kinase p110 α | |
| Phospholipase Cγ1 | |
| 14-3-3 η and ε | |
| Bax | |
| p53 | |
| RexB/NSP | |
| Phosphatidylinositol transfer protein | |
| α 4 phosphoprotein [79] | |
| Transcription factors | IRF-1 [7] |
| c-Myc [18] | |
| c-Fos (not in Nb2 cells [18]) | |
| MRG1-related protein | |
| E2F-1 [78] | |
| Zfx | |
| Heat shock, stress response and chaperones | Cyclophilin (B) |
| TCP-1 ε and η | |
| Hsp70 (not expressed in Nb2 cells) | |
| Hsp70-like = Nb29 [41] | |
| Hsp27 | |
| Hsp86 | |
| β-actin | |
| α2-tubulin | |
| Rdnuc (Golgi-associated protein) [11] | |
| Myosin heavy chain | |
| Focal adhesion kinase (FAK) | |
| Metabolism (energy) | Phosphoglycerate kinase |
| Enolase α | |
| Aldehyde dehydrogenase | |
| ATP synthase β subunit | |
| Protein and RNA synthesis, modifications and degradation | |
| Ribosomal proteins | L3, L12, L13A, new ribosomal protein L15 type (unknown 4-27) |
| S8, S13 | |
| Glycosylation factor | Itm1 |
| Elongation factor | EF-2 [77] |
| Inter-compartment transport and trafficking | CRM-1 = exportin 1 |
| Sec-22 | |
| Glycine transporter | |
| Unknown 4-58 (SH3, Rab GAP, TBC domain: putative nuclear pore protein) | |
| Unknown function(s) | FGF-responsive Non/p54nrb |
| Unknowns T22, T34, 4-2, 4-4, 4-11, 4-15, 4-20, 4-49, 4-59, BO1 | |
| Unknowns 6-2, 6-3, 6-4, 6-9, 6-10, 6-12 |
Functional classification of cell-cycle-regulated transcripts
We identified 70 differentially expressed genes in proliferating Nb2 cells. Although this number is not negligible, it is of course not exhaustive, as the number of genes involved in cell-cycle modifications could be as high as several hundred. However, estimations of the number of genes modulated using other proliferation and cell-cycle models, such as yeast [24,25,26] or human fibroblasts [27,28], are equally limited. We compared these different models, adding new information generated from large-scale differential screening techniques (microarrays and DNA chips). On the basis of these analyses, approximately 7% of transcripts from yeast (416 out of 6,220) and 6% from normal human fibroblasts (517 out of 8,600) display cell-cycle-dependent fluctuations. All the yeast cell-cycle-regulated transcripts are not, however, regulated in vertebrates, and vice versa.
These transcripts were classified into 10 different functional categories as shown in Table 2: cell cycle (cyclins and cell-cycle regulators); nucleotide metabolism, and DNA replication and repair; chromatin structure; cytoskeleton, cell surface antigens, adhesion molecules and signaling molecules (involved in apoptosis, survival and/or proliferation); heat shock, stress response and chaperones; metabolism (energy); protein and RNA synthesis, modifications and degradation; inter-compartment transport and trafficking; and unknown function(s).
These functional categories agree with previous observations concerning cell-cycle-regulated transcripts in various eukaryotic models. Indeed, cell-cycle-dependent mRNA fluctuations have been observed for genes involved in many cellular processes, including control of mRNA transcription [29], responsiveness to external stimuli [30] and subcellular localization of proteins [31]. Genetic studies have revealed that the activity of cell-cycle regulatory proteins is required for normal DNA repair [32], meiosis [33] and multicellular development [34,35]. These observations suggest that, in eukaryotic cells, diverse biological events depend on maintenance of this periodicity.
Expression abnormalities
The loss of appropriate cell-cycle regulation leads to genomic instability [36] and is believed to have a role in the etiology of both hereditary and spontaneous cancers [37,38,39,40]. In Nb2-11C cells, several growth-related genes that display abnormalities in their expression patterns were observed. These abnormalities may be the cause or the consequence of the tumor phenotype of Nb2 cells.
Using the candidate gene approach (Figure 3a), striking expression abnormalities were observed. For example, Nb2 cells display an abnormal response to heat shock. Indeed, whereas the hsp70-like mRNA is upregulated following lactogen stimulation [41], no expression of the inducible hsp70 (GenBank X74271) gene was detected. As components of the heat-shock response are involved in normal cell-cycle-progression, the abnormalities observed in Nb2 cells may have important consequences for their growth. Furthermore, in comparison with mammalian models of cell-cycle progression, expression abnormalities of immediate-early genes are observed in Nb2 cells. Indeed, the expression of c-fos remains undetectable in our model (Table 2) as well as in starved Nb2 cells, which resume proliferation after prolactin stimulation [18]. In contrast, the expression of EGR-1 (also termed Zif268 / KROX24 / ETR103 / NGFIA / TIS8 / GOS30) is constitutive in Nb2 cells. This peculiarity, observed using the candidate gene approach (Figures 3b,4), was confirmed by northern blot (Figure 3c). The gene has been shown in numerous model systems to have induction kinetics similar to c-fos, characterized by a rapid transient expression requiring de novo transcription between 15 and 30 minutes after the mitogenic stimulus [42,43,44,45,46,47].
Figure 3.
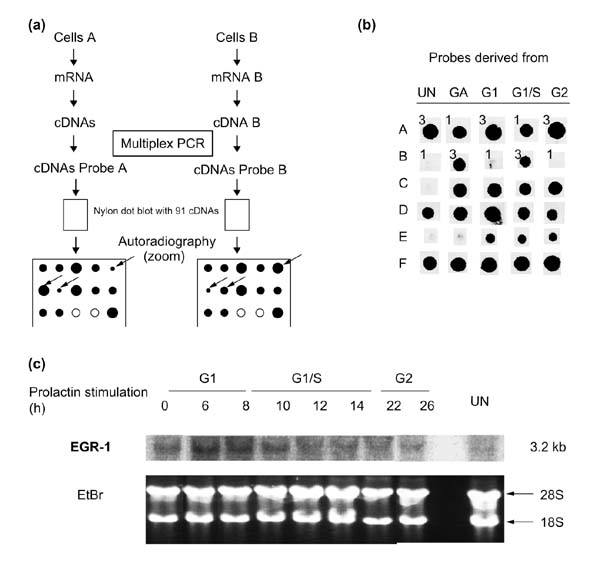
Analysis of candidate gene expression. (a) General principles. Messenger RNAs from the different cell populations (cells A and B) are reverse transcribed. Multiplex PCR is then performed using specific primer pairs to amplify the cDNAs of interest. The resulting mixture of PCR products is radiolabeled and these complex probes are used to hybridize identical membranes spotted with the candidate gene cDNA targets. After autoradiography, the intensities of the hybridization signals are compared and quantified. Arrows indicate the positions of differentially expressed genes. The absence of hybridization (open circles) indicates that the candidate gene is not expressed. (b) Efficiency of the technique and examples of differentially expressed genes. The expression of different candidate genes was compared in either unsynchronized (UN), growth-arrested (GA), G1 phase (G1), G1/S transition (G1/S) or G2 phase (G2) cultures of Nb2 cells. The efficiency of the technique was controlled using equivalent amounts of rabbit α and β globin cDNAs, which were included on the nylon membranes along with the candidate gene targets. The two globin cDNAs were added in different amounts (50 or 150 ng) to each cDNA population before co-amplification. For each population tested, filters were hybridized with both globin probes, but only representative hybridization signals are shown, for either α (Panel A) or β (Panel B) globin. Numbers 1 and 3 represent the relative amount of the control rabbit globin cDNAs added, and are reflected in the differences in the intensity of the hybridization signals. Thus, a threefold difference in the quantity of a particular transcript in the initial population generates a clear difference in the intensity of the corresponding hybridization signals. Rows C, D, E and F are examples of the results obtained with ganglioside synthase GD3, EGR-1, FAK p125 and Stat3, respectively. Except for Stat3, which is not differentially expressed in probes UN, GA, G1, G1/S and G2, the three other genes showed a clear differential expression. (c) Northern blot analysis showing the constitutive expression of EGR-1 during Nb2 cell-cycle progression. Growth-arrested Nb2 cells were stimulated with ovine prolactin and collected after various periods of stimulation corresponding to different stages of the cell cycle (G1, G1/S and G2). The expression of EGR-1 was evaluated by northern blot using 10 μg of total RNA from the various times following prolactin stimulation. Ethidium bromide (EtBr) staining of the gel is shown as a control (18S and 28S rRNA).
Figure 4.
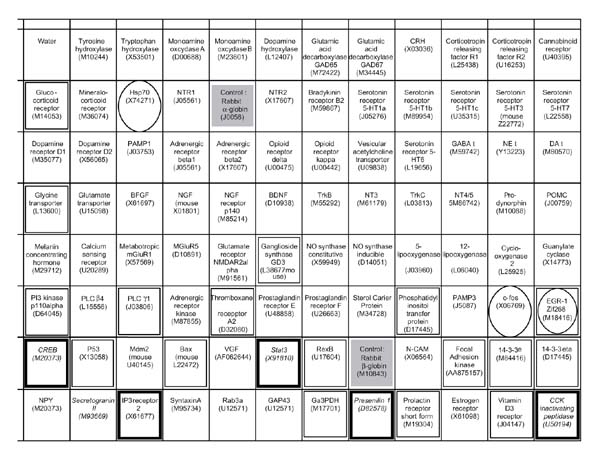
Schematic representation of rat candidate genes on a nylon filter. Squares with names and accession numbers represent the places where the cDNAs were spotted. The solid gray boxes correspond to the controls (rabbit α and β globin). The boxes enclosed in a thick black square represent differentially expressed genes in Nb2 cells; the boxes enclosed in a thin black square represent genes that are repressed, but not differentially in Nb2 cells; and those enclosed in an oval correspond to expression abnormalities in Nb2 cells.
Discussion
New putative signaling molecules in Nb2 cells
In this study we have identified new regulated genes encoding potential signaling molecules. Genes encoding proteins involved in cell survival, apoptosis, proliferation and/or cell-cycle progression may or may not have cell-cycle-dependent expression. Most of these proteins (listed in Tables 1,2) are known to be activated by post-translational mechanisms; little is known, however, about their regulation at the transcriptional level.
In our experiments, for example, PI 3-kinase, PLCγ1 and ganglioside synthase GD3 share identical expression profiles, characterized by lower expression in unsynchronized than in synchronized Nb2 cells. This indicates that they may share similar regulation mechanisms. In high-density Nb2 cell cultures [48], secreted growth factors maybe involved in this negative regulation. PI 3-kinase and/or PLCγ1 have been implicated in cell-cycle progression, proliferation, survival, transformation and apoptosis in different cellular models [49,50,51,52]. Thus, ganglioside synthase may also take part in similar processes in immune cells. Indeed, ganglioside synthase GD3 is highly expressed in various human cancer cell lines, is upregulated in activated T lymphocytes [53], and has been implicated in Fas-mediated apoptosis [23]. It may therefore be of interest to determine whether prolactin is able to activate, directly or indirectly, the activity of GD3 and the Fas signaling pathways.
Expression profiles of genes for Bax, p53, 14-3-3 ε and FAK are characterized by increased mRNA expression during the G1, G1/S and G2 phases in comparison to the growth-arrested or unsynchronized Nb2 cells (Table 1). These kinetics suggest that prolactin may have a direct effect on the transcription of these genes. Indeed, in myeloid cells [54] and in proliferating prostate cells [55], FAK expression is induced by various cytokines. This molecule is located at the signaling crossroads of cell growth and attachment, and is involved in dynamic cytoskeletal rearrangements [56]. In Nb2 cells, prolactin has been shown to increase bax mRNA expression in 8 hours [20]. The thromboxane A2 receptor, which is highly expressed in immature thymocytes, has also been shown to mediate DNA fragmentation and apoptosis [57]. It is possible that, in Nb2 cells, prolactin could also counteract thromboxane-induced apoptosis, as is the case for glucocorticoids.
It is not known at present whether these expression profiles are common to all dividing mammalian cells or only to a particular subclass of immune-system cells. Alternatively, these profiles could be the consequence of the genetic abnormalities displayed by Nb2 cells. It is of interest that the expression of the p38 MAP kinase gene is modulated in Nb2 cells [21] but not in normal human fibroblasts [27,28], suggesting that this regulation is specific to the T-cell lineage. Different arguments exist in favor of the existence of cross-talk between the JAK/STAT and p38 MAP kinase pathways, at both the translational and transcriptional levels. As the expression of ganglioside synthase GD3 is restricted to the brain and the hematopoietic lineage, the regulation of the transcript and the involvement of the protein in the regulation of survival and apoptosis may also be shared by these tissues. In contrast, the cell-cycle-dependent expression of Ant-2, observed in both Nb2 cells (our results) and human fibroblasts [27,28], suggests that this feature is common to all dividing mammalian cells. These speculations require further experimentation at the protein level for confirmation.
Expression abnormalities
We report for the first time, to our knowledge, the constitutive expression of the transcription factor EGR-1 in synchronized proliferating cells. EGR-1 has a role in differentiation and development, in normal growth and in virus-induced growth and immortalization (reviewed in [58]). Many of these effects may be related to complex cooperative and competitive mechanisms between the three transcription factors Sp1, EGR-1 and Wt1, which often have overlapping binding sites in target promoters.
Several arguments suggest that EGR-1 may act as a tumor suppressor [59,60,61], and that its anti-oncogenic function could be due to the transcriptional induction of the gene for transforming growth factor β1 (TGFβ1), which suppresses growth by an autocrine mechanism in the late G1 phase of the cell cycle [62,63]. Exogenous TGFβ inhibits Nb2 cell growth [64], suggesting that these cells are still sensitive to the anti-proliferative action of TGFβ, but are unable to synthesize or activate TGFβ on their own, despite their constitutive EGR-1 expression. It is possible that, in Nb2 cells, the anti-proliferative effect induced by the constitutive expression of EGR-1 is suppressed by other abnormalities such as a deficiency in the production of active TGFβ.
Other studies, however, argue in favor of the existence of anti-apoptotic and/or pro-proliferative properties of EGR-1 [65,66,67,68]. In this context, the constitutive expression of EGR-1 in Nb2 cells suggests that this transcription factor may have both anti- and pro-proliferative effects, as previously described for other proteins such as p53 [69]. These dual and antagonistic functions may constitute a protective mechanism against tumor formation. In this model, three oncogenic abnormalities would have to occur in order to generate continuous tumor growth: immortalization; activation of all the transduction pathways required for proliferation; and suppression of all the anti-proliferative and apoptotic properties resulting from proto/anti-oncogene modifications.
Further studies are required to confirm the integrity of the EGR-1 protein and its constitutive expression in Nb2 cells and to understand the relationships between the EGR-1 target genes and their signaling pathways.
Comparison of various differential screening techniques
We have compared the advantages and drawbacks of four differential screening techniques. Of the four approaches, differential display analysis presents several advantages: it is easy, rapid, does not require large amounts of biological material, and it allows the comparison of multiple transcriptomes in a single experiment. The occurrence of false-positive clones is, however, quite high. In our experimental conditions, we isolated 20 potential positive cDNAs; only two, however, presented a relatively distinct cell-cycle-modulated expression profile. The problem with this technique lies in the low reproducibility of the PCR reaction and the occurrence of non-differential PCR products, which are recovered together with differentially expressed transcripts from the acrylamide gel. Several methods (such as reverse northern blots) have been proposed to circumvent this problem, albeit with rather limited success. Another disadvantage is that the identification of cDNA clones may be difficult if the model system studied has not previously been used in an extended EST sequencing program.
RDA and SSH are based on the same principle. These techniques are easy to use and enable the rapid generation of RDA or SSH subtractive libraries. The proportion of false-positive cDNAs can be less than 10% (our data), but if the differences between the two libraries are discrete, this number is increased. RDA or SSH cDNA clones correspond to sequences positioned in the middle portions of transcripts (generally coding regions), and the sequencing of each cDNA clone enables their identification independently of the model used. These techniques have two principal disadvantages: only two different transcriptomes can be compared in one experiment, and these approaches are far from being exhaustive. Although they should facilitate the detection of low-level transcripts, this is often not the case, as non-optimal conditions appear preferentially to select cDNAs corresponding to highly expressed transcripts.
The screening of an organized library can be compared with the use of DNA arrays, and the detection of a wide range of differential transcripts. This approach theoretically allows the screening of transcripts corresponding to unknown genes. False-positive cDNAs are relatively rare (10-20%) if the amount of DNA fixed on the high-density nylon filters is strictly controlled. The detection threshold for these techniques, however, does not allow the detection of weakly expressed differential transcripts and remains a major limitation.
The principle of the analysis of weakly expressed candidate genes is derived from that of macroarrays. In that case, the detection threshold is increased by a step of moderate PCR amplification for each candidate gene in the complex probes, but allows only a semi-quantitative detection of differentially expressed candidate genes. Internal controls are included to monitor the efficiency of the technique.
Cell-cycle-regulated transcripts in mammals
To facilitate the comparative analysis of cell-cycle-regulated transcripts, we classified them into ten different functional categories (Table 2). This classification is in agreement with the target genes of growth-related transcription factors. Indeed, putative c-Myc target genes are involved in the cell cycle, apoptosis, DNA metabolism and dynamics, energy metabolism and macromolecular synthesis [70]. Nevertheless, as transcriptional gene activation or inhibition result from a complex multifactorial cis and trans regulation, it is necessary to integrate the various components of this regulation and of post-transcriptional modifications to explain the origin of differential expression.
The current functional classification is too restrictive and does not take into account the interactions between these functional categories. For example, the regulation of proto-oncogene and cytokine mRNAs and proteins is particularly complex. The regulatory processes involved include transcriptional control, nuclear export and import of transcripts and proteins, translation, heat-shock pathways (Hsc70-Hsp70), and the ubiquitin- and proteasome-mediated degradation of mRNA and proteins [71]. Interestingly, transcripts of many proteins involved in these processes (such as Hsp proteins, translation factors, and CRM-1) were found to be induced by prolactin in Nb2 cells in this study, as well as in other models. Moreover, the abnormal heat-shock response described in Nb2 cells may perturb this regulation and have important consequences for tumor progression.
Comparison of cell-cycle regulated transcripts in yeast and higher eukaryotes
Data are still lacking for an extensive comparison of cell-cycle-modulated transcripts in unicellular (yeast) and multicellular organisms (essentially mammals) and for deduction of evolutionarily conserved features. Several conclusions can be drawn, however. In almost all the functional subclasses detailed in Table 2, homologous proteins are regulated at the transcriptional level in both yeast and mammals. For example, the yeast cyclins, Cdc proteins, histones and their mammalian homologs are usually regulated in the same fashion during the cell cycle. These observations have been well documented in numerous studies [72] and suggest that the functions of the corresponding proteins and the regulation of their transcripts are both conserved. A list of yeast cell-cycle-regulated transcripts that have known mammalian homologs has been established [73]. In this list, 26 out of 99 of the yeast cell-cycle-regulated transcripts correspond to proteins involved in nucleotide synthesis, DNA replication and repair. Within the cytoskeleton subclass, the transcripts for yeast MYO3 and its mammalian homolog, myosin I heavy chain, both peak at the G2/M phase of the cell cycle, whereas TUB3 and mammalian tubulin γ peak in G1. Proteins in several classes and subclasses do not have yeast homologs. For example, 'adhesion molecules, extracellular matrix and cellular surface antigens' are specific to multicellular organisms.
There are numerous cellular and physiological differences between yeast and multicellular organisms, and some of the molecular divergences observed may reflect these differences. In unicellular organisms the growth priorities are to proliferate as long as enough nutrients are available. In contrast, in multicellular organisms the integrity of the organism is paramount, and individual cell behavior is highly controlled. Therefore proliferation is basically prohibited, and occurs in a cell lineage only if the environment sends the appropriate combination of signals to unlock all the growth-inhibition mechanisms. Despite these differences, cell-cycle checkpoints located just before and at the end of mitosis are essential and similar in all organisms. These two controls ensure that all the DNA has been correctly replicated before the cell enters mitosis and that the condensed chromosomes are properly aligned on the division spindle before anaphase. Thus, all proteins potentially involved in such controls may be regulated similarly from yeasts to animals and plants.
The present study illustrates how data from various large-scale differential screening analyses can be integrated into specific and/or global biological studies. Such comparisons of gene expression profiles are of value in understanding general expression profiles of all dividing cells and in analyzing differences between unicellular and multicellular organisms. They can also identify new signaling molecules and explain how different signals and transduction pathways could regulate the proliferation of different cell types. They can help elucidate the function of proteins, and finally they can identify abnormal patterns of gene expression in transformed and tumor cells. With the increasing amount of information being generated from microarrays and DNA chips, the potential value of comparative analyses will be all the greater.
Materials and methods
Cell culture
Suspension cultures of lactogen-dependent Nb2-11C lymphoma cells were grown in RPMI 1640 medium containing 10% fetal calf serum (FCS), 10% lactogen-free horse serum, 0.1 mM β-mercaptoethanol, 2 mM L-glutamine, 5 mM HEPES pH 7.3, and penicillin-streptomycin (50 lU/ml and 50 μg/ml, respectively) at 37°C with 5% CO2. Cultures were rendered quiescent by transferring cells into starvation medium (cell density about 1.5 × 105 cells/ml), deficient in FCS and β-mercaptoethanol, for 24 hours. Under these conditions, about 80-85% of the cells were arrested in the G0/G1 phase (Figure 1a). Ovine prolactin (Sigma) was added at a concentration of 20 ng/ml to starvation medium to reinitiate growth (Figure 1b). Cells were collected after various periods of prolactin stimulation and total RNA was extracted. Unsynchronized Nb2 cells were collected from high-density cultures (106 cells/ml).
RNA extraction, poly(A)+ RNA preparation, cDNA synthesis, northern blot and reverse northern blot analyses
For each cell population to be compared, RNA was prepared by acid guanidinium-thiocyanate-phenol-chloroform extraction [74] and poly(A)+ RNA was isolated using magnetic oligodT (Dynabeads, Dynal). Double-stranded cDNA was transcribed using a commercial kit (Boehringer). Northern blots were performed using the formaldehyde/formamide procedure and reverse northern blots as described in [75]. The Vacugene transfer system (Pharmacia), nylon filters (Hybond N+, Amersham) and the hybridization solution (ExpressHyb, Clontech) were used following the manufacturer's instructions. The cDNAs were radiolabeled using the Readyprime kit (Amersham).
mRNA differential display, representational difference analysis (RDA), subtractive suppressive hybridization (SSH) and organized library screening
Total RNA treated with DNase I was prepared from Nb2 cells treated for 0, 2, 4, 6, 8, 12 or 24 h with prolactin, and used in mRNA differential display using the GenHunter kit (GeneHunter Corporation, TN, USA). Poly(A)+ mRNA extracted from Nb2 cells treated with prolactin for 12 h was used for RDA according to the protocol described in [13]. SSH [14] was performed using poly(A)+ RNA obtained by mixing mRNA from Nb2 cells stimulated with ovine prolactin for 2, 4, 6 and 8 h. An organized rat brain library was screened as described in [15].
Preparation of complex probes, multiplex PCR and differential screening of candidate genes
In this technique, nylon filters dotted with candidate genes are hybridized with complex cDNA probes from different cell populations, to compare expression levels (see, for example [76]). Originally developed and validated by SANOFI-Recherche (unpublished data), the nylon filters contain 91 rat candidate genes (PCR products), including signaling molecules and transcription factors (Figure 4). These cDNAs were selected with the idea of defining a panel of genes whose expression is likely to be modulated in response to a proliferative signal. To increase the sensitivity of the approach, the mRNAs of interest are co-amplified by reverse transcription polymerase chain reaction (RT-PCR), using primers specific for the 91 candidate genes (the multiplex PCR step), before their use as hybridization probes (Figure 3a). Under these conditions, moderate PCR amplification allows the detection of weakly expressed genes and the evaluation of their differential expression in a semi-quantitative manner. mRNAs are amplified by RT-PCR, and the number of PCR cycles is adapted to their relative abundance in the population tested (16, 21, 24 or 26 cycles are performed for the detection of relatively abundant, moderately expressed, weakly expressed and very weakly expressed transcripts, respectively). For each cell population analyzed, the multiplex PCR products are then mixed, radiolabeled by random priming with α-32P-labeled dCTP, and used as hybridization probes against the 91 candidate genes. Hybridization signals are quantified by densitometry (Visiomic) for each of the different populations. During the development of this technique, control studies involving repeated hybridizations with replicate filters showed minimum variation of signal response (data not shown).
The efficiency of the technique was controlled by including rabbit α and β globin cDNAs on the nylon membranes along with the candidate genes. Defined amounts (50 or 150 ng) of the globin cDNAs were also added to the cDNAs mixtures before the multiplex PCR step. As expected, when these different amounts of rabbit globin cDNAs were added to the complex cDNAs, spots of different intensities were obtained (Figure 3b), indicating that this approach could detect at least a threefold difference in mRNA expression between two cell populations.
Figure 4 shows the list of the candidate genes analyzed and indicates their expression pattern in Nb2 cells.
Sequencing
The potential positive clones isolated by differential display, RDA, SSH or screening of the organized library were sequenced with a dye terminator kit using the ABI Prism system (Perkin-Elmer).
Bioinformatics
To identify the sequenced cDNAs, BLAST and UniGene from NCBI were used [77]. For comparison, we have also consulted databases of transcripts differentially expressed during cell-cycle progression in human fibroblasts [27,28] and in S. cerevisiae [24,25].
Acknowledgments
Acknowledgements
We thank C. Coridun for excellent secretarial assistance and Sébastien Jeay for help with the cell-cycle analysis. Christine Bole-Feysot received studentships from the Ligue National contre Ie Cancer and the Fondation pour la Recherche Médicale.
References
- Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19:225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- Yu-Lee LY. Molecular actions of prolactin in the immune system. . Proc Soc Exp Biol Med. 1997;215:35–52. doi: 10.3181/00379727-215-44111. [DOI] [PubMed] [Google Scholar]
- Clevenger CV, Freier DO, Kline JB. Prolactin receptor signal transduction in cells of the immune system. J Endocrinol. 1998;157:187–197. doi: 10.1677/joe.0.1570187. [DOI] [PubMed] [Google Scholar]
- Richards JF, Beer CT, Bourgeault C, Chen K, Gout PW. Biochemical response of lymphoma cells to mitogenic stimulation by prolactin. . Mol Cell Endocrinol. 1982;26:41–49. doi: 10.1016/0303-7207(82)90005-3. [DOI] [PubMed] [Google Scholar]
- Gertler A, Walker A, Friesen HG. Enhancement of human growth hormone-stimulated mitogenesis of Nb2 node lymphoma cells by 12-O-tetradecanoyl-phorbol-13-acetate. Endocrinology. 1985;116:1636–1644. doi: 10.1210/endo-116-4-1636. [DOI] [PubMed] [Google Scholar]
- Yu-Lee LY. Prolactin stimulates transcription of growth-related genes in Nb2 T lymphoma cells. Mol Cell Endocrinol. 1990;68:21–28. doi: 10.1016/0303-7207(90)90165-5. [DOI] [PubMed] [Google Scholar]
- Stevens AM, Wang YF, Sieger KA, Lu HF, Yu-Lee LY. Biphasic transcriptional regulation of the interferon regulatory factor- 1 gene by prolactin: involvement of gamma-interferon-activated sequence and Stat-related proteins. Mol Endocrinol. 1995;9:513–525. doi: 10.1210/mend.9.4.7659094. [DOI] [PubMed] [Google Scholar]
- Hosokawa Y, Onga T, Nakashima K. Induction of D2 and D3 cyclin-encoding genes during promotion of the G1/S transition by prolactin in rat Nb2 cells. Gene. 1994;147:249–252. doi: 10.1016/0378-1119(94)90075-2. [DOI] [PubMed] [Google Scholar]
- Buckley AR, Buckley DJ, Leff MA, Hoover DS, Magnuson NS. Rapid induction of pim-1 expression by prolactin and interleukin-2 in rat Nb2 lymphoma cells. Endocrinology. 1995;136:5252–5259. doi: 10.1210/endo.136.12.7588268. [DOI] [PubMed] [Google Scholar]
- Gilks CB, Porter SD, Barker C, Tsichlis PN, Gout PW. Prolactin (PRL)-dependent expression of a zinc finger protein-encoding gene, Gfi-1, in Nb2 lymphoma cells: constitutive expression in autonomous sublines. . Endocrinology. 1995;136:1805–1808. doi: 10.1210/endo.136.4.7895694. [DOI] [PubMed] [Google Scholar]
- Morris SM, Yu-Lee LY. Expression of RNUDC, a potential nuclear movement protein, in mammalian cells: localization to the Golgi apparatus. . Exp Cell Res. 1998;238:23–32. doi: 10.1006/excr.1997.3822. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res . 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. . Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret E, Ferran EA, Marinx O, Liauzun P, Dumont X, Fournier J, Kaghad M, Ferrara P, Caput D. Improved differential screening approach to analyse transcriptional variations in organized cDNA libraries. . Gene. 1998;208:103–115. doi: 10.1016/s0378-1119(97)00658-6. [DOI] [PubMed] [Google Scholar]
- O'Neal KD, Schwarz LA, Yu-Lee LY. Prolactin receptor gene expression in lymphoid cells. Mol Cell Endocrinol. 1991;82:127–135. doi: 10.1016/0303-7207(91)90023-l. [DOI] [PubMed] [Google Scholar]
- Hosokawa Y, Yang M, Kaneko S, Tanaka M, Nakashima K. Prolactin induces switching of T-cell receptor gene expression from alpha to gamma in rat Nb2 pre-T lymphoma cells (1). Biochem Biophys Res Commun. 1996;220:958–962. doi: 10.1006/bbrc.1996.0514. [DOI] [PubMed] [Google Scholar]
- Andrews GK, Varma S, Ebner KE. Regulation of expression of c-fos and c-myc in rat lymphoma Nb-2 cells. Biochim Biophys Acta. 1987;909:231–236. doi: 10.1016/0167-4781(87)90082-0. [DOI] [PubMed] [Google Scholar]
- Clevenger CV, Thickman K, Ngo W, Chang WP, Takayama S, Reed JC. Role of Bag-1 in the survival and proliferation of the cytokine-dependent lymphocyte lines, Ba/F3 and Nb2. Mol Endocrinol . 1997;11:608–618. doi: 10.1210/mend.11.5.9925. [DOI] [PubMed] [Google Scholar]
- Leff MA, Buckley DJ, Krumenacker JS, Reed JC, Miyashita T, Buckley AR. Rapid modulation of the apoptosis regulatory genes, bcl-2 and bax by prolactin in rat Nb2 lymphoma cells. Endocrinology. 1996;137:5456–5462. doi: 10.1210/endo.137.12.8940371. [DOI] [PubMed] [Google Scholar]
- Nemeth E, Bole-Feysot C, Tashima LS. Suppression subtractive hybridization (SSH) identifies prolactin stimulation of p38 MAP kinase gene expression in Nb2 T lymphoma cells: molecular cloning of rat p38 MAP kinase. . J Mol Endocrinol. 1998;20:151–156. doi: 10.1677/jme.0.0200151. [DOI] [PubMed] [Google Scholar]
- Gollob JA, Schnipper CP, Murphy EA, Ritz J, Frank DA. The functional synergy between IL-12 and IL-2 involves p38 mitogen-activated protein kinase and is associated with the augmentation of STAT serine phosphorylation. J Immunol. 1999;162:4472–4481. [PubMed] [Google Scholar]
- De Maria R, Rippo MR, Schuchman EH, Testi R. Acidic sphingomyelinase (ASM) is necessary for fas-induced GD3 ganglioside accumulation and efficient apoptosis of lymphoid cells. J Exp Med . 1998;187:897–902. doi: 10.1084/jem.187.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeast Cell Cycle Analysis Project. http://genome-www.stanford.edu/cellcycle
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JF, Trent JM, Staudt LM, Hudson JJ, Boguski MS, et al. The transcriptional program in the response of human fibroblasts to serum. Science . 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- The transcriptional program in the response of human fibroblasts to serum. http://genome-www.stanford.edu/serum [DOI] [PubMed]
- Oehlen LJ, McKinney JD, Cross FR. Ste12 and Mcm1 regulate cell cycle-dependent transcription of FAR1. Mol Cell Biol. 1996;16:2830–2837. doi: 10.1128/mcb.16.6.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehlen B, Cross FR. Signal transduction in the budding yeast Saccharomyces cerevisiae. . Curr Opin Cell Biol. 1994;6:836–841. doi: 10.1016/0955-0674(94)90053-1. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Weinert T. Yeast checkpoint controls and relevance to cancer. . Cancer Surv. 1997;29:109–132. [PubMed] [Google Scholar]
- Verlhac MH, Kubiak JZ, Weber M, Geraud G, Colledge WH, Evans MJ, Maro B. Mos is required for MAP kinase activation and is involved in microtubule organization during meiotic maturation in the mouse. . Development. 1996;122:815–822. doi: 10.1242/dev.122.3.815. [DOI] [PubMed] [Google Scholar]
- Thomas PQ, Brown A, Beddington RS. Hex: a homeobox gene revealing peri-implantation asymmetry in the mouse embryo and an early transient marker of endothelial cell precursors. Development. 1998;125:85–94. doi: 10.1242/dev.125.1.85. [DOI] [PubMed] [Google Scholar]
- Dong JZ, Dunstan DI. Expression of abundant mRNAs during somatic embryogenesis of white spruce [Picea glauca (Moench) Voss]. Planta . 1996;199:459–466. doi: 10.1007/BF00195740. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. . Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hunter T, Pines J. Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- Wang XH, Whyzmuzis CA, An S, Chen Y, Wu JM, Schneidau TA, Mallouh C, Tazaki H. Regulation of cell growth and the c-myc proto-oncogene expression by phorbol ester 12-0-tetradecanoyl phorbol-13-acetate (TPA) in the androgen-independent human prostatic JCA-1 cells. Biochem Mol Biol Int. 1994;34:47–53. [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- Hall M, Peters G. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv Cancer Res. 1996;68:67–108. doi: 10.1016/s0065-230x(08)60352-8. [DOI] [PubMed] [Google Scholar]
- deToledo SM, Murphy LJ, Hatton TH, Friesen HG. Regulation of 70-kilodalton heat-shock-like messenger ribonucleic acid in vitro and in vivo by prolactin. Mol Endocrinol. 1987;1:430–434. doi: 10.1210/mend-1-6-430. [DOI] [PubMed] [Google Scholar]
- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. . Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- Christy B, Nathans D. Functional serum response elements upstream of the growth factor-inducible gene zif268. . Mol Cell Biol. 1989;9:4889–4895. doi: 10.1128/mcb.9.11.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XM, Koski RA, Gashler A, McKiernan M, Morris CF, Gaffney R, Hay RV, Sukhatme VP. Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. . Mol Cell Biol. 1990;10:1931–1939. doi: 10.1128/mcb.10.5.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darland T, Samuels M, Edwards SA, Sukhatme VP, Adamson ED. Regulation of Egr-1 (Zfp-6) and c-fos expression in differentiating embryonal carcinoma cells. Oncogene. 1991;6:1367–1376. [PubMed] [Google Scholar]
- Haber DA, Buckler AJ. WT1: a novel tumor suppressor gene inactivated in Wilms' tumor. New Biol. 1992;4:97–106. [PubMed] [Google Scholar]
- Madden SL, Rauscher FJ. Positive and negative regulation of transcription and cell growth mediated by the EGR family of zinc-finger gene products. Ann N Y Acad Sci. 1993;684:75–84. doi: 10.1111/j.1749-6632.1993.tb32272.x. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Xu J, Erwin RA, Larner AC, Rui H. A lymphoma growth inhibitor blocks some but not all prolactin-stimulated signaling pathways. . J Biol Chem. 1999;274:14699–14705. doi: 10.1074/jbc.274.21.14699. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gluck S, Zhang L, Moran MF. Requirement for phospholipase C-gamma1 enzymatic activity in growth factor-induced mitogenesis. Mol Cell Biol. 1998;18:590–597. doi: 10.1128/mcb.18.1.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Court DW, Kim HK, Park JB, Rhee SG, Rhim JS, Kung HF. Overexpression of phosphoinositide-specific phospholipase Cgamma in NIH 3T3 cells promotes transformation and tumorigenicity. . Carcinogenesis. 1998;19:177–185. doi: 10.1093/carcin/19.1.177. [DOI] [PubMed] [Google Scholar]
- Serve H, Yee NS, Stella G, Sepp-Lorenzino L, Tan JC, Besmer P. Differential roles of PI3-kinase and Kit tyrosine 821 in Kit receptor-mediated proliferation, survival and cell adhesion in mast cells. . EMBO J. 1995;14:473–483. doi: 10.1002/j.1460-2075.1995.tb07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuwa N, Fukui Y, Takuwa Y. Cyclin D1 expression mediated by phosphatidylinositol 3-kinase through mTOR-p70(S6K)-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts. Mol Cell Biol. 1999;19:1346–1358. doi: 10.1128/mcb.19.2.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Okada M, Haraguchi M, Furukawa K, Lloyd KO, Shiku H. Expression of alpha 2,8-sialyltransferase (GD3 synthase) gene in human cancer cell lines: high level expression in melanomas and up-regulation in activated T lymphocytes. Glycoconj J. 1995;12:894–900. doi: 10.1007/BF00731251. [DOI] [PubMed] [Google Scholar]
- Kume A, Nishiura H, Suda J, Suda T. Focal adhesion kinase upregulated by granulocyte-macrophage colony-stimulating factor but not by interleukin-3 in differentiating myeloid cells. Blood. 1997;89:3434–3442. [PubMed] [Google Scholar]
- Tremblay L, Hauck W, Aprikian AG, Begin LR, Chapdelaine A, Chevalier S. Focal adhesion kinase (pp125FAK) expression, activation and association with paxillin and p50CSK in human metastatic prostate carcinoma. . Int J Cancer. 1996;68:164–171. doi: 10.1002/(sici)1097-0215(19961009)68:2<169::aid-ijc4>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Zachary I, Rozengurt E. Focal adhesion kinase (p125FAK): a point of convergence in the action of neuropeptides, integrins, and oncogenes. . Cell. 1992;71:891–894. doi: 10.1016/0092-8674(92)90385-p. [DOI] [PubMed] [Google Scholar]
- Ushikubi F, Aiba Y, Nakamura K, Namba T, Hirata M, Mazda O, Katsura Y, Narumiya S. Thromboxane A2 receptor is highly expressed in mouse immature thymocytes and mediates DNA fragmentation and apoptosis. J Exp Med. 1993;178:1825–1830. doi: 10.1084/jem.178.5.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Calogero A, Ragona G, Adamson E, Mercola D. EGR-1, the reluctant suppression factor: EGR-1 is known to function in the regulation of growth, differentiation, and also has significant tumor suppressor activity and a mechanism involving the induction of TGF-beta1 is postulated to account for this suppressor activity. Crit Rev Oncog. 1996;7:101–125. [PubMed] [Google Scholar]
- Huang RP, Liu C, Fan Y, Mercola D, Adamson ED. Egr-1 negatively regulates human tumor cell growth via the DNA-binding domain. . Cancer Res. 1995;55:5054–5062. [PubMed] [Google Scholar]
- Liu C, Yao J, de Belle I, Huang RP, Adamson E, Mercola D. The transcription factor EGR-1 suppresses transformation of human fibrosarcoma HT1080 cells by coordinated induction of transforming growth factor-beta1, fibronectin, and plasminogen activator inhibitor-1. J Biol Chem . 1999;274:4400–4411. doi: 10.1074/jbc.274.7.4400. [DOI] [PubMed] [Google Scholar]
- Huang RP, Fan Y, de Belle I, Niemeyer C, Gottardis MM, Mercola D, Adamson ED. Decreased Egr-1 expression in human, mouse and rat mammary cells and tissues correlates with tumor formation. Int J Cancer. 1997;72:102–109. doi: 10.1002/(sici)1097-0215(19970703)72:1<102::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Liu C, Adamson E, Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci USA. 1996;93:11831–11836. doi: 10.1073/pnas.93.21.11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- Rayhel EJ, Prentice DA, Tabor PS, Flurkey WH, Geib RW, Laherty RF, Schnitzer SB, Chen R, Hughes JP. Inhibition of Nb2 T-lymphoma cell growth by transforming growth factor-beta. Biochem J. 1988;253:295–298. doi: 10.1042/bj2530295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhatme VP. The Egr transcription factor family: from signal transduction to kidney differentiation. Kidney Int. 1992;41:550–553. doi: 10.1038/ki.1992.79. [DOI] [PubMed] [Google Scholar]
- Vara PM, Shore SK, Dhanasekaran N. Activated mutant of G alpha 13 induces Egr-1, c-fos, and transformation in NIH 3T3 cells. . Oncogene. 1994;9:2425–2429. [PubMed] [Google Scholar]
- Molnar G, Crozat A, Pardee AB. The immediate-early gene Egr-1 regulates the activity of the thymidine kinase promoter at the G0-to-G1 transition of the cell cycle. Mol Cell Biol. 1994;14:5242–5248. doi: 10.1128/mcb.14.8.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle I, Huang RP, Fan Y, Liu C, Mercola D, Adamson ED. p53 and Egr-1 additively suppress transformed growth in HT1080 cells but Egr-1 counteracts p53-dependent apoptosis. Oncogene. 1999;18:3633–3642. doi: 10.1038/sj.onc.1202696. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. . Nature. 1991;351:453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol . 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- Cell Cycle Regulated Transcripts with Known Mammalian Homologs. http://genomics.stanford.edu/yeast_cell_cycle/mammalian_homologs.html
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Martin-Laurent F, Franken P, Gianinazzi S. Screening of cDNA fragments generated by differential RNA display. Anal Biochem . 1995;228:182–184. doi: 10.1006/abio.1995.1337. [DOI] [PubMed] [Google Scholar]
- Pietu G, Alibert O, Guichard V, Lamy B, Bois F, Leroy E, Mariage-Sampson R, Houlgatte R, Soularue P, Auffray C. Novel gene transcripts preferentially expressed in human muscles revealed by quantitative hybridization of a high density cDNA array. Genome Res. 1996;6:492–503. doi: 10.1101/gr.6.6.492. [DOI] [PubMed] [Google Scholar]
- National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov
- Hosokawa Y, Yang M, Kaneko S, Tanaka M, Nakashima K. Synergistic gene expressions of cyclin E, cdk2, cdk5 and E2F-1 during the prolactin-induced G1/S transition in rat Nb2 pre-T lymphoma cells. . Biochem Mol Biol Int. 1995;37:393–399. [PubMed] [Google Scholar]
- Too CK. Differential expression of elongation factor-2, alpha4 phosphoprotein and Cdc5-like protein in prolactin-dependent/independent rat lymphoid cells. Mol Cell Endocrinol. 1997;131:221–232. doi: 10.1016/s0303-7207(97)00112-3. [DOI] [PubMed] [Google Scholar]
- Richards JF. Ornithine decarboxylase activity in tissues of prolactin-treated rats. Biochem Biophys Res Commun. 1975;63:292–299. doi: 10.1016/s0006-291x(75)80042-8. [DOI] [PubMed] [Google Scholar]
- Axtell SM, Truong TM, O'Neal KD, Yu-Lee LY. Characterization of a prolactin-inducible gene, clone 15, in T cells. Mol Endocrinol . 1995;9:312–318. doi: 10.1210/mend.9.3.7776977. [DOI] [PubMed] [Google Scholar]
- Too CK, Knee R, Pinette AL, Li AW, Murphy PR. Prolactin induces expression of FGF-2 and a novel FGF-responsive NonO/p54nrb-related mRNA in rat lymphoma cells. Mol Cell Endocrinol. 1998;137:187–195. doi: 10.1016/s0303-7207(97)00240-2. [DOI] [PubMed] [Google Scholar]
- Wilson TM, Yu-Lee LY, Kelley MR. Coordinate gene expression of luteinizing hormone-releasing hormone (LHRH) and the LHRH-receptor after prolactin stimulation in the rat Nb2 T-cell line: implications for a role in immunomodulation and cell cycle gene expression. Mol Endocrinol . 1995;9:44–53. doi: 10.1210/mend.9.1.7760850. [DOI] [PubMed] [Google Scholar]
- Puissant C, Mitev V, Lemnaouar M, Manceau V, Sobel A, Houdebine LM. Stathmin gene expression in mammary gland and in Nb2 cells. . Biol Cell. 1995;85:109–115. doi: 10.1016/0248-4900(96)85271-3. [DOI] [PubMed] [Google Scholar]


