Abstract
By the use of high-throughput molecular technologies, the number of genes and proteins potentially relevant to testicular germ cell tumor (TGCT) and other diseases will increase rapidly. In a recent transcriptional profiling, we demonstrated the overexpression of GRB7 and JUP in TGCTs, and confirmed the reported overexpression of CCND2. We also have recent evidences for frequent genetic alterations of FHIT and epigenetic alterations of MGMT. To evaluate whether the expression of these genes is related to any clinicopathological variables, we constructed a tissue microarray with 510 testicular tissue cores from 279 patients diagnosed with TGCT, covering various histological subgroups and clinical stages. By immunohistochemistry, we found that JUP, GRB7, and CCND2 proteins were rarely present in normal testis, but frequently expressed at high levels in TGCT. Additionally, all premalignant intratubular germ cell neoplasias were JUP-immunopositive. MGMT and FHIT were expressed by normal testicular tissues, but at significantly lower frequencies in TGCT. Except for CCND2, the expressions of all markers were significantly associated with various TGCT subtypes. In summary, we have developed a high-throughput tool for the evaluation of TGCT markers, and utilized this to validate five candidate genes whose protein expressions were indeed deregulated in TGCT.
Keywords: candidate genes, molecular tumorigenesis, protein expression, testicular germ cell tumor, tissue microarray
Introduction
Testicular germ cell tumors (TGCTs) of adolescent and young adult males are classified into two main histological subtypes, seminomas and nonseminomas [1], and both types develop from premalignant intratubular germ cell neoplasia unclassified (ITGCN; other used terms for these lesions are carcinoma in situ, gonocytoma in situ, intratubular malignant germ cell, and testicular intraepithelial neoplasia) [2]. Whereas the seminomas resemble ITGCN cells, but do not constrain within the tubules and are quite proliferative, the nonseminomas develop through a pluripotent embryonal carcinoma stage, which may differentiate into cells and tissue types of all three primary germ layers at various stages of differentiation (somatically differentiated teratomas and extraembryonally differentiated choriocarcinomas and yolk sac tumors). Thus, tumor development in the testis mimics embryogenesis and makes TGCT an interesting model also for developmental biology.
TGCT genomes are usually hypotriploid or hypertriploid with extensive chromosome losses and gains [3]. Virtually all TGCTs have extra copies of chromosome arm 12p, often seen as an isochromosome [4–6]. There are also other chromosomal copy number alterations occurring at high frequencies, implying the existence of genes within them with relevance to TGCT development. Furthermore, epigenetically deregulated gene expression through aberrant CpG island methylation seems to be common in TGCTs [7].
In three recent reports on the genetics and epigenetics of TGCT, we have gained evidence for specific genes involved in the development of TGCT [8–10]. A cDNA microarray study, mainly focusing on the frequently overrepresented chromosome arm 17q [11], revealed growth factor receptor-bound protein 7 (GRB7) and junction plakoglobin (JUP; γ-catenin) as transcriptionally overexpressed in TGCT [8]. This study also confirmed the overexpression of cyclin D2 (CCND2) [12–15]. By a candidate gene approach, we found epigenetic alterations of the DNA repair gene O6-methylguanine-DNA methyltransferase (MGMT) and frequent allelic imbalances in the chromosome band 10q26 that harbors this gene [9]. Although in a limited series, methylation of the MGMT promoter was found to be associated with lack of protein expression [9]. Furthermore, we reported the fragile histidine triad (FHIT) gene, located within the commonly deleted region on 3p14, to have aberrant splice variants and downregulated expression in TGCT [10].
All these studies reported novel genes relevant to the development of TGCT, but because of limited sample sizes (range 14–70 TGCTs), only a few conclusions could be drawn in relation to clinicopathological variables.
Tissue microarrays facilitate the validation of candidate genes/proteins as a larger series of samples is evaluated, and thus give statistically strong data to associations between genotypes or phenotypes and clinicopathological variables [16–18]. We therefore constructed a tissue microarray from archival blocks of a large series of primary TGCTs of various clinical stages, including all histological subtypes, as well as ITGCN and normal testicular tissues. We have evaluated the in situ protein expressions of the candidates JUP, GRB7, CCND2, MGMT, and FHIT in this set of testicular tissue samples and correlated the results with clinical and pathological variables.
Materials and Methods
Tumor Material and the Tissue Microarray Technology
A tissue microarray block was constructed with 510 testicular tissue cores punched from formalin-fixed and paraffin-embedded tissue blocks from 279 individuals (278 orchiectomy specimens from TGCT patients and one testicular autopsy from a person with no known history of cancer). The distribution of histological subtypes is shown in Table 1. One to five tissue cores of different histological subtypes from each TGCT/patient were transferred into the array block, reflecting the number of histological components. Fifty of the tissue cores were replicates of the same histological subtype taken from a different site of the same tumor, and were included for validation of heterogeneity and consistency of staining. According to the Royal Marsden Staging System (stage I: nonmetastatic TGCT; stages II–IV: metastatic TGCT) [19], there were 174 patients classified as stage I, 53 as stage II, 13 as stage III, and 38 as stage IV. Patients without clinically demonstrated metastases underwent retroperitoneal lymph node dissection, or followed a surveillance program. Patients with metastases received cisplatin-based chemotherapy followed, in the majority of cases, by resection of residual masses. The patients underwent orchiectomy between 1981 and 1999, and all patients were followed up until death or May 2002. All TGCT samples are available from the archives of the Department of Pathology of The Norwegian Radium Hospital. The study was approved by the Regional Committee for Medical Research Ethics (S-00201, 150800).
Table 1.
Histological Subtypes of 510 Testicular Tissue Cores in the Tissue Microarray.
| Histology | Individuals (n) | Tissue Cores (n) |
| Normal testis | 22 | 28 |
| Intratubular germ cell neoplasia | 21 | 21 |
| Seminoma | 167 | 184 |
| Embryonal carcinoma | 99 | 102 |
| Choriocarcinoma | 16 | 16 |
| Yolk sac tumor | 62 | 69 |
| Teratoma | 75 | 90 |
| Total | 279* | 510 |
The total number of individuals is lower than the sum of each histological subtype as there often are tissue cores of several histological subtypes provided from each orchiectomy specimen.
Sections of up to 10 tissue blocks from each orchiectomy specimen were stained with hematoxylin and eosin, and light microscopically examined by an expert pathologist on germ cell tumors (V.A.). The best areas for tissue punching were marked. The tissue microarray was assembled using a robotic tissue microarrayer. Briefly, cylindrical tissue cores with 0.6 mm diameter were transferred from the donor archival tissue blocks and arrayed into an empty recipient paraffin block, building up the tissue microarray block [16].
The Instrumedics (Instrumedics, Hackensack, NJ) tape transfer method was used to transfer 4-µm sections of the tissue microarray block to glass slides. Hematoxylin and eosin-stained tissue microarray sections were evaluated to check for consistency with the originally assigned histology. Histological classification was performed according to World Health Organization recommendations [1]. The distinction of ITGCN from normal tissue was assisted by immunohistochemical staining of a parallel section using antibodies targeting germ cell and placental alkaline phosphatases (PLAP) extensively present in ITGCN but not in normal spermatogenic germ cells [2,20].
Immunohistochemistry
Tissue microarray sections were stained with the biotin-streptavidin peroxidase method (Supersensitive Immunodetection System, LP000-UL; BioGenex, San Raman, CA) and OptiMax Plus Automated Cell Staining System (BioGenex). One tissue microarray section for each antibody was deparaffinized and rehydrated, and high-temperature antigen retrieval was performed by microwave oven at 900 W. The slides were then incubated with 1% hydrogen peroxide (H2O2) for 10 minutes to block endogenous peroxidase activity before incubation with polyclonal antibodies GRB7 (N-20, sc-607, 1:100, 2 µg/ml IgG; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), CCND2 (C-17, sc-181, 1: 400, 0.5 µg/ml IgG; Santa Cruz Biotechnology, Inc.), MGMT (C-20, sc-8825, 1:200, 1 µg/ml IgG; Santa Cruz Biotechnology, Inc.), FHIT (ZP54, 1:100, 5 µg/ml IgG; Zymed Laboratories, Inc., South San Francisco, CA), and monoclonal antibodies JUP (clone 15, 1:300, 0.8 µg/ml IgG2a; Nota Bene Scientific ApS, Hellebæk, Denmark) and PLAP (clone 8A9, 1:20, IgG1k; Novocastra Laboratories Ltd., Newcastle, UK) for 30 minutes at room temperature. Afterward, the sections were incubated for 20 minutes with multilink biotinylated antiimmunoglobulins (1:30; BioGenex) and for 20 minutes with streptavidin peroxidase (1:30; BioGenex). Finally, the sections were stained for 5 minutes with 0.05% of the peroxidase substrate 3′3-diaminobenzidine tetrahydrochloride (DAB) freshly prepared in 0.05 M Tris-HCl buffer at pH=7.6 containing 0.01% H2O2, before being counterstained with hematoxylin, dehydrated, and mounted.
Immunohistochemistry Scoring
For scoring the immunohistochemical staining, we used a binary scoring of only “positive” and “negative” categories to avoid making pseudoquantitative measures of individual tissue cores. Upon deciding the cutoff criteria for individual antibodies, we considered both general consensus for immunohistochemistry scoring and previously published scoring systems.
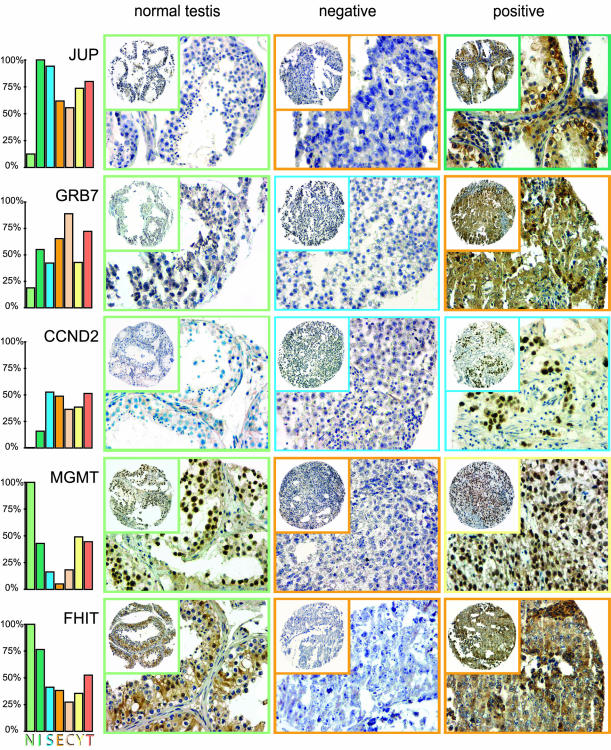
JUP immunostaining was membranous and/or cytoplasmic (Figure 1). Cases with any membranous and/or moderate to strong cytoplasmic staining were scored as positive. GRB7 immunostaining was membranous and/or cytoplasmic. Cases with moderate or strong staining in the relevant cells were scored as positive. CCND2 immunostaining was nuclear, although some cytoplasmic staining was seen in negative normal testicular tissues. Cases with staining of more than 5% of the relevant nuclei were considered positive. MGMT immunostaining was nuclear, and cases with staining of more than 5% of the relevant nuclei were considered positive. FHIT immunostaining was cytoplasmic and the cases were evaluated according to the previously published scoring system for FHIT immunohistochemistry [10,21]. This system uses a composite score, which is calculated by multiplication of the intensity (1, weak/absent; 2, moderate; 3, strong) and the fraction of positive cells (1, <10%; 2, 10–50%; 3, >50%), where cases with composite scores of four or above were regarded as FHIT-positive.
Figure 1.
TGCT tissue microarray, immunohistochemical staining. For each of the five analyzed proteins, representative tissue cores of one normal testicular parenchyma, one negative neoplasm, and one positive neoplasm are shown. The colored squares designate the histological subtype of each sample specified by the color code of the histograms. The histograms, again, indicate the frequencies of positive cases for each histological subtype. N=normal spermatocytic germ cells; I = intratubular germ cell neoplasia unclassified (carcinoma in situ); S = seminoma; E = embryonal carcinoma; C = choriocarcinoma; Y = yolk sac tumor; T = teratoma.
A tumor was considered positive when one or more of the tumor tissue cores from that specific tumor were positive. Equally, when more than one tissue core of a specific histological component of a tumor was present on the array, that specific component was considered positive if at least one of the samples was scored as positive.
Control Experiments
All antibodies were purchased from commercial companies (see above), which have experimental evidence for their specificity and detection of correct-sized proteins. We hybridized each of the antibodies to a Western blot with cell extracts from a panel of 17 cell lines cultured from various types of tumor. Briefly, from each cell line, 25 µg of total protein lysate was separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto Immobilin P-membranes (Millipore, Bedford, MA). The membrane was blocked and incubated with various antibodies overnight at 4°C according to the recommendations of the manufacturers. After washing, the immunoreactive proteins were visualized using horseradish peroxidase-conjugated secondary antibodies and the ECL Western blotting detection system (Amersham-Pharmacia Biotech, Amersham, UK). Bands at correct sizes were detected for all five antibodies, but additional bands at lower intensities and unknown origin were seen for the antibodies against CCND2 and MGMT.
For three of the antibodies (GRB7, CCND2, and MGMT), peptide antigenes were also available (Santa Cruz Biotechnology, Inc.), and we performed peptide blocking reactions on separate tissue microarray sections. Furthermore, we performed negative controls of all antibodies by replacement of primary polyclonal antibodies with normal rabbit IgG at the same concentration as the polyclonal antibodies and by replacement of primary monoclonal antibodies with mouse myeloma proteins of the same subclass and concentration as the monoclonal antibodies. Both the peptide blocking and the antibody replacement reactions gave satisfactory results. Furthermore, test tissue microarrays containing 73 tissue cores with normal and malignant tissues from 10 different organs were used for the optimization of immunohistochemistry conditions, and this served as additional positive and negative controls because the expressions of the five proteins were known beforehand for several of the tissue types.
Statistical Analysis
Comparisons of different groups were statistically tested with two-sided Fisher's exact tests.
Results
In total, the immunohistochemical analyses of JUP, GRB7, CCND2, MGMT, and FHIT resulted in 433, 414, 428, 434, and 418 scored testicular tissue cores, from 256, 254, 260, 261, and 255 individuals, respectively. The frequencies of positive staining for each antibody and histological subtype are shown in Figure 1.
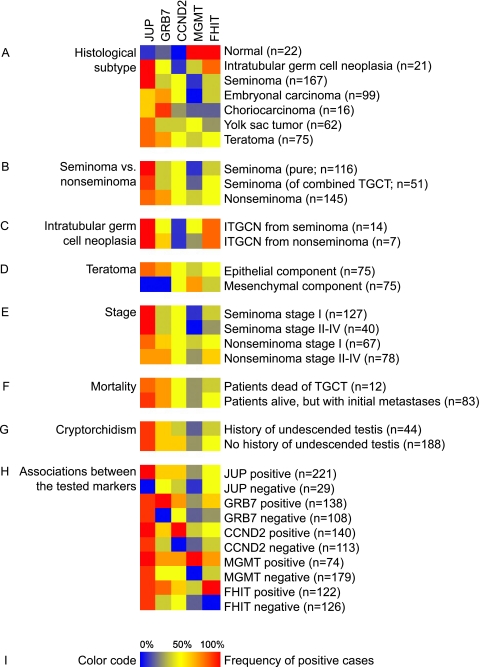
The frequencies of positive tissues according to histological subtypes are illustrated in Figure 2A. For JUP, only 13% of the normal tissues stained positive, compared to 100% and 94% for ITGCN and seminomas (P = 1 x 10-5 and P = 1 x 10-10, respectively). The frequency of JUP-positives in nonseminomas was 77%, which is significantly lower than that for ITGCN and seminomas (Figure 2B; P = 6 x 10-4 and P = 6 x 10-6, respectively).
Figure 2.
Expression profiles of five TGCT candidate genes according to various clinical and pathological subgroups. The 510 testicular tissue cores, derived from healthy and tumor tissues of 279 individuals, were subgrouped according to several criteria. Each row represents one subgroup of TGCT, and the colored squares illustrate the frequencies of immunopositive cases for the different markers. Parenthesized numbers specify the number of analyzable tumors/patients from each subgroup. The frequencies of immunopositive cases are summarized according to (A) the histological subgroup, (B) seminoma versus nonseminoma, (C) intratubular germ cell neoplasia in proximity of seminoma versus nonseminoma, (D) the teratoma component, (E) clinical stage, (F) mortality, and (G) history of undescended testis. Panel (H) presents the associations between the tested markers. Panel (I) presents the color code for the frequencies of immunopositive cases.
For GRB7, 19% of the normals were scored positive, compared to 55% of the ITGCN and 56% of the TGCTs (P = .04 and P = 4 x 10-3, respectively). The frequency of GRB7 immunoreactivity in seminomas (42%) was significantly different from that in nonseminomas (66%; P = 1 x 10-4).
For CCND2, all normal tissues were negative, whereas 16% of the ITGCN samples and 55% of the TGCTs were positive (P = 2 x 10-6; normal to invasive tumor). The frequency of CCND2 immunoreactivity was similar in seminomas and nonseminomas (P = 1.00).
For MGMT, all normal tissues had positive spermatogenic cells, whereas the percentages of positives among ITGCN, seminomas, and embryonal carcinomas were 47%, 16%, and 6%, respectively (P = 6 x 10-5, 1 x 10-13, and 8 x 10-17 when compared to normal testicular tissues). However, among yolk sac tumors and teratomas, 49% and 44% were positive, respectively, which signifies a significant reexpression of MGMT upon differentiation from embryonal carcinoma (P = 4 x 10-9 and P = 2 x 10-7).
For FHIT, all normal tissues and 76% of ITGCN were positive. Both seminomas (41%) and embryonal carcinomas (38%) were positive in significantly fewer cases than both normal tissues (P = 3 x 10-6 and P = 2 x 10-6) and ITGCN (P = 9 x 10-3 and P = 6 x 10-3).
When we compared pure seminomas with the seminoma components from combined tumors, no significant differences were seen in the expression patterns of the five analyzed proteins (Figure 2B). The various antibodies also revealed comparable staining in ITGCN samples from seminomas and nonseminomas (Figure 2C). Within the teratomas, the epithelial components were significantly more frequently positive for JUP and GRB7 than the mesenchymal components (Figure 2D).
None of the markers was significantly associated with clinical stage or mortality (Figure 2, E and F). Among TGCTs from patients with history of undescended testis (cryptorchidism; Figure 2G), 40% were positive for CCND2, as compared to 60% among TGCTs from patients with no history of cryptorchidism (P = .03).
The immunostaining results evaluated for several combinations (Figure 2H) and the strongest associations were seen between FHIT-positives and tumors positive for GRB7 and MGMT (P = 6 x 10-8 and P = 3 x 10-4), followed by associations between CCND2-positives and tumors positive for MGMT and GRB7 (P = .002 and P = .006).
Replicate tissue cores (i.e., same histological subtypes from same tumors) were scored identically in 77% of the cases [range 71% (for FHIT) to 80% (for GRB7)].
Discussion
As the human genome gets unraveled and high-throughput molecular technologies are utilized, the number of genes with putative relation to various diseases, including TGCT, increases dramatically [3]. Hence, there is a need for validation of new putative disease markers, but so far, studies on TGCT have analyzed too few samples to really pinpoint significant associations to clinicopathological variables.
By the present study, we have taken advantage of the tissue microarray technology [16–18], and by transferring more than 500 cylindrical testicular tissue cores into a single recipient block, we developed a tool enabling us to analyze candidate testicular cancer genes in a large series of samples of all histological subtypes and stages, linked to a database with relevant clinical, pathological, and genetic information. We have used this tool to examine the protein expression of five TGCT candidate genes. Four of these (JUP, GRB7, MGMT, and FHIT) were recently targeted by us [8–10], and the fifth is the CCND2 candidate gene on chromosome arm 12p [8,12–15].
JUP and GRB7 showed high expression in TGCT but not in normal testicular tissues in a cDNA microarray study focusing on chromosome arm 17q [8], which is overrepresented in every second TGCT [11]. By transcriptional profiling using DNA microarrays, many genes are usually analyzed in a relatively small sample set, often identifying a molecular signature of the tumor in question, but with weak statistics regarding the individual genes. However, by analyzing the two candidate genes JUP and GRB7 further on the TGCT tissue microarray, we are confident about their general overexpression in TGCT and also on their protein levels, and evidence was provided for their differential expression across various histological subtypes of TGCT.
JUP belongs to the catenin family and may have oncogenic potential through its function in the WNT signaling pathway [22,23]. Tissue microarray data demonstrated that JUP protein is rarely expressed in normal spermatogenic germ cells, even though it is expressed in virtually all ITGCN and seminomas, and in most nonseminomas. However, it remains to be elucidated whether the induction of JUP expression is an initial event in the development of ITGCN, or if JUP is already expressed in fetal gonocytes, the germ cell precursors of which ITGCN is believed to originate from [2,24]. The fact that the WNT pathway is involved in embryogenesis, which again is mimicked by testicular tumorigenesis, makes components of this pathway interesting candidates for examination. The JUP binding partner E-cadherin is expressed in embryonal carcinomas [25,26] and the JUP homolog β-catenin is expressed in both normal and malignant testicular tissues [26], but the impact of WNT signaling in TGCT remains poorly understood.
GRB7 encodes an adaptor protein that, through its SH2 domain, interacts with the cytoplasmic domain of several tyrosine kinase growth factor receptors, including ERBB2, KIT, PDGFR, RET, and INSR [27–31], as well as with cytoplasmic tyrosine kinases [32,33]. GRB7 also has a RAS-associating-like domain [34] and plays a role in cell migration [35,36]. Tissue microarray results for GRB7 confirm that positive immunostaining is more frequent in ITGCN, seminomas, and nonseminomas than in normal testicular tissues, but with the highest frequency in nonseminomas. But still, seminoma components within combined TGCTs were not more often positive than pure seminomas. Within teratomas, epithelial parts were generally positive, whereas mesenchymal components were not. In breast, esophageal, and gastric cancers, GRB7 is often coamplified and cooverexpressed with ERBB2 [27,37–39]. Although we do not have copy number data for these two genes, the combined CGH and cDNA microarray analyses of TGCT showed that this chromosome region is overrepresented [11] but with overexpression only of GRB7 [8], suggesting that the GRB7 protein interacts with another main target other than ERBB2 in these cells.
CCND2 is located at chromosome arm 12p, and several studies have noted its high expression in TGCT [8,12–15]. This most likely reflects DNA sequence copy number gains seen in virtually all TGCTs, and often as i(12p) [4,6,40]. However, CCND2 can also be induced downstream of several molecular pathways such as RAS and WNT signaling [41,42]. We noted that 56% of the TGCTs in our series was immunopositive for CCND2, which is somewhat lower than the frequency found by Bartkova et al. [14] (n = 31, 81%). In ITGCN, the frequency of CCND2-positives was intermediate between the always-negative normals and the TGCT samples. One might speculate whether we underestimate the frequency of CCND2-positives in ITGCN, as there are fewer ITGCN nuclei in each tissue core than, for instance, seminoma nuclei in a seminoma tissue core. However, as parallel sections were stained with antibodies against germ cell and placental alkaline phosphatases extensively present in ITGCN but absent in normal spermatogenic germ cells, we saw that there were usually about 50 intratubular malignant germ cells in each ITGCN tissue core. Among the invasive tumors, we confirmed that the CCND2 expression is not associated with histological subtype (P = 1.0), which has been previously reported for the mRNA level [15]. CCND2 mRNA expression has been shown to correlate with the mRNA expression of its protein binding partner CDK4 [15] and, in the present study, we demonstrated that its protein expression correlated to those of GRB7 and MGMT. Interestingly, we found in our series that TGCTs of patients with history of cryptorchidism had a lower frequency of CCND2 immunoreactivity. However, this association is of borderline statistical significance, and a biological impact would be an enigma.
MGMT is a DNA repair gene, which was recently demonstrated to be frequently inactivated in TGCTs by promoter hypermethylation [9,43,44]. In the present study, we have shown also that the amount of protein product is downregulated, in particular in seminomas and embryonal carcinomas. However, the MGMT protein seems to be re-expressed upon further differentiation of embryonal carcinoma into choriocarcinomas, yolk sac tumors, and teratomas. The observed hypermethylation of the MGMT promoter fits well with such a reversible silencing mechanism. The existence of other regulatory mechanisms for MGMT expression is also evident, as the expression is lost in most seminomas, a subgroup that, only infrequently, is methylated in the MGMT promoter.
FHIT was also newly identified as relevant to TGCT development [10]. We confirmed that the FHIT protein is downregulated in half of the TGCTs compared to normal testicular tissues. The downregulation seems to take place when ITGCN is transformed into invasive TGCT (P = .009). The immunoreactivity of FHIT was strongly associated with those of GRB7 and MGMT. However, here we have failed to confirm the associations proposed by the initial study [10] between reduced FHIT expression and metastasis (present study, P = .8) and that mesenchymal components of teratomas have more frequently reduced expression compared to epithelial components (present study, P = .5). Ten whole mount sections of TGCTs that also were present on the tissue microarray were analyzed for FHIT staining, yielding the same score in 8 of 10 cases. Hence, it is likely that the conflicting conclusions of this and the previous FHIT study were not due to the different technologies, but rather due to the limited sample size and borderline significance levels of the initial study.
For all tested markers, the frequencies of immunoreactive cases were similar for pure seminomas and seminoma components of combined TGCTs. Thus, this gives evidence for seminomas of both groups to be evaluated together in the same category in molecular studies of TGCT. Additionally, this speaks in favor of seminomas developing through the same molecular-pathological pathway, irrespective of whether it is pure or in combination with nonseminoma components.
The tissue microarray technology has been criticized because the analysis of a small tissue core is not able to reflect the situation of a larger heterogeneous tumor. We therefore included 50 replicate tissue punches with identical histology but from a different site in the tumor. Duplicate tissue cores were scored differently in as much as 24% of the cases (average of the five antibodies; data not shown). This underlines the fact that tissue microarrays are not a tool for the diagnosis of individual tumors, but because of the possibility to analyze huge sample sets, real associations between tested markers and clinicopathological variables yield very strong statistical evidence. Therefore, the tissue microarray technology is powerful in the discovery of new markers with associations to subgroups of samples.
In summary, we have constructed a TGCT tissue microarray on which we have evaluated the protein expression of five candidate testicular cancer genes. We found that JUP was upregulated and MGMT was downregulated upon initiation of ITGCN, and that the upregulation of CCND2, downregulation of FHIT, and further downregulation of MGMT were related to the development of invasive tumors. GRB7 is upregulated and JUP is downregulated in the transition into embryonal carcinoma, whether embryonal carcinomas develop directly from ITGCN or through a seminoma stage. Additionally, MGMT is reexpressed during further differentiation of embryonal carcinomas. Hence, we have demonstrated that our tissue microarray enables high-throughput evaluation of TGCT markers, and we have utilized this tool to validate five TGCT candidate genes whose protein expressions were indeed deregulated.
Acknowledgements
We acknowledge Liv Inger Håseth for mining the tissue archive, and Ellen Hellesylt for assistance in immunohistochemistry.
Abbreviations
- CCND2
cyclin D2
- FHIT
fragile histidine triad
- GRB7
growth factor receptorbound protein 7
- ITGCN
intratubular germ cell neoplasia unclassified (other commonly used terms for ITGCN are carcinoma in situ and intratubular malignant germ cells)
- JUP
junction plakoglobin
- MGMT
O6-methylguanine-DNA methyltransferase
- PLAP
germ cell and placental alkaline phosphatases
- TGCT
testicular germ cell tumor
Footnotes
This work was supported by grants from the Norwegian Cancer Society (R.A.L. and R.I.S.) and the Research Council of Norway (R.A.L. and R.I.S.).
References
- 1.Mostofi FK, Sesterhenn IA. World Health Organization International Histological Classification of Tumours: Histological Typing of Testis Tumours. 2nd ed. Berlin: Springer-Verlag; 1998. [Google Scholar]
- 2.Rajpert-De Meyts E, Bartkova J, Samson M, Hoei-Hansen CE, Frydelund-Larsen L, Bartek J, Skakkebæk NE. The emerging phenotype of the testicular carcinoma in situ germ cell. APMIS. 2003;111:267–278. doi: 10.1034/j.1600-0463.2003.11101301.x. [DOI] [PubMed] [Google Scholar]
- 3.Skotheim RI, Lothe RA. The testicular germ cell tumour genome. APMIS. 2003;111:136–151. doi: 10.1034/j.1600-0463.2003.11101181.x. [DOI] [PubMed] [Google Scholar]
- 4.Atkin NB, Baker MC. Specific chromosome change, i(12p), in testicular tumours? Lancet. 1982;2:1349. doi: 10.1016/s0140-6736(82)91557-4. [DOI] [PubMed] [Google Scholar]
- 5.Rodriguez E, Mathew S, Reuter V, Ilson DH, Bosl GJ, Chaganti RS. Cytogenetic analysis of 124 prospectively ascertained male germ cell tumors. Cancer Res. 1992;52:2285–2291. [PubMed] [Google Scholar]
- 6.Rodriguez E, Houldsworth J, Reuter VE, Meltzer P, Zhang J, Trent JM, Bosl GJ, Chaganti RS. Molecular cytogenetic analysis of i(12p)-negative human male germ cell tumors. Genes Chromosomes Cancer. 1993;8:230–236. doi: 10.1002/gcc.2870080405. [DOI] [PubMed] [Google Scholar]
- 7.Smiraglia DJ, Szymanska J, Kraggerud SM, Lothe RA, Peltomäki P, Plass C. Distinct epigenetic phenotypes in seminomatous and nonseminomatous testicular germ cell tumors. Oncogene. 2002;21:3909–3916. doi: 10.1038/sj.onc.1205488. [DOI] [PubMed] [Google Scholar]
- 8.Skotheim RI, Monni O, Mousses S, Fosså SD, Kallioniemi O-P, Lothe RA, Kallioniemi A. New insights into testicular germ cell tumorigenesis from gene expression profiling. Cancer Res. 2002;62:2359–2364. [PubMed] [Google Scholar]
- 9.Smith-Sørensen B, Lind GE, Skotheim RI, Fosså SD, Fodstad Ø, Stenwig A-E, Jakobsen KS, Lothe RA. Frequent promoter hypermethylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene in testicular cancer. Oncogene. 2002;21:8878–8884. doi: 10.1038/sj.onc.1205978. [DOI] [PubMed] [Google Scholar]
- 10.Kraggerud SM, Åman P, Holm R, Stenwig AE, Fosså SD, Nesland JM, Lothe RA. Alterations of the fragile histidine triad gene, FHIT, and its encoded products contribute to testicular germ cell tumorigenesis. Cancer Res. 2002;62:512–517. [PubMed] [Google Scholar]
- 11.Kraggerud SM, Skotheim RI, Szymanska J, Eknæs M, Fosså SD, Stenwig AE, Peltomäki P, Lothe RA. Genome profiles of familial/bilateral and sporadic testicular germ cell tumors. Genes Chromosomes Cancer. 2002;34:168–174. doi: 10.1002/gcc.10058. [DOI] [PubMed] [Google Scholar]
- 12.Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, Robker RL, Richards JS, McGinnis LK, Biggers JD, Eppig JJ, Bronson SJ, Elledge SJ, Weinberg RA. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- 13.Houldsworth J, Reuter V, Bosl GJ, Chaganti RS. Aberrant expression of cyclin D2 is an early event in human male germ cell tumorigenesis. Cell Growth Differ. 1997;8:293–299. [PubMed] [Google Scholar]
- 14.Bartkova J, Rajpert-De Meyts E, Skakkebæk NE, Bartek J. D-type cyclins in adult human testis and testicular cancer: relation to cell type, proliferation, differentiation, and malignancy. J Pathol. 1999;187:573–581. doi: 10.1002/(SICI)1096-9896(199904)187:5<573::AID-PATH289>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt BA, Rose A, Steinhoff C, Strohmeyer T, Hartmann M, Ackermann R. Up-regulation of cyclin-dependent kinase 4/cyclin D2 expression but down-regulation of cyclin-dependent kinase 2/cyclin E in testicular germ cell tumors. Cancer Res. 2001;61:4214–4221. [PubMed] [Google Scholar]
- 16.Kononen J, Bubendorf L, Kallioniemi A, Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 17.Moch H, Schraml P, Bubendorf L, Mirlacher M, Kononen J, Gasser T, Mihatsch MJ, Kallioniemi OP, Sauter G. High-throughput tissue microarray analysis to evaluate genes uncovered by cDNA microarray screening in renal cell carcinoma. Am J Pathol. 1999;154:981–986. doi: 10.1016/S0002-9440(10)65349-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Köchli OR, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi OP, Sauter G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendry WF, Barrett A, McElwain TJ, Wallace DM, Peckham MJ. The role of surgery in the combined management of metastases from malignant teratomas of testis. Br J Urol. 1980;52:38–44. doi: 10.1111/j.1464-410x.1980.tb02917.x. [DOI] [PubMed] [Google Scholar]
- 20.Hofmann MC, Millán JL. Developmental expression of alkaline phosphatase genes; reexpression in germ cell tumours and in vitro immortalized germ cells. Eur Urol. 1993;23:38–45. doi: 10.1159/000474568. [DOI] [PubMed] [Google Scholar]
- 21.Greenspan DL, Connolly DC, Wu R, Lei RY, Vogelstein JT, Kim YT, Mok JE, Munoz N, Bosch FX, Shah K, Cho KR. Loss of FHIT expression in cervical carcinoma cell lines and primary tumors. Cancer Res. 1997;57:4692–4698. [PubMed] [Google Scholar]
- 22.Kolligs FT, Kolligs B, Hajra KM, Hu G, Tani M, Cho KR, Fearon ER. Gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes Dev. 2000;14:1319–1331. [PMC free article] [PubMed] [Google Scholar]
- 23.Barker N, Clevers H. Catenins, Wnt signaling and cancer. Bioessays. 2000;22:961–965. doi: 10.1002/1521-1878(200011)22:11<961::AID-BIES1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Skakkebæk NE, Berthelsen JG, Giwercman A, Muller J. Carcinoma-in-situ of the testis: possible origin from gonocytes and precursor of all types of germ cell tumours except spermatocytoma. Int J Androl. 1987;10:19–28. doi: 10.1111/j.1365-2605.1987.tb00161.x. [DOI] [PubMed] [Google Scholar]
- 25.Heidenreich A, Sesterhenn IA, Mostofi FK, Moul JW. Prognostic risk factors that identify patients with clinical stage I nonseminomatous germ cell tumors at low risk and high risk for metastasis. Cancer. 1998;83:1002–1011. [PubMed] [Google Scholar]
- 26.Saito T, Katagiri A, Watanabe R, Tanikawa T, Kawasaki T, Tomita Y, Takahashi K. Expression of E-cadherin and catenins on testis tumor. Urol Int. 2000;65:140–143. doi: 10.1159/000064859. [DOI] [PubMed] [Google Scholar]
- 27.Stein D, Wu J, Fuqua SA, Roonprapunt C, Yajnik V, D'Eustachio P, Moskow JJ, Buchberg AM, Osborne CK, Margolis B. The SH2 domain protein GRB-7 is co-amplified, overexpressed and in a tight complex with HER2 in breast cancer. EMBO J. 1994;13:1331–1340. doi: 10.1002/j.1460-2075.1994.tb06386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey A, Liu X, Dixon JE, Di Fiore PP, Dixit VM. Direct association between the Ret receptor tyrosine kinase and the Src homology 2-containing adapter protein Grb7. J Biol Chem. 1996;271:10607–10610. doi: 10.1074/jbc.271.18.10607. [DOI] [PubMed] [Google Scholar]
- 29.Yokote K, Margolis B, Heldin CH, Claesson-Welsh L. Grb7 is a downstream signaling component of platelet-derived growth factor alpha- and beta-receptors. J Biol Chem. 1996;271:30942–30949. doi: 10.1074/jbc.271.48.30942. [DOI] [PubMed] [Google Scholar]
- 30.Thömmes K, Lennartsson J, Carlberg M, Rönnstrand L. Identification of Tyr-703 and Tyr-936 as the primary association sites for Grb2 and Grb7 in the c-Kit/stem cell factor receptor. Biochem J. 1999;341:211–216. [PMC free article] [PubMed] [Google Scholar]
- 31.Kasus-Jacobi A, Bereziat V, Perdereau D, Girard J, Burnol AF. Evidence for an interaction between the insulin receptor and Grb7. A role for two of its binding domains, PIR and SH2. Oncogene. 2000;19:2052–2059. doi: 10.1038/sj.onc.1203469. [DOI] [PubMed] [Google Scholar]
- 32.Keegan K, Cooper JA. Use of the two hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene. 1996;12:1537–1544. [PubMed] [Google Scholar]
- 33.Han DC, Guan JL. Association of focal adhesion kinase with Grb7 and its role in cell migration. J Biol Chem. 1999;274:24425–24430. doi: 10.1074/jbc.274.34.24425. [DOI] [PubMed] [Google Scholar]
- 34.Wojcik J, Girault JA, Labesse G, Chomilier J, Mornon JP, Callebaut I. Sequence analysis identifies a ras-associating (RA)-like domain in the N-termini of band 4./JEF domains and in the Grb7/10/14 adapter family. Biochem Biophys Res Commun. 1999;259:113–120. doi: 10.1006/bbrc.1999.0727. [DOI] [PubMed] [Google Scholar]
- 35.Manser J, Roonprapunt C, Margolis B. C. elegans cell migration gene mig-10 shares similarities with a family of SH2 domain proteins and acts cell nonautonomously in excretory canal development. Dev Biol. 1997;184:150–164. doi: 10.1006/dbio.1997.8516. [DOI] [PubMed] [Google Scholar]
- 36.Han DC, Shen TL, Guan JL. Role of Grb7 targeting to focal contacts and its phosphorylation by focal adhesion kinase in regulation of cell migration. J Biol Chem. 2000;275:28911–28917. doi: 10.1074/jbc.M001997200. [DOI] [PubMed] [Google Scholar]
- 37.Akiyama N, Sasaki H, Ishizuka T, Kishi T, Sakamoto H, Onda M, Hirai H, Yazaki Y, Sugimura T, Terada M. Isolation of a candidate gene, CAB1, for cholesterol transport to mitochondria from the c-ERBB-2 amplicon by a modified cDNA selection method. Cancer Res. 1997;57:3548–3553. [PubMed] [Google Scholar]
- 38.Tanaka S, Mori M, Akiyoshi T, Tanaka Y, Mafune K, Wands JR, Sugimachi K. Coexpression of Grb7 with epidermal growth factor receptor or Her2/erbB2 in human advanced esophageal carcinoma. Cancer Res. 1997;57:28–31. [PubMed] [Google Scholar]
- 39.Kauraniemi P, Bärlund M, Monni O, Kallioniemi A. New amplified and highly expressed genes discovered in the ERBB2 amplicon in breast cancer by cDNA microarrays. Cancer Res. 2001;61:8235–8240. [PubMed] [Google Scholar]
- 40.Suijkerbuijk RF, Sinke RJ, Meloni AM, Parrington JM, van Echten J, de Jong B, Oosterhuis JW, Sandberg AA, Geurts van Kessel A. Overrepresentation of chromosome 12p sequences and karyotypic evolution in i(12p)-negative testicular germ-cell tumors revealed by fluorescence in situ hybridization. Cancer Genet Cytogenet. 1993;70:85–93. doi: 10.1016/0165-4608(93)90173-j. [DOI] [PubMed] [Google Scholar]
- 41.Dey A, She H, Kim L, Boruch A, Guris DL, Carlberg K, Sebti SM, Woodley DT, Imamoto A, Li W. Colony-stimulating factor-1 receptor utilizes multiple signaling pathways to induce cyclin D2 expression. Mol Biol Cell. 2000;11:3835–3848. doi: 10.1091/mbc.11.11.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Megason SG, McMahon AP. A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development. 2002;129:2087–2098. doi: 10.1242/dev.129.9.2087. [DOI] [PubMed] [Google Scholar]
- 43.Koul S, Houldsworth J, Mansukhani MM, Donadio A, McKiernan JM, Reuter VE, Bosl GJ, Chaganti RS, Murty VV. Characteristic promoter hypermethylation signatures in male germ cell tumors. Mol Cancer. 2002;1:8. doi: 10.1186/1476-4598-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honorio S, Agathanggelou A, Wernert N, Rothe M, Maher ER, Latif F. Frequent epigenetic inactivation of the RASSF1A tumour suppressor gene in testicular tumours and distinct methylation profiles of seminoma and nonseminoma testicular germ cell tumours. Oncogene. 2003;22:461–466. doi: 10.1038/sj.onc.1206119. [DOI] [PubMed] [Google Scholar]