Abstract
The maspin gene is not expressed in normal human pancreas, but its expression is acquired during human pancreatic carcinogenesis. In other normal human cells and their malignant counterparts, maspin expression is controlled through the epigenetic state of its promoter. In studies presented herein, we used bisulfite genomic sequencing and chromatin immunoprecipitation studies to show that maspin-negative pancreas cells have a methylated maspin promoter, and that the associated H3 and H4 histones are hypoacetylated. In contrast to normal pancreas, four of six human pancreatic carcinoma cell lines investigated displayed activation of maspin expression. This activation of maspin expression in pancreatic carcinoma cells was linked to demethylated promoters and hyperacetylation of the associated H3 and H4 histones. In addition, 5-aza-2′-deoxycytidine treatments activated maspin expression in the two maspin-negative pancreatic carcinoma cell lines, suggesting a causal role for cytosine methylation in the maintenance of a transcriptionally silent maspin gene. Thus, human pancreatic carcinoma cells acquire maspin expression through epigenetic derepression of the maspin locus, and in so doing appear to co-opt a normal cellular mechanism for the regulation of this gene.
Keywords: histone acetylation, DNA methylation, tumor suppressor gene, cancer, gene expression
Introduction
The concept that DNA methylation might play in role in the establishment and/or maintenance of tissue-specific gene expression was first put forward by Holliday [1] and Riggs [2]. Recently, we described an example of a human gene that provides a strong evidence of this hypothesis [3]. This gene, maspin (SERPINB5), is a member of the serpin superfamily whose expression is regulated at the level of transcription in a cell type-specific manner [3–5]. Most epithelial cells display abundant an expression of maspin, whereas mesenchymal cells do not express maspin, with the notable exception of corneal stromal cells [6].
The loss of maspin expression appears to be an early event in human breast carcinogenesis and can be detected even in ductal carcinoma in situ [7]. Although the precise cellular and biochemical activities of maspin are currently unknown, it has been shown that forced reexpression of maspin, or exogenous maspin protein, can inhibit certain characteristics of invasive and malignant human breast carcinoma cell populations [4,8]. In other human cancers such as ovarian, lung, and pancreatic cancers, maspin expression is gained in the carcinoma cells compared to their normal cells of origin [9–12]. Because maspin has a metastasis suppressor function in human breast cells, the gain of expression observed in these other tumor types is paradoxical.
Because the primary mechanism governing the loss of maspin expression resides at the level of transcriptional inactivation, much effort has been focused on understanding what controls maspin transcriptional activity. Recent studies directed toward understanding the transcriptional regulation of maspin have revealed that epigenetic control is an important mechanism that directs maspin mRNA expression [3,13,14]. The epigenetic determinants associated with the control of maspin expression occur in the 5′ regulatory region of the maspin gene and involve cytosine methylation, histone acetylation, and chromatin architecture [3,13,14]. Interestingly, these epigenetic differences participate in the normal cell type-restricted expression of this gene and participate in the establishment of the active versus repressed states of transcriptional activity [3].
We previously reported that maspin is aberrantly methylated and epigentically silenced in human breast carcinoma cells compared to normal human mammary epithelial cells (HMECs) [13]. Recent reports indicate that, in contrast to breast cancer, maspin expression is gained during pancreatic carcinogenesis; normal human pancreas is negative for maspin expression [11,12]. However, during human pancreatic carcinogenesis, transformed pancreatic cells acquire maspin expression. Similar results have been reported for maspin expression during ovarian carcinogenesis as well [9]. We hypothesized that the mechanism underlying the gain of expression in human pancreatic carcinoma cells is epigenetic in nature.
In this report, we demonstrate that gain of maspin expression in human pancreatic carcinoma cell lines is regulated by the same epigenetic mechanisms, but in the opposite direction, of maspin expression during breast carcinogenesis. Thus, whether or not maspin transcription is silenced or activated during tumor progression of a specific cell type, human tumor cells appear to co-opt a normal mechanism to regulate the transcriptional activity of this gene. Taken together, these results indicate that the cell type-specific methylation pattern of the maspin promoter that exists in normal nonexpressing cells can be replicated in tumorigenic cells whose normal counterparts transcribe maspin, and that this methylation pattern is associated with its transcriptional silencing.
Materials and Methods
Normal Human Pancreas
Normal human pancreas cells were obtained from transplant donors where the pancreas was considered unsuitable for transplantation, or no recipient was available. Pancreatic specimens were discarded and not used in this study if the donor had any history of pancreatic disease. All specimens were obtained at The University of Iowa Hospital and Clinics. The protocol to use the normal human pancreas samples was approved by The University of Iowa Institutional Review Board for Human Subjects on February 12, 2001.
Cell Lines and Cell Culture
Normal HMECs were obtained from Clonetics (San Diego, CA) and grown according to the manufacturer's instructions. The immortalized but nontumorigenic human breast epithelial cell line MCF10A was obtained from the American Type Culture Collection (Manassas, VA) and were maintained in RPMI 1640 containing 10% fetal bovine serum supplemented with 50 µg/ml gentamicin. All other human breast cancer cell lines (MDA-MB-453, MDA-MB-157, MDAMB-468, MDA-MB-435, and MDA-MB-231) as well as human pancreatic carcinoma cell lines (BxPC3, AsPC-1, Capan-1, Capan-2, MiaPaCa-2, and Panc-1) were obtained from the American Type Culture Collection and were maintained in RPMI 1640 containing 10% fetal bovine serum supplemented with 50 µg/ml gentamicin. For 5-aza-2′-deoxycytidine (5-aza-dC) reactivation studies in pancreatic cancer cell lines, cells were grown at low density in six-well plates and were treated with 50 µM 5-aza-dC (Sigma, St. Louis MO) in 1 x phosphate-buffered saline on days 0 and 2. On day 4, total RNA was isolated from the different treatment groups, and analyzed by reverse transcription polymerase chain reaction (RT-PCR) as described below.
RT-PCR Assays for Gene Expression Analysis
Total cellular RNA was isolated using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA) and quantitated by absorption measurements at 260 nm. RT-PCR was performed using RTG RT-PCR beads (Pharmacia, Piscataway, NJ). To assess maspin expression, 250 ng of total RNA was used, and to assess glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression, 125 ng of total RNA was used. The RT reaction was primed with random hexamers and incubated at 42°C for 1 hour followed by a chill at 4°C. Maspin-specific PCR was performed by adding 25 pmol of each maspin mRNA-specific primer prior to thermal cycling. The maspin upstream primer is 5′-CTGACAACAGTGTGAACGAC-3′ and the downstream primer is 5′-CAAGCCTTGGGATCAATCATCT-3′, and correspond to nt 446 to 465 and nt 838 to 860 of the maspin cDNA (accession no. NM002639). GAPDH expression was assessed using GAPDH-specific primers obtained from R&D Systems (Minneapolis, MN). PCR conditions for maspin and GAPDH were the same except that 35 cycles of PCR were performed for maspin analysis and 32 cycles were performed for GAPDH. The parameters used were: 95°C for 5 minutes followed by the stated number of cycles of 94°C for 1 minute; 56°C for 30 seconds, and 72°C for 1 minute, ending with a final extension at 72°C for 5 minutes and a quick chill to 4°C. For analysis, 20% of the respective PCR products was separated through a 3% agarose gel, stained with ethidium bromide, and imaged using the Eagle Eye II Still Video System (Stratagene, La Jolla, CA).
Sodium Bisulfite Genomic Sequencing of the Maspin Promoter
Genomic DNA was isolated using the DNeasy Tissue Kit (Qiagen) and quantitated spectrophotometrically. Five micrograms of genomic DNA was modified with sodium bisulfite under conditions previously described [15]. The maspin promoter [16] was amplified from the bisulfite-modified DNA by two rounds of PCR using nested primers specific to the bisulfite-modified sequence of the maspin CpG island. The first-round primers are as follows: primer U2, 5′-AAAAGAATGGAGATTAGAGTATTTTTTGTG-3′; primer D2, 5′-CCTAAAATCACAATTATCCTAAAAAATA-3′. The second-round primers are as follows: primer U3, 5′-GAAATTTGTAGTGTTATTATTATTATA-3′; primer D3, 5′-AAAAACACAAAAACCTAAATATAAAAA-3′. Both rounds of PCR were performed under the same parameters, with 1% of the first-round PCR product serving as the template in the second-round PCR. PCR amplification was performed under the following conditions: 94°C for 4 minutes followed by five cycles of 94°C for 1 minute, 56°C for 2 minutes, 72°C for 3 minutes, then 35 cycles of 94°C for 30 seconds, 56°C for 2 minutes, 72°C for 1.5 minutes, and ending with a final extension of 72°C for 6 minutes. The resultant PCR product was cloned into a TA vector according to the manufacturer's instructions (pGEM-T-Easy cloning kit; Promega, Madison, WI). Ten positive recombinants were isolated using a Qiaprep Spin Plasmid Miniprep Kit (Qiagen) according to the manufacturer's instructions and sequenced on an ABI automated DNA sequencer. The methylation status of individual CpG sites was determined by comparison of the sequence obtained with the known maspin sequence. The number of methylated CpGs at a specific site was divided by the number of clones analyzed (minimum of 10 in all cases) to yield a percent methylation for each site.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitations using the acetyl-histone H3 and H4 antibodies were performed according to the manufacturer's instructions (Upstate Biotech, Lake Placid, NY) with slight modifications [17]. Cells were rinsed in 1 x HBSS with 0.1% EDTA and treated with 1% formaldehyde for 10 minutes at 37°C to form DNA-protein cross-links. The cells were rinsed in ice-cold 1 x HBSS with 0.1% EDTA containing protease inhibitors (1 mM PMSF, 1 µg/ml aprotinin, 1 µg/ml pepstatin A), scraped, and collected by centrifugation at 4°C. Cells were then resuspended in a PIPES buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP40) containing protease inhibitors and incubated for 10 minutes on ice. Cells were then collected by centrifugation and resuspended in a sodium dodecyl sulfate lysis buffer containing protease inhibitors and incubated on ice for 10 minutes. The DNA-protein complexes were sonicated to lengths between 200 and 1000 bp as determined by gel electrophoresis. Samples were centrifuged at 14,000 rpm at 4°C to spin out cell debris, then the supernatant was diluted 10-fold with ChIP dilution buffer containing protease inhibitors. One tenth of the sample was set aside for input control, and the remaining sample was then precleared with either Protein A Sepharose (Amersham Pharmacia Biotech, Piscataway, NJ).
Following preclearing, the samples were split into thirds, with two of the three samples treated with anti-acetylhistones H3 and H4, whereas the third sample was left as minus antibody (-Ab) control. All samples were rotated overnight at 4°C. The chromatin-antibody complexes were collected using Protein A Sepharose and then sequentially washed with the manufacturer's low-salt, high-salt, and LiCl buffers, then twice with TE buffer (10 mM Tris-HCl, 1 mM EDTA).
The chromatin-antibody complexes were eluted and the DNA-protein cross-links were reversed with 400 mM NaCl at 65°C for 4 hours for all samples, including the input DNA control. All samples were treated with proteinase K, and the acetyl-histone H3 and H4-enriched fractions of genomic DNA were recovered by phenol/chloroform extractions and ethanol precipitations, which were later quantitated using a BioPhotometer (Eppendorf Scientific, Westbury, NY). Quantitative real-time PCR was used to analyze ChIP DNA, using the ABI Prism 7000 sequence detector (Applied Biosystems, Foster City, CA). PCR amplification of the MASPIN promoter was done utilizing TaqMan primer/probes, whereas the GAPDH promoter was amplified using conventional PCR primers (primer sequences are available upon request). Amplifications were done as outlined in Applied Biosystems, TaqMan Universal PCR Master Mix, and SYBR Green PCR Master Mix protocols for MASPIN and GAPDH, respectively. MASPIN was amplified using the printed universal conditions for 40 cycles, whereas GAPDH was amplified using the following conditions; 95°C for 10 minutes followed by 40 cycles of 94°C for 1 minutes, and 68°C for 30 seconds, and 72°C for 1 minute.
For each experiment, the threshold bar was set within the linear range of the PCR amplification. For the majority of the experiments, the data were analyzed with the threshold set at 0.1. The resulting Ct and Rn files were exported to Microsoft Excel for data and graphical analysis. Ct is the number of PCR cycles necessary to reach fluorescence intensity (an indirect measure of PCR product) within the linear range of PCR amplification. Quantification was determined by applying the comparative Ct method, as described in the ABI 7000 sequence detection user guide and others. Briefly, fold enrichment was calculated by subtracting the Ct value of the ChIP DNA from the Ct value of the input DNA fraction, and by using this value as the power that 2 is raised to (i.e., 2Ct(Input)-Ct(ChIP)).
Results
Normal Human Pancreas Cells Do Not Express Maspin mRNA
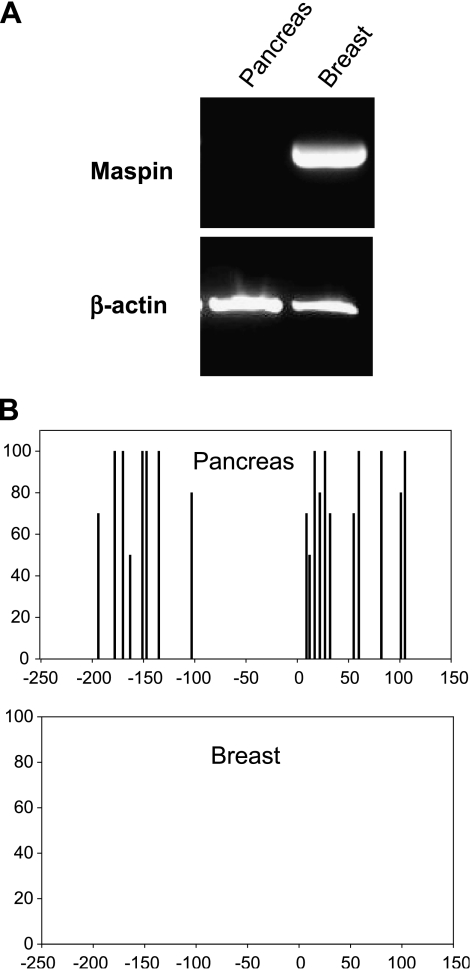
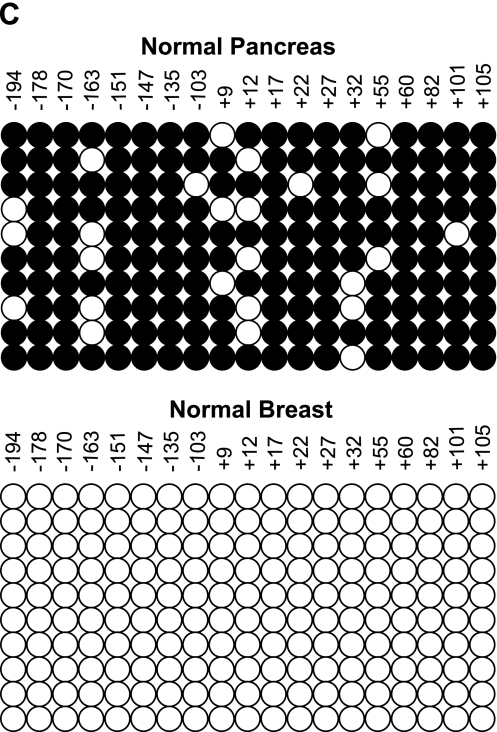
Maspin mRNA was measured in normal human pancreas and HMECs by RT-PCR as shown in Figure 1A. The human pancreas was previously shown to be negative for maspin protein by immunohistochemistry [11] and our findings that maspin mRNA is also undetectable in normal human pancreas confirm and extend those earlier findings. Maspin mRNA was abundantly expressed in normal HMECs as has been previously described [4,13]. From these expression patterns and from our previous experience with methylation-associated silencing of the maspin gene in normal human tissues [3], we hypothesized that CpG cytosines in the maspin promoter region would be methylated in normal human pancreas. Figure 1B shows histograms of CpG methylation across the maspin promoter regions in normal human breast and pancreas cells. In breast cells that are maspin-positive, the promoter is unmethylated as has been previously described [13], whereas in maspin-negative pancreas cells, the promoter is extensively methylated at all 19 CpG sites analyzed. These findings are in precise agreement with the previously described role for DNA methylation in the cell type-specific expression of maspin [3]. Figure 1C displays an alternative representation of the distribution of methylated cytosines in a number of clones to indicate that maspin is not an imprinted locus in pancreas cells. These findings support previous findings regarding the cell type-specific regulation of this genetic locus, and elucidate yet another normal cell type (i.e., pancreas) wherein the genomic methylation status of the maspin promoter was strictly and inversely correlated with maspin mRNA expression.
Figure 1.
Lack of maspin mRNA in normal pancreatic epithelial cells is tightly associated with promoter methylation status. (A) RNA was isolated from normal pancreas tissue and normal mammary cells (HMECs). RT-PCR was performed for maspin mRNA expression and PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining. Normal pancreas showed no maspin expression whereas normal mammary cells showed robust maspin expression; β-actin was used as a control. (B) DNA from both tissues was sodium bisulfite-modified to determine the methylation status of the maspin promoter. Summary of 5-methylcytosine levels obtained by sodium bisulfite genomic sequencing of the maspin promoter. Cytosine methylation frequency histograms are shown for normal pancreas and HMECs. The x-axis is nucleotide position relative to the transcription start and the y-axis is the percent cytosine methylation. (C) Methylation status of the individual alleles determined by bisulfite sequencing of cloned PCR products. Each row of circles represents the cytosine methylation pattern obtained from individual clones of the maspin promoter. The position of each CpG site relative to transcription start is shown. Open circles indicate unmethylated CpG sites; filled circles indicate methylated CpG sites.
Maspin Expression Is Acquired During Human Pancreatic Carcinogenesis
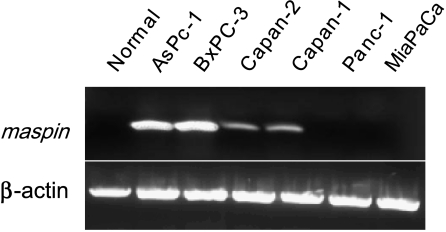
Maass et al. first reported that human pancreatic cancer cells showed increased maspin expression relative to normal human pancreas [11]. This somewhat surprising finding has fueled recent speculations as to the role of maspin as a tumor suppressor [18]. Nevertheless, to confirm and extend the findings by Maass et al., we performed an RNA expression analysis on normal human pancreas and six human pancreas carcinoma cell lines. Our results, shown in Figure 2, indicate that although maspin mRNA is undetectable in normal human pancreas, four of six human pancreatic cancer cell lines examined expressed maspin mRNA. All six of the pancreas carcinoma cell lines possess mutant p53 [19], which suggests that although p53 has been shown to be a positive regulator of maspin expression in human breast and prostate cells [20,21], it is unlikely to be playing a part in maspin expression among this panel of human pancreas cancer cell lines. Thus, we sought to determine what other mechanisms could account for the acquisition of maspin mRNA expression during pancreatic carcinogenesis.
Figure 2.
Maspin expression is activated during pancreatic carcinogenesis. RNA was isolated from normal pancreas tissues and six pancreatic cancer cell lines, and RT-PCR was performed for maspin expression and ′-actin; PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining. Maspin expression was gained in four of six pancreatic carcinoma cell lines.
The Maspin Promoter Becomes Unmethylated in Pancreatic Carcinoma Cells That Have Acquired Maspin Expression
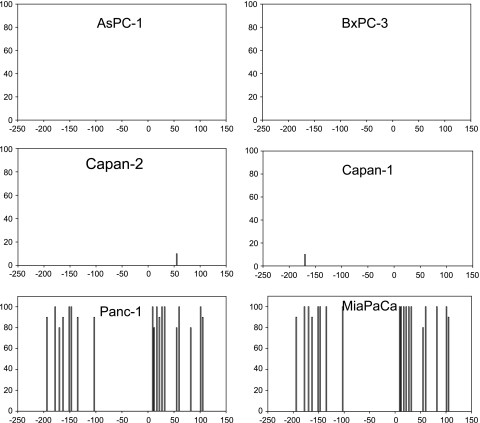
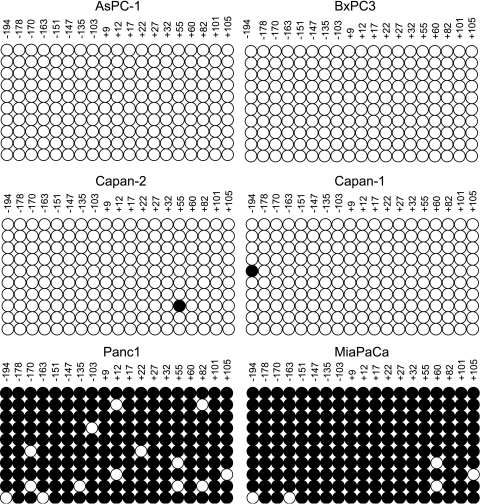
To test the hypothesis that demethylation of the maspin promoter region could be responsible for the activation of maspin expression seen in the human pancreatic cancer cell lines, we performed bisulfite analysis on the maspin 5′ regulatory region in normal and malignant human pancreas cells. The results of this experiment are shown in Figure 3, and clearly indicate that the acquisition of maspin expression in the human pancreatic cancer cells is strictly correlated with demethylation of the maspin promoter in these cell lines. Whereas the maspin-negative pancreatic carcinoma cells (Panc-1 and MiaPaCa) displayed a methylated CpG pattern across the maspin promoter comparable to that seen in normal human pancreas, the maspin-positive human pancreatic cancer cells were unmethylated in the maspin promoter region. Figure 4 displays a different representation of the distribution of methylated cytosines in 10 sequenced clones from each cell line to indicate that maspin is not an imprinted gene and that acquisition of maspin expression is not allele-specific.
Figure 3.
Activation of maspin expression is associated with the demethylation of its promoter in pancreatic cancer cells. Bisulfite sequencing was used to analyze cytosine methylation in six pancreatic cancer cell lines. The four unmethylated cell lines (AsPC-1, BxPC-3, Capan-2, and Capan-1) all showed activation of maspin gene expression. In contrast, two maspin-negative cell lines (Panc-1 and MiaPaCA) were heavily methylated and exhibited no maspin expression. Summary of 5-methylcytosine levels obtained by sodium bisulfite genomic sequencing of the maspin promoter. Cytosine methylation frequency histograms are shown for normal pancreas and HMECs. The x-axis is nucleotide position relative to the transcription start and the y-axis is the percent cytosine methylation.
Figure 4.
Maspin promoter demethylation during transcriptional activation is not allele-specific. The methylation status of the individual alleles was determined by bisulfite sequencing of 10 cloned PCR products. Each row of circles represents the cytosine methylation pattern obtained from an individual clone of the maspin promoter. The position of each CpG site relative to transcription start is shown. Open circles indicate unmethylated CpG sites; filled circles indicate methylated CpG sites.
Maspin Expression Is Activated in Maspin-Negative Pancreatic Cancer Cells by 5-aza-dC, a DNA Demethylating Agent
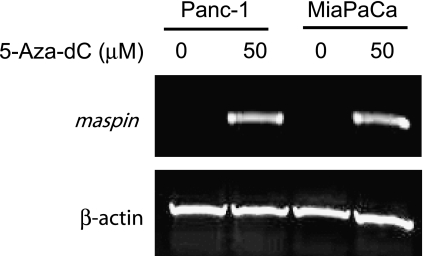
To test whether cytosine methylation in the maspin promoter was functionally linked to its silencing in these human pancreatic cancer cells, we treated the cells with 5-aza-dC, a demethylating drug that has been shown to induce maspin expression in human breast carcinoma cells as well as maspin-negative human fibroblasts and kidney cells [3,13]. The maspin-negative Panc-1 and MiaPaCa cells were grown for 4 days in the presence or absence of 50 µM 5-aza-dC and then analyzed for maspin mRNA expression. Figure 5 shows that 5-aza-dC caused a robust induction of maspin expression in both maspin-negative cell lines, further indicating the importance of cytosine methylation in maintaining the transcriptionally repressed state of maspin in maspin-negative normal and tumor cell types.
Figure 5.
Maspin expression can be activated in maspin-negative pancreatic carcinoma cell lines with 5-aza-dC. Panc-1 and MiaPaCA cells lines with methylated maspin promoters and no maspin expression were treated with 50 µM 5-aza-dC for 48 hours. RNA was isolated, RT-PCR was performed, and PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining.
Activation of Maspin Expression in Human Pancreatic Cancer Cells Is Linked to Hyperacetylation of Histones H3 and H4 Associated with the Maspin Promoter
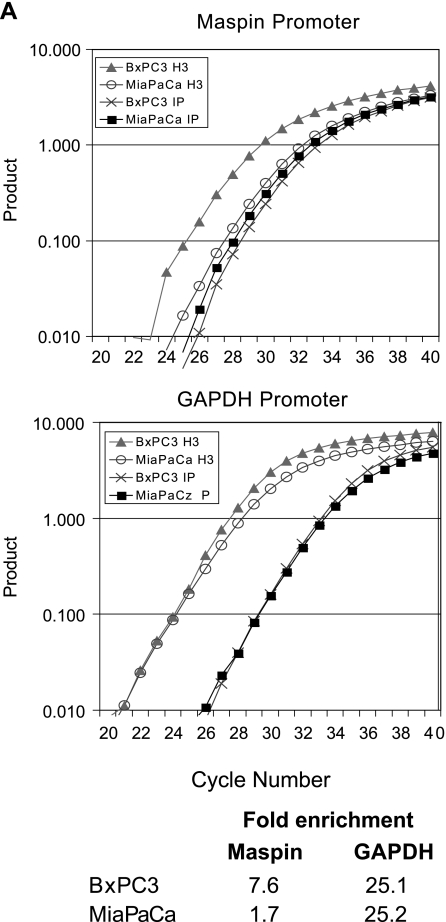
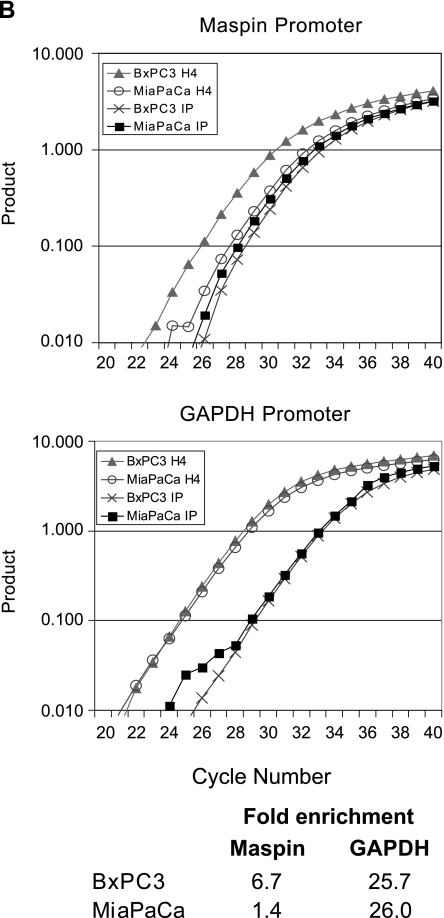
To assess whether chromatin structure at the maspin promoter participates in the regulation of this gene, we performed chromatin immunoprecipitation experiments with antibodies directed against acetylated histones H3 and H4 and amplified the maspin promoter. The acetylation status of these nucleosome constituents is tightly correlated to cytosine methylation status and gene activity [22–24]. Thus, we hypothesized that demethylated maspin-positive pancreatic carcinoma cells would remodel their maspin promoter in an active chromatin structure with hyperacetylated histones, whereas the maspin-negative pancreas cells would maintain their methylated maspin promoter in a chromatin structure with hypoacetylated histones. Our results, shown in Figure 6, indicate that the maspin promoter is associated with hypoacetylated histones in maspin-negative MiaPaCa pancreatic cancer cells. However, the maspin promoter is associated with hyperacetylated histones in maspin-positive BxPC3 human pancreatic cancer cells. These results suggest that the maspin promoter becomes not only demethylated, but the associated histones H3 and H4 become hyperacetylated during the acquisition of maspin expression during pancreatic carcinogenesis. These findings provide further evidence for strong epigenetic component to the regulation of this gene in normal human tissues, and provide a likely mechanism for the gain of maspin expression observed in not only human pancreatic cancer, but also ovarian and lung cancers [9–11], as well as the well-documented loss of maspin expression in human breast cancer.
Figure 6.
Histone acetylation state is linked to DNA methylation state at the maspin promoter in pancreatic cells. Acetylation of histones H3 (A) and H4 (B) in the maspin and GAPDH promoters was analyzed using a quantitative chromatin immunoprecipitation assay. Gene promoter-specific real-time PCR was carried out on DNA from the immunoprecipitated chromatin as well as input DNA. A representative real-time PCR graph and the fold enrichment of histone acetylation are shown. Assessment of histone acetylation state of the ubiquitously expressed GAPDH was performed as a positive control.
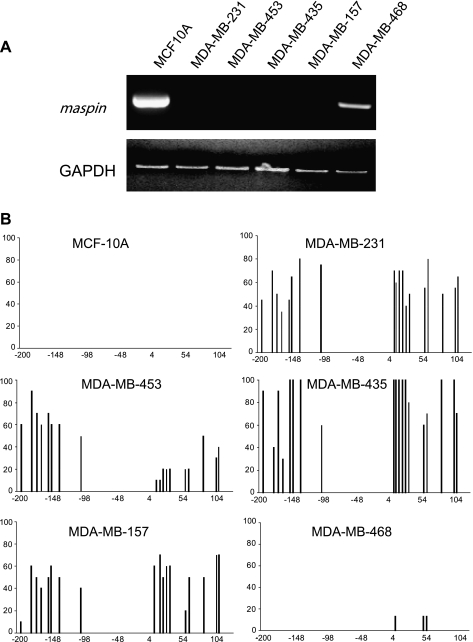
The Maspin Promoter Becomes Aberrantly Methylated in Mammary Carcinoma Cells That Have Lost Maspin Expression
The dense methylation of the maspin promoter seen in normal maspin-negative normal pancreas tissues is similar to the methylation state encountered in breast cancer cells where the maspin gene is often silenced. As an example and extension of cytosine methylation profiles obtained in human breast cells, we show in Figure 7 the maspinpositive nontumorigenic but immortalized HMEC line MCF-10A, as well as the several additional human breast cancer cell lines that have lost maspin expression in association with aberrant methylation of the maspin promoter. Four of five of these human breast cancer cell lines showed loss of maspin expression that was correlated without exception to maspin promoter methylation. Only the MDAMB-468 human breast cancer cell line was maspin-positive and its promoter was unmethylated. These results are consistent with and extend previous reports indicating that maspin inactivation in breast carcinogenesis is a frequent event and that silencing of maspin expression is often associated with aberrant cytosine methylation of the maspin promoter. Taken together, these results indicate that the cell type-specific methylation pattern of the maspin promoter that exists in normal nonexpressing cells can be replicated in tumorigenic cells whose normal counterparts transcribe maspin, and that this methylation pattern is associated with its transcriptional silencing.
Figure 7.
The loss of maspin expression in the majority of breast cancer cell lines is associated with aberrant methylation of its promoter. (A) RT-PCR was performed on normal breast and five breast cancer cell lines not previously analyzed. Normal breast cells were maspin-positive, but four of five breast cancer cell lines were maspin-negative, consistent with previous observations. (B) Bisulfite sequence analysis of maspin promoter methylation in normal breast cells and breast cancer cells. Cytosine methylation histograms show that maspin promoter methylation in breast cancer cells is associated with the attenuation of maspin expression.
Discussion
A critical mechanism that governs the differential transcriptional activity of the maspin tumor suppressor gene in normal human cells types is a transition in chromatin architecture around the maspin promoter from an open and active state to a heterochromatic state that is transcriptionally silentdddddd. The structural changes in the maspin promoter leading to gene silencing involve multiple enzyme-catalyzed modifications at the level of the nucleosome including cytosine methylation by DNA methyltransferases (DNMTs) and histone acetylation by histone acetyltransferases (HATs). Overall, these indicators of chromatin structure are highly predictive of the level of maspin expression within any given human cell type; when the maspin promoter is unmethylated and hyperacetylated, the gene is actively expressed; however, when the maspin promoter is hypermethylated and hypoacetylated, the gene is not expressed. These chromatin structural modifications have been demonstrated as a major mechanism for the establishment and/or maintenance of the maspin phenotype in many types of normal human cells [3].
In breast cancer cells, this control is relaxed, producing an epigenetic switch in the maspin promoter from an open and on state to a closed and off state, resulting in the silencing of maspin expression [13,21]. Surprising recent work has found maspin expression to be activated in certain forms of human cancers compared to the corresponding normal cells from which they are presumed to have arisen, and include pancreatic, ovarian, lung, and gastric cancers [9–12,25]. We sought to determine if the epigenetic switch at the maspin promoter in pancreatic cancer worked from off to on, the direction opposite to that seen in breast cancer, and to determine if this switch was responsible for maspin activation. We chose to examine pancreatic cancers as they were among the first human cancer types reported to display this paradoxical activation of maspin expression [11].
Our analysis of human pancreatic cancer cells shows a strict correlation between acquisition of maspin expression and demethylation of the maspin promoter and hyperacetylation of the associated histones H3 and H4. In normal maspin-negative pancreas cells, the maspin promoter was densely methylated, whereas the epigenetic state (DNA methylation and histone acetylation) of the maspin promoter in the maspin-positive pancreatic cancer cells resembled the state of the maspin promoter in normal maspin-positive cells (e.g., HMECs). By comparison, in breast cancer cells, where the maspin gene is inappropriately silenced, the epigenetic state of the maspin promoter resembled that seen in normal maspin-negative cells (e.g., pancreas). Together, these results indicate that tumor cells can replicate the DNA methylation and histone acetylation patterns specific to other cell types, and that this epigenetic switch is tightly associated with its transcriptional state.
The biological significance of increased maspin expression in pancreatic carcinoma cells and other human cancers where maspin is activated is unclear. Nevertheless, these reports have challenged the role of maspin as a tumor suppressor and/or suppressor of metastasis; however, it should be pointed out that these were correlative studies where a role for maspin in the disease process was not determined. In contrast, direct functional studies in breast cancer cells have clearly shown that maspin suppresses aggressive tumor characteristics, such as cell adhesion, motility, and angiogenic properties [4,26–28].
Thus, whether maspin activation is a participant in the carcinogenic process in pancreatic cancer, or whether it is a bystander event is unknown. The loss of maspin promoter methylation may be a nonspecific alteration that accompanies the complex and genome-wide epigenetic changes that occur in the genomes of cancer cells [29,30], and therefore may provide no selective advantage to the cancer cell. Another possible explanation for these apparently contradictory results is that the maspin protein has different functions in different cell types depending on the spectrum of other maspin interacting proteins that are expressed within a given cell type or its surroundings. Finally, the subcellular localization of maspin may play a critical role in determining its function. Indeed, there have been recent reports that maspin is expressed in the nuclei of certain cell types [31]. Whether this inappropriate activation of maspin expression imparts any significant phenotypic changes on the tumor, or whether its activation is an epiphenomenon that reflects loss of methylation homeostasis remains to be determined.
In summary, we show that pancreatic cancer cells acquire maspin expression through demethylation of the maspin promoter and hyperacetylation of the associated histones. These results show that the maspin promoter in pancreatic cancer cells undergoes an epigenetic switch from the off to on position. In comparison, breast cancer cells undergo a switch from on to off at the same locus. In both cases, the maspin promoter took on the epigenetic appearance of a different normal cell type. We speculate that the same epigenetic mechanisms that control maspin expression in normal cell types are co-opted during carcinogenesis. This view would be consistent with current concepts of the plasticity of the differentiated state, the overall and regional states of differentiation within a tumor (i.e., well differentiated or poorly differentiated), and the nature of the mechanisms underlying changes in gene expression patterns during transdifferentiation, including epithelial-mesenchymal transition [32,33].
Acknowledgements
We thank Marilyn Hinckhouse for supplying pancreatic carcinoma cell lines. We also thank S. Vaught and Kevin Knudtson for technical assistance.
Footnotes
This work was supported, in part, by National Institutes of Health grants to F.E.D. and B.W.F., as well as an institutional grant from the Holden Comprehensive Cancer Center at The University of Iowa. M.O. and K.K. were supported by training grants from the National Cancer Institute.
References
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- 4.Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 5.Sager R, Sheng S, Pemberton P, Hendrix MJ. Maspin: a tumor suppressing serpin. Curr Top Microbiol Immunol. 1996;213:51–64. doi: 10.1007/978-3-642-61107-0_4. [DOI] [PubMed] [Google Scholar]
- 6.Ngamkitidechakul C, Burke JM, O'Brien WJ, Twining SS. Maspin: synthesis by human cornea and regulation of in vitro stromal cell adhesion to extracellular matrix. Invest Ophthalmol Vis Sci. 2001;42:3135–3141. [PubMed] [Google Scholar]
- 7.Maass N, Teffner M, Rosel F, Pawaresch R, Jonat W, Nagasaki K, Rudolph P. Decline in the expression of the serine proteinase inhibitor maspin is associated with tumour progression in ductal carcinomas of the breast. J Pathol. 2001;195:321–326. doi: 10.1002/path.948. [DOI] [PubMed] [Google Scholar]
- 8.Sheng S, Pemberton PA, Sager R. Production, purification, and characterization of recombinant maspin proteins. J Biol Chem. 1994;269:30988–30993. [PubMed] [Google Scholar]
- 9.Sood AK, Fletcher MS, Gruman LM, Coffin JE, Jabbari S, Khalkhali-Ellis Z, Arbour N, Seftor EA, Hendrix MJC. The paradoxical expression of maspin in ovarian carcinoma. Clin Cancer Res. 2002;8:2924–2932. [PubMed] [Google Scholar]
- 10.Heighway J, Knapp T, Boyce L, Brennand S, Field JK, Betticher DC, Ratschiller D, Gugger M, Donovan M, Lasek A, Rickert P. Expression profiling of primary non-small cell lung cancer for target identification. Oncogene. 2002;21:7749–7763. doi: 10.1038/sj.onc.1205979. [DOI] [PubMed] [Google Scholar]
- 11.Maass N, Hojo T, Ueding M, Luttges J, Kloppel G, Jonat W, Nagasaki K. Expression of the tumor suppressor gene Maspin in human pancreatic cancers. Clin Cancer Res. 2001;7:812–817. [PubMed] [Google Scholar]
- 12.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- 13.Domann FE, Rice JC, Hendrix MJ, Futscher BW. Epigenetic silencing of maspin gene expression in human breast cancers. Int J Cancer. 2000;85:805–810. doi: 10.1002/(sici)1097-0215(20000315)85:6<805::aid-ijc12>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Maass N, Biallek M, Rosel F, Schem C, Ohike N, Zhang M, Jonat W, Nagasaki K. Hypermethylation and histone deacetylation lead to silencing of the maspin gene in human breast cancer. Biochem Biophys Res Commun. 2002;297:125–128. doi: 10.1016/s0006-291x(02)02136-8. [DOI] [PubMed] [Google Scholar]
- 15.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Maass N, Magit D, Sager R. Transactivation through Ets and Ap1 transcription sites determines the expression of the tumor-suppressing gene maspin. Cell Growth Differ. 1997;8:179–186. [PubMed] [Google Scholar]
- 17.Rice JC, Futscher BW. Transcriptional repression of BRCA1 by aberrant cytosine methylation, histone hypoacetylation and chromatin condensation of the BRCA1 promoter. Nucleic Acids Res. 2000;28:3233–3239. doi: 10.1093/nar/28.17.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domann FE, Futscher BW. Editorial: maspin as a molecular target for cancer therapy. J Urol. 2003;169:1162–1164. doi: 10.1097/01.ju.0000058368.53821.68. [DOI] [PubMed] [Google Scholar]
- 19.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendation to users. Hum Mutat. 2002;19:607–614. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 20.Zou Z, Gao C, Nagaich AK, Connell T, Saito S, Moul JW, Seth P, Appella E, Srivastava S. p53 regulates the expression of the tumor suppressor gene maspin. J Biol Chem. 2000;275:6051–6054. doi: 10.1074/jbc.275.9.6051. [DOI] [PubMed] [Google Scholar]
- 21.Oshiro MM, Watts GS, Wozniak RJ, Junk DJ, Munoz-Rodriguez JL, Domann FE, Futscher BW. Mutant p53 and aberrant cytosine methylation cooperate to silence gene expression. Oncogene. 2003;22:3624–3634. doi: 10.1038/sj.onc.1206545. [DOI] [PubMed] [Google Scholar]
- 22.Fischle W, Wang Y, Allis CD. Histone and chromatin crosstalk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 23.Ng HH, Bird A. DNA methylation and chromatin modification. Curr Opin Genet Dev. 1999;9:158–163. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 24.Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 25.Son HJ, Sohn TS, Song SY, Lee JH, Rhee JC. Maspin expression in human gastric adenocarcinoma. Pathol Int. 2002;52:508–513. doi: 10.1046/j.1440-1827.2002.01381.x. [DOI] [PubMed] [Google Scholar]
- 26.Sheng S, Carey J, Seftor EA, Dias L, Hendrix MJ, Sager R. Maspin acts at the cell membrane to inhibit invasion and motility of mammary and prostatic cancer cells. Proc Natl Acad Sci USA. 1996;93:11669–11674. doi: 10.1073/pnas.93.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seftor RE, Seftor EA, Sheng S, Pemberton PA, Sager R, Hendrix MJ. Maspin suppresses the invasive phenotype of human breast carcinoma. Cancer Res. 1998;58:5681–5685. [PubMed] [Google Scholar]
- 28.Zhang M, Volpert O, Shi YH, Bouck N. Maspin is an angiogenesis inhibitor. Nat Med. 2000;6:196–199. doi: 10.1038/72303. [DOI] [PubMed] [Google Scholar]
- 29.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 30.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 31.Maass N, Rolver L, Ziebart M, Nagasaki K, Rudolph P. Maspin locates to the nucleus in certain cell types. J Pathol. 2002;197:273–274. [Google Scholar]
- 32.Boyer B, Valles AM, Edme N. Induction and regulation of epithelial-mesenchymal transitions. Biochem Pharmacol. 2000;60:1091–1099. doi: 10.1016/s0006-2952(00)00427-5. [DOI] [PubMed] [Google Scholar]
- 33.Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5:101–106. doi: 10.1186/bcr578. [DOI] [PMC free article] [PubMed] [Google Scholar]