Abstract
The purpose of this study was to investigate the presence of the anaerobic intestinal spirochetes Brachyspira aalborgi and Brachyspira pilosicoli in the feces of captive nonhuman primates (n = 35) from 19 species housed at the Zoological Gardens, Perth, Western Australia. Both spirochete species are known to infect human beings. DNA was extracted from freshly collected feces with a commercially available QIAamp DNA stool minikit and subjected to PCR protocols amplifying portions of the 16S rRNA genes of the two spirochete species. The feces were also subjected to selective culture for the spirochetes. Subsequently, feces from 62 other captive animals or birds representing 39 species at the zoo were examined by PCR to determine whether they were reservoirs of infection. Six fecal samples from individuals from four primate species (two vervet monkeys, two Tonkean macaques, one Japanese macaque, and one hamadryas baboon) tested positive in the B. aalborgi PCR. B. aalborgi was not detected by PCR in any of the other animal or bird species tested, and B. pilosicoli was not detected in the primates or any of the other animals or birds. B. aalborgi was isolated from both PCR-positive vervet monkeys. This is the first time that B. aalborgi has been isolated from nonhuman primates and the first time that it has been isolated from the feces of any species.
The term intestinal spirochetosis (IS) was originally used to describe a condition in which a dense fringe of spirochetal bacteria was observed attached by one cell end to the colorectal epithelium of humans, forming a “false brush border” (5). Since that first description, the clinical significance of this colonization has been debated, but a number of studies have linked the condition to symptoms including chronic diarrhea and rectal bleeding (3, 5, 19). Clinical symptoms have sometimes improved following antimicrobial treatment that has removed the spirochetes (6, 18). Two species of fastidious anaerobic spirochetes have been associated with IS: Brachyspira aalborgi (7-9, 13) and Brachyspira pilosicoli (23-25). Fecal culture has been used to show that colonization with B. pilosicoli is widespread among people living in developing countries (1, 25), but in developed countries it is common only in homosexual males (23). The spirochete also colonizes many animal species, including pigs and dogs, and birds (20, 24, 25). In contrast, recent studies using PCR on DNA extracted from colorectal biopsy specimens or direct in situ hybridization on biopsy specimens have demonstrated that B. aalborgi is the most common cause of IS among patients in Western countries, with B. pilosicoli only occasionally being detected (8, 10, 12-14). Both species of spirochete are slow-growing anaerobes with specialized growth requirements, but B. aalborgi is even more fastidious than B. pilosicoli. To date there have only been three reports of B. aalborgi having been isolated, in all cases directly from human colorectal biopsy specimens (7-9). B. aalborgi has never been isolated from feces, although its presence has been detected in feces using PCR (15).
A spirochetal colonization of the large intestine which is morphologically identical to human IS has been observed in a number of nonhuman primate species, including colony-raised rhesus monkeys (Macaca mulatta) (4, 21, 22), wild-caught baboons (Papio spp.) (17), and wild-caught vervet monkeys (Cercopithecus aethiops) (2). To date the only study which attempted to identify such organisms was undertaken by Duhamel and colleagues, who observed spirochetes attached to the colonic mucosa among a group of 10 colony-raised rhesus monkeys and two crab-eating monkeys (Macaca fascicularis) housed in a research institute (4). Using selective-culture techniques, they isolated B. pilosicoli from five of the rhesus monkeys. They then applied PCR to DNA extracted from formalin-fixed colonic biopsy specimens and identified B. pilosicoli DNA in two of these culture-positive monkeys. In addition, B. aalborgi DNA was detected in seven of the rhesus monkeys (including two that were also positive for B. pilosicoli) and in both crab-eating monkeys.
The aims of the present study were to determine whether captive nonhuman primates in a public zoological collection were colonized with intestinal spirochetes and to determine which spirochete species might be involved. Other animal and bird species in the collection were also tested for the presence of B. aalborgi and B. pilosicoli to determine whether and to what extent such captive species on the same site might be infected. In view of the fastidious growth requirements of the spirochetes, direct PCRs were conducted on DNA extracted from feces. For comparative purposes, selective culture designed to isolate B. aalborgi and B. pilosicoli was also undertaken on the fecal samples from the nonhuman primates.
MATERIALS AND METHODS
Fecal samples.
A total of 97 fresh fecal samples from 59 animal and bird species were collected by zookeepers at the Zoological Gardens in Perth, Western Australia. Thirty-five of the samples were from 19 nonhuman primate species (Table 1), each species of which lived in a separate enclosure. The remaining samples included 18 from 14 species of birds, 7 from 7 species of reptiles, and 37 from 18 species of marsupials and mammals. The animals were observed, and the fecal samples were collected from the floors of the cages after they had been deposited. A single sample was collected from each animal. The samples were stored at 4°C and transported to Murdoch University for processing within 24 h of collection.
TABLE 1.
Species and numbers of nonhuman primates sampled
| Species | Common name | No. of samples
|
|
|---|---|---|---|
| Tested | Positive in B. aalborgi PCRa | ||
| Prosimians | |||
| Lemur catta | Ring-tailed lemur | 2 | 0 |
| Eulemur fulvus albifrons | White-fronted lemur | 2 | 0 |
| Varecia variegata | Black and white ruffed lemur | 2 | 0 |
| Catarrhines | |||
| Hominidae | |||
| Pongo pygmaeus abelii | Orangutan | 5 | 0 |
| Hylobatidae | |||
| Hylobates concolor leucogenys | White-cheeked gibbon | 1 | 0 |
| Hylobates moloch | Silvery gibbon | 1 | 0 |
| Cercopithecidae (Old World monkeys) | |||
| Saguinus oedipus | Cotton-topped tamarin | 2 | 0 |
| Presbytis senex | Western purple-faced langur | 2 | 0 |
| Cercopithecus aethiops | Vervet | 3 | 2 |
| Macaca fuscata | Japanese macaque | 1 | 1 |
| Colobus guereza | Colobus monkey | 2 | 0 |
| Macaca nigra | Crested macaque | 1 | 0 |
| Macaca tonkeana | Tonkean macaque | 2 | 2 |
| Papio hamadryas | Hamadryas baboon | 1 | 1 |
| Platyrrhines (New World monkeys) | |||
| Saguinus imperator | Emperor tamarin | 2 | 0 |
| Aotus trivirgatus | Owl monkey | 1 | 0 |
| Cebus capucinus | White-fronted capuchin | 1 | 0 |
| Callithrix jacchus | Common marmoset | 2 | 0 |
| Leontopithecus rosalia | Golden lion tamarin | 2 | 0 |
No fecal samples positive for B. pilosicoli by PCR.
Control spirochete strains and media.
Control spirochete strains were obtained from the collection held at the Reference Center for Intestinal Spirochetes, Murdoch University. B. aalborgi 513AT and B. pilosicoli P43/6/78T were revived from frozen stock and propagated anaerobically in an atmosphere of 94% H2 and 6% CO2 at 37°C on nonselective Trypticase soy agar (TSA) (BBL, Cockeysville, Md.) containing 10% (vol/vol) defibrinated ovine blood for up to 15 days for B. aalborgi. Viable cells were scraped from the agar and suspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) to a concentration of 1011 cells per ml.
PCRs.
The PCRs used were based on those described for detection of the spirochete species in human colorectal biopsy specimens and feces (13-15). The target for B. aalborgi PCR amplification was a 471-bp section of the 16S rRNA gene equivalent to bp 172 to 675 of the 16S rRNA gene of Escherichia coli, while for B. pilosicoli it was a 439-bp section of the 16S rRNA gene equivalent to bp 204 to 676 of the 16S rRNA gene of E. coli.
Throughout the study, standard procedures were followed to prevent possible contamination of the PCRs. A biosafety cabinet was used for handling feces. For pipetting, dedicated pipettes with filtered tips were used. DNA extraction, PCR setup, and amplification were undertaken in three separate rooms. The amplification mixtures (25 μl) contained 1× PCR buffer, 0.55 U of Tth Plus DNA polymerase, 1.5 mM MgCl2, 5 nmol of each deoxynucleoside triphosphate (Amersham Pharmacia Biotech AB, Uppsala, Sweden), and 12.5 pmol of each primer. Thermocycling was as follows: denaturation for 4 min 30 s at 94°C, followed by 33 cycles of denaturation at 94°C for 30 s, annealing at 46°C for B. aalborgi or 51°C for B. pilosicoli, and primer extension at 72°C for 30 s and a final extension at 72°C for 5 min. The PCR products were subjected to electrophoresis in 1.5% agarose gels in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA) for 25 min at 110 V, stained by immersion for 10 min in ethidium bromide at a concentration of 0.001 mg/ml in distilled water, and then viewed over UV light.
PCR sensitivities.
The sensitivities of the PCRs were estimated by seeding human feces previously shown to be negative for B. aalborgi and B. pilosicoli by culture and PCR with known concentrations of B. aalborgi or B. pilosicoli cells. A range of 500-μl serial dilutions of the 1011-cell/ml stock, from 1011 to 101 cells/ml, were added to 0.2 g of feces, and each mixture was vortexed until homogenous. Negative controls consisted of 0.2 g of feces with 500 μl of sterile phosphate-buffered saline. DNA was extracted from the feces by use of the QIAamp DNA stool minikit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. This extraction procedure was also subsequently used with the test samples from the zoological collection.
Sequencing of PCR products.
All amplified products from the primate feces were sequenced with a commercially available cycle sequencing kit (ABI PRISM dye terminator cycle sequencing ready reaction kit; Applied Biosystems Inc.) according to the manufacturer's instructions. The sequence data obtained were aligned and compared with 16S ribosomal DNA (rDNA) sequences of B. aalborgi 513AT (GenBank accession no. Z22781) and B. pilosicoli P43/6/78T (GenBank accession no. U23032) by using SeqEd, version 1.0.3 (Applied Biosystems Inc.; 1995).
Isolation.
For isolation of B. aalborgi, fecal samples from the primates were plated to TSA containing 10% (vol/vol) defibrinated ovine blood, spectinomycin (400 μg/ml), colistin (25 μg/ml), and polymyxin (5 μg/ml) and cultured in an atmosphere of 94% H2 and 6% CO2 at 37°C for 26 days. For isolation of B. pilosicoli, the plates contained 5% ovine blood, spectinomycin (400 μg/ml), colistin (25 μg/ml), and vancomycin (25 μg/ml) and were incubated for up to 10 days. Spirochete growth on the B. aalborgi plates was subcultured to the same plate type, as well as to the same plate type in which the polymyxin was replaced with vancomycin (25 μg/ml). Contamination was reduced on the latter plate type, and after 21 days the spirochetes were subcultured from these plates to TSA without antibiotics. Growth then occurred on these plates within 15 days. Cell picks were made from the plates and subjected to PCR, and the product sequences were compared with those obtained directly from the fecal samples. Growth from the plates was also transferred into Kunkle's medium, a prereduced anaerobic Trypticase soy broth (BBL) containing 2% fetal calf serum and 0.002% ethanolic cholesterol (11).
API- ZYM.
The enzymatic reactions of the isolates obtained were tested with the commercial API- ZYM system (Analytab Products, Marcy-l'Etoile, France), as previously described for B. aalborgi (7, 9). Bacteria were pelleted by centrifugation from Kunkle's broth, resuspended in sterile distilled water to 107 cells/ml, and placed in the test cupules, and the strips were read after 4 h of incubation at 37°C. To test for indole production, 2 ml of broth culture (∼108 cells/ml) was extracted with 1 ml of xylene and then 4 drops of Kovács' reagent were added. Development of a red or purple color at the surface indicated a positive culture. Growth of Brachyspira hyodysenteriae strain B78T was used as a positive control.
Electron microscopy.
For electron microscopy, the spirochetes were scraped from the subculture plates and resuspended in distilled water. Drops of the suspensions were placed on Formvar-coated reinforced grids and stained with phosphotungstic acid (3%, pH 7.2). Grids were examined in a Philips CM100 Biotwin transmission electron microscope.
RESULTS
PCR detection limits.
The lower limit of detection for the B. aalborgi-specific PCR was between 2.5 × 104 and 2.5 × 105 cells per g of seeded human feces, equivalent to 2.5 × 102 to 2.4 × 103 cells per PCR. For the B. pilosicoli PCR, the detection level was between 2.5 × 105 and 2.5 × 106 cells per g of human feces, or 2.5 × 103 to 2.5 × 104 cells per PCR.
PCR.
When the PCRs were applied to DNA extracted from the feces collected at the zoo, 6 of the 35 (17.1%) fecal samples from the 19 species of primates examined were positive in the B. aalborgi PCR. The positive samples came from 4 of 8 species of Old World monkeys (comprising a total of 14 individuals) (Table 1). None of the samples from other animal or bird species were positive in this PCR, and none of the samples from primates or other species were positive in the B. pilosicoli PCR.
Sequence analysis.
Results of the analysis of the sequences over 433 bp of the 16S rDNA B. aalborgi PCR product for the six positive reactions and comparisons with the corresponding sequences of type strains of B. aalborgi and B. pilosicoli are shown in Table 2. All detected products had from 96.7 to 97.8% sequence identity with B. aalborgi 513AT and only 94.2 to 94.7% identity with B. pilosicoli P43/6/78T. The sequences from the three macaques were identical and were the most closely related to that of the type strain from humans (97.8% identity). Products from the two vervet monkeys were 99.8% identical and had 98.5% sequence identity to the product from the baboon.
TABLE 2.
Percent similarities between the 16S rDNA PCR product sequences obtained from the six positive samples and B. aalborgi type strain 513AT and comparisons with equivalent position in B. pilosicoli type strain P43/6/78T
| Sample; source | % Similarity over 433 bp for:
|
|
|---|---|---|
| B. aalborgi 513AT | B. pilosicoli P43/6/78T | |
| Z13; Japanese macaque | 97.8 | 94.2 |
| Z18; Tonkean macaque 1 | 97.8 | 94.7 |
| Z23; Tonkean macaque 2 | 97.8 | 94.7 |
| Z28; Hamadryas baboon | 96.9 | 94.7 |
| Z12; vervet 1 | 96.9 | 94.7 |
| Z17; vervet 2 | 96.7 | 94.7 |
| B. aalborgi 513AT | 100 | 94.0 |
Isolation and characterization.
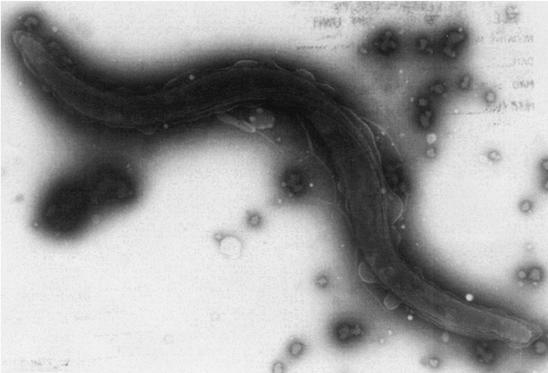
After 26 days of culture, followed by subculture of initial growth for 21 days, isolates of B. aalborgi were obtained from the two PCR-positive vervet monkeys (Z12 and Z17). By phase-contrast microscopy, spirochetes were also seen in the primary inoculum on the initial isolation plates inoculated with feces from the Japanese macaque (Z13) and one Tonkean macaque (Z23), but these could not be isolated. Growth from the vervet monkeys appeared as a thin haze on the plates, with moderate hemolysis around the growth. The sequence of 16S rDNA PCR product generated from these isolates was identical to those obtained directly from the feces of these animals. In API-ZYM, the two isolates had identical profiles, with the only activities being strong β-galactosidase activity and weak phosphohydrolase and alkaline phosphatase activities. In Table 3 these activities are contrasted with those reported for human Swedish isolate W1 and strain 513AT (9). The only consistent differences from the two human-derived strains were that the vervet isolates lacked the reported weak esterase-lipase and acid phosphatase activities. Under the electron microscope, both isolates had a morphology similar to that previously described for human isolates of B. aalborgi (7). The cells were sigmoidal with tapered ends, were 2 to 6 μm long and 0.2 μm wide, and had four periplasmic flagella inserted at each cell end (Fig. 1).
TABLE 3.
Biochemical reactivity of the vervet monkey isolates and of two strains of B. aalborgi from humans
| Strainc or isolates | Reactiona with enzymeb:
|
|||||
|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 11 | 12 | 14 | |
| Vervet isolates | 1 | 0 | 0 | 0 | 2 | 5 |
| B. aalborgi 513AT | 1 | 0 | 1 | 2 | 2 | 5 |
| B. aalborgi W1 | 0 | 1 | 1 | 2 | 2 | 5 |
Reactions in the API-ZYM system. Numerals indicate level of reaction: 0, negative reaction; 1, weak positive reaction; 5, strong positive reaction.
Enzyme numbers: 2, alkaline phosphatase; 3, esterase; 4, esterase-lipase; 11, acid phosphatase; 12, naphthol-AS-BI-phosphohydrolase; 14, β-galactosidase. The other 13 enzymes in the API-ZYM panel were negative for all strains, as were the indole reactions.
Results for strain 513AT and for Swedish strain W1 from humans are from reference 9.
FIG. 1.
Transmission electron microscope appearance of negatively stained B. aalborgi isolate Z12 from a vervet monkey.
DISCUSSION
The PCRs that were used for detection of intestinal spirochetes had some limitations. For example, the detection limits were established by using human feces, and these may be different from the limits in feces from nonhuman primates or other animal species. The thresholds for detection of both spirochete species were quite high, and the failure to detect B. pilosicoli DNA from feces collected at the zoo may have been due to the poor detection limit of the B. pilosicoli PCR compared to that of the B. aalborgi PCR. Nevertheless, the failure to detect B. pilosicoli by PCR or by isolation in primates and other species examined was consistent with the results of a previous survey of this zoological collection (16). In that study, feces from 204 animals, including primates, were subjected to selective culture for B. pilosicoli, but only a single isolate was obtained from a domestic pig.
Sequencing the 16S rDNA products from the B. aalborgi PCRs demonstrated that all the spirochetes detected in the primates were more closely related to B. aalborgi (96.7 to 97.8% identity) than to B. pilosicoli (94.2 to 94.7% identity). Previously, a comparison of partial 16S rDNA sequences (from a diagnostic PCR product) from the four available human isolates of B. aalborgi showed that these have 99.5% identity (8). Hence the spirochetes detected here apparently diverge somewhat from typical human isolates of B. aalborgi. It would not be appropriate to present a phylogenetic analysis of these nonhuman primate isolates based on the small amount of 16S rRNA gene sequence currently available. It is clear that the nonhuman primate isolates have in vitro growth characteristics, morphologies, and biochemical reactivities similar to those of the human isolates and are best considered B. aalborgi. Further work to clarify the exact relationships between the human and nonhuman primate isolates is required.
This is the second study that has used PCR to detect B. aalborgi in nonhuman primates, and it is the first to achieve this by using DNA extracted directly from feces. In the previous PCR study, B. aalborgi (and B. pilosicoli) DNA was detected in colonic biopsy specimens from colony-raised primates with histological evidence of IS (4). The identification of B. aalborgi in animals in a public zoological collection in the present study raises the question of the origin of these organisms and their potential for zoonotic spread. A survey of feces from a selection of other animals and birds in the collection failed to identify any other reservoirs of infection. The apparent sensitivity of detection of the B. aalborgi PCR was not particularly good, and the possibility that additional primates or other species could have been colonized at an undetectable level cannot be excluded. All the primates were captive bred, and in the preceding 17 years the only primate acquisitions were two Tonkean macaques, obtained from the Rotterdam zoo. While two Tonkean macaques were positive for B. aalborgi in this study, it was not recorded whether these were the same individuals that were introduced. There was no direct contact between the various primate species in the different enclosures, and the sequence variation among the PCR products indicated that there were several strains of the organism present in different primate species. Overall, these results indicate that B. aalborgi can persist among groups of individuals for prolonged periods (decades), suggesting that they are part of the autochthonous microflora in these animals. Nevertheless, as in humans, they may perhaps act as opportunistic pathogens under certain circumstances (19). Although none of the animals showed evidence of disease, the presence of histological IS and/or colitis was not investigated. Even if these strains of B. aalborgi have a commensal relationship with nonhuman primates, it is possible that they could be transmitted to humans and cause disease. This zoonotic potential requires further study.
The approximate 50% colonization rate in Old World monkeys found here was in contrast to the failure to detect B. aalborgi in prosimians (three species, six individuals) or platyrrhines (New World monkeys; six species, eight individuals). Interestingly, there have been no previous reports of intestinal spirochetes colonizing prosimians or New World monkeys. This could either be because too few animals have been examined or because they are not naturally colonized. In contrast, Old World monkeys commonly have been shown to have histological IS, with this being reported in rhesus monkeys (4, 21, 22), baboons (17), crab-eating monkeys (4), and vervets (2). These results could be interpreted as suggesting that B. aalborgi has coevolved with the catarrhines (the group including the Old World monkeys, gibbons, great apes, and humans), following an initial colonization after their separation (geographic and evolutionary) from the New World monkeys some 35 to 37 million years ago. Colonization of human beings with B. aalborgi has been recorded in many parts of the world (12), presumably reflecting later human migration from the Old World. Although in this study colonization was not detected in other members of the catarrhines, including five orangutans (family Pongidae) and two gibbons from different species (family Hylobatidae), these captive individuals may simply not have been exposed to the organism. It would be of considerable interest to look for the presence of B. aalborgi in other captive and wild members of the last two families, as well as in New World monkeys. The catarrhines are all omnivorous and monogastric, and the similarity of their gastrointestinal systems, perhaps including similar colonic epithelial receptors and intestinal microenvironments, might facilitate colonization by B. aalborgi.
In view of the present successful isolation of B. aalborgi from feces (the first time that this has been achieved for this spirochete species), it may be possible to obtain sufficient isolates for a much more comprehensive molecular analysis of the species, such that the comparative phylogeny of B. aalborgi and members of the catarrhines can be studied. The variation identified here in the 16S rDNA PCR products from the different primate species encourages this hypothesis. The availability of isolates will also allow future studies on the pathogenicity of the organisms, provided that a suitable animal model can be developed.
Acknowledgments
This work was supported by a Special Research Grant from Murdoch University.
Thanks are due to Leif Cocks and the keepers and staff at Perth Zoological Gardens for collection of the fecal specimens analyzed in the study.
REFERENCES
- 1.Barrett, S. P. 1990. Intestinal spirochaetes in a gulf Arab population. Epidemiol. Infect. 104:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowley, H. M., and R. R. Hill. 1985. “Intestinal spirochaetosis” of the vervet monkey. Onderstepoort J. Vet. Res. 52:47-50. [PubMed] [Google Scholar]
- 3.Douglas, J. G., and V. Crucioli. 1981. Spirochaetosis: a remediable cause of diarrhoea and rectal bleeding? Br. Med. J. 283:1362.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duhamel, G. E., R. O. Elder, N. Muniappa, M. R. Mathiesen, V. J. Wong, and R. P. Tarara. 1997. Colonic spirochetal infections in non-human primates that were associated with Brachyspira aalborgi, Serpulina pilosicoli, and unclassified flagellated bacteria. Clin. Infect. Dis. 25:S186-S188. [DOI] [PubMed] [Google Scholar]
- 5.Harland, W. A., and F. D Lee. 1967. Intestinal spirochaetosis. Br. Med. J. 3:718-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heine, R. G., P. B. Ward, A. S. J. Mikosza, V. Bennet-Wood, R. M. Robins-Browne, and D. J. Hampson. 2001. Brachyspira aalborgi infection in four Australian children. J. Gastroenterol. Hepatol. 16:872-875. [DOI] [PubMed] [Google Scholar]
- 7.Hovind-Hougen, K., A. Birch-Andersen, R. Henrik-Nielsen, M. Orholm, J. O. Pedersen, P. S. Teglbjærg, and E. H. Thaysen. 1982. Intestinal spirochaetosis: morphological characterization and cultivation of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J. Clin. Microbiol. 16:1127-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen, T. K., M. Boye, P. Ahrens, B. Korsager, P. S. Teglbjærg, C. F. Lindboe, and K. M/oller. 2001. Diagnostic examination of human intestinal spirochetosis by fluorescent in situ hybridization for Brachyspira aalborgi, Brachyspira pilosicoli, and other species of the genus Brachyspira (Serpulina). J. Clin. Microbiol. 39:4111-4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraaz, W., B. Pettersson, U. Thunberg, L. Engstrand, and C. Fellström. 2000. Brachyspira aalborgi infection diagnosed by culture and 16S ribosomal DNA sequencing using human colonic biopsy specimens. J. Clin. Microbiol. 38:3555-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraaz, W., U. Thunberg, B. Pettersson, and C. Fellstrom. 2001. Human intestinal spirochetosis diagnosed with colonoscopy and analysis of partial 16S rDNA sequences of involved spirochetes. Anim. Health. Res. Rev. 2:111-116. [PubMed] [Google Scholar]
- 11.Kunkle, R. A., D. L. Harris, and J. M. Kinyon. 1986. Autoclaved liquid medium for propagation of Treponema hyodysenteriae. J. Clin. Microbiol. 24:669-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikosza, A. S. J., and D. J. Hampson. 2001. Human intestinal spirochetosis: Brachyspira aalborgi and/or Brachyspira pilosicoli? Anim. Health Res. Rev. 2:83-91. [PubMed] [Google Scholar]
- 13.Mikosza, A. S. J., T. La, C. J. Brooke, C. F. Lindboe, P. B. Ward, R. G. Heine, J. G. Guccion, W. B. de Boer, and D. J. Hampson. 1999. PCR amplification from fixed tissue indicates frequent involvement of Brachyspira aalborgi in human intestinal spirochetosis. J. Clin. Microbiol. 37:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mikosza, A. S. J., T. La, W. B. de Boer, and D. J. Hampson. 2001. The comparative prevalence of Brachyspira (Serpulina) pilosicoli and Brachyspira aalborgi as the etiologic agents of histologically identified intestinal spirochetosis in Australia. J. Clin. Microbiol. 39:347-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikosza, A. S. J., T. La, K. R. Margawani, C. J. Brooke, and D. J. Hampson. 2001. PCR detection of Brachyspira aalborgi and Brachyspira pilosicoli in human faeces. FEMS Microbiol. Lett. 197:167-170. [DOI] [PubMed] [Google Scholar]
- 16.Oxberry, S. L., D. J. Trott, and D. J. Hampson. 1998. Serpulina pilosicoli, water birds and water: potential source of infection for humans and other animals. Epidemiol. Infect. 121:219-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson, G. R., P. N. Skelton-Stroud, and C. R. Orford. 1985. Observations by light microscopy and transmission electron microscopy on intestinal spirochaetosis in baboons (Papio spp.). J. Med. Microbiol. 19:359-366. [DOI] [PubMed] [Google Scholar]
- 18.Peghini, P. L., J. G. Guccion, and A. Sharma. 2000. Improvement of chronic diarrhea after treatment for intestinal spirochetosis. Dig. Dis. Sci. 45:1006-1010. [DOI] [PubMed] [Google Scholar]
- 19.Rodgers, F. G., C. Rodgers, A. P. Shelton, and C. J. Hawkey. 1986. Proposed pathogenic mechanism for the diarrhea associated with human intestinal spirochetes. Am. J. Clin. Pathol. 86:679-682. [DOI] [PubMed] [Google Scholar]
- 20.Stephens, C. P., and D. J. Hampson. 2002. Experimental infection of broiler breeder hens with the intestinal spirochaete Brachyspira (Serpulina) pilosicoli causes reduced egg production. Avian Pathol. 31:169-175. [DOI] [PubMed] [Google Scholar]
- 21.Takeuchi, A., and J. A. Zeller. 1972. Scanning electron microscopic observations on the surface of the normal and spirochaete-infested colonic mucosa of the rhesus monkey. J. Ultrastruct. Res. 40:313-324. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi, A., H. R. Jervis, H. Nakazawa, and D. M. Robinson. 1974. Spiral-shaped organisms on the surface colonic epithelium of the monkey and man. Am. J. Clin. Nutr. 27:1287-1296. [DOI] [PubMed] [Google Scholar]
- 23.Trivett-Moore, N. L., G. L. Gilbert, C. L. H. Law, D. J. Trott, and D. J. Hampson. 1998. Isolation of Serpulina pilosicoli from rectal biopsy specimens showing evidence of intestinal spirochetosis. J. Clin. Microbiol. 36:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trott, D. J., T. B. Stanton, N. S. Jensen, G. E. Duhamel, J. L. Johnson, and D. J. Hampson. 1996. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int. J. Syst. Bacteriol. 46:206-215. [DOI] [PubMed] [Google Scholar]
- 25.Trott, D. J., B. G. Combs, S. L. Oxberry, A. S. J. Mikosza, I. D. Robertson, M. Passey, J. Taime, R. Sehuko, and D. J. Hampson. 1997. The prevalence of Serpulina pilosicoli in humans and domestic animals in the eastern highlands of Papua New Guinea. Epidemiol. Infect. 119:369-379. [DOI] [PMC free article] [PubMed] [Google Scholar]