Abstract
A multiplex PCR assay was designed to amplify the Aeromonas hydrophila and A. veronii bv. sobria hemolysin and aerolysin genes. The assay was evaluated by using 121 clinical isolates and 7 reference strains of Aeromonas spp., and these were divided into five genotypes on the basis of the results of the multiplex PCR. The five genotypes were characterized as type 1 for those carrying the ahh1 gene only (36% of isolates), type 2 for those carrying the asa1 gene only (8.5% of isolates), type 3 for those carrying both the ahh1 and the asa1 genes (4% of isolates), type 4 for those carrying the ahh1 gene and the A. hydrophila aerA (aerolysin) gene (37.5% of isolates), and type 5 for those in which no hemolysin genes were detected (14% of isolates). The most common single hemolysin gene carried among all the Aeromonas isolates examined was ahh1, with 99 of 128 (77%) of isolates testing positive for this gene either alone or in combination with other hemolysin genes. Phenotypic expression of toxins was evaluated in a Vero cell culture cytotoxicity assay. These results indicated that there is a statistically significant correlation between the cytotoxin titers and the hemolysin genotype. Isolates belonging to genotype 4 (carrying both the ahh1 gene and the aerolysin and hemolysin aerA genes) expressed higher cytotoxin titers than isolates of the other genotypes (P < 0.001). These isolates were more cytotoxic in cell culture and may have greater clinical significance.
Aeromonas hydrophila and A. veronii bv. sobria (otherwise known as A. sobria) are gram-negative bacteria of the family Vibrionaceae that are often found in association with hemorrhagic septicemia in cold-blooded animals including fish, reptiles, and amphibians (4). However, these organisms have also been implicated as primary pathogens in cases of acute diarrheal disease in immunocompetent humans of all age groups (1, 12, 13, 34). Indeed, Poirier et al. (35) reported a case of pneumonia of nosocomial origin caused by A. hydrophila that complicated toxic coma, while both A. hydrophila and A. sobria have been isolated from humans with sepsis, peritonitis, urinary tract infections, and severe muscle degeneration (12). Hemolytic-uremic syndrome, a life-threatening condition normally associated with infections due to Escherichia coli O157:H7, has also been associated with A. hydrophila infection (6, 23, 37); and a cytotoxin with homology to Shiga toxin 1 has been identified in both A. hydrophila and A. caviae (18).
A number of virulence factors derived from A. hydrophila and A. sobria have been proposed in an effort to explain the pathogenesis of infections due to these organisms. Toxins with hemolytic, cytotoxic, and enterotoxic activities have been described in many Aeromonas spp. (12, 32); and while a number of toxins are produced by different species, single isolates often carry the genes encoding multiple toxins. Mutagenesis studies indicated that the hemolytic activity of A. hydrophila is related to both the hemolysin and the aerolysin genes. In addition, a gene encoding the cytolytic enterotoxin (Act) from A. hydrophila has been sequenced and shown to possess hemolytic, cytolytic, and enterotoxic activities (11). In an isogenic investigation in which a ligated ileal loop assay was used in a mouse model, it was concluded that the three toxins studied, the cytotoxic enterotoxin (act gene product), the heat-labile cytotonic enterotoxin (alt gene product), and the heat-stable cytotonic enterotoxin (ast gene product), all contributed to A. hydrophila-induced gastroenteritis. The Act enterotoxin resulted in the highest degree of fluid accumulation (38). Sequence analysis revealed that the act gene shared 89 and 93% DNA and amino acid homologies, respectively, with the A. hydrophila aerA (aerolysin) gene (11, 38). The biological functionality of this gene has been mapped to specific amino acid residues within the protein (14). Distinct hemolysins (hemolysin and aerolysin) have been detected in A. hydrophila by both immunologic and genetic methods (20, 22, 26). Beta-hemolytic isolates of Aeromonas were found to cause significantly more fluid accumulation in the ileal loops of experimentally infected rabbits than the alpha-hemolytic and nonhemolytic isolates, regardless of their species designation (40). Nacescu et al. (29) postulated a correlation between the pathogenic potential and the hemolytic activity of Aeromonas species. A majority of the A. hydrophila and A. sobria isolates were highly hemolytic, whereas only 11% of the A. caviae isolates were capable of lysing sheep erythrocytes (30). Mutagenesis of A. hydrophila isolates that carry two hemolysin genes (hlyA and aerA) revealed that the hemolytic activity of the isolate on horse blood agar was eliminated only following double mutations in the hlyA and aerA genes (44). A subsequent study suggested that all virulent A. hydrophila isolates carried both the hlyA and the aerA genes (19).
By using methods involving PCR and restriction fragment length polymorphism analysis, the virulence genes of Aeromonas spp. were grouped as aerolysins-hemolysins, cytolytic enterotoxins, or cytotonic enterotoxins (25). A PCR method for the amplification of the aerolysin gene was shown to detect β-hemolysin-positive A. hydrophila isolates from patients with diarrhea (36). Control isolates of hemolytic A. sobria, isolates of nonhemolytic Aeromonas spp., and A. caviae isolates did not produce an amplification product with these primers, even when these isolates were capable of producing cytotoxins or enterotoxins (5, 36). PCR techniques for the detection of two distinct hemolysin genes of A. sobria have also been developed (39). Alignment of known cytotoxic enterotoxin-aerolysin genes was used to detect sequences conserved throughout this family of genes, and PCR primers whose sequences are complementary to these sequences should detect all known cytotoxin genes (25). This PCR protocol in combination with PCR and restriction fragment length polymorphism analysis or PCR amplicon sequence analysis should partially permit estimation of the distribution of gene variants within different Aeromonas spp. and should assist in assessing the virulence of isolates carrying defined subsets of cytotoxin genes (25).
While a number of PCR methods reported previously appear to be effective for detection of a subset of cytotoxin-hemolysin genes, the multiplex PCR assay developed in the present study was designed to amplify both the A. hydrophila and the A. sobria hemolysin and aerolysin genes. The assay was evaluated with 121 clinical isolates and 7 reference strains of Aeromonas spp., and the relationship between the hemolysin genotypes and the virulence phenotypes of clinical isolates was also analyzed. In addition, a scheme for classifying Aeromonas spp. into five distinct genotypes is proposed.
MATERIALS AND METHODS
Bacterial isolates and culture media.
A total of 121 clinical isolates of Aeromonas spp. from sporadic cases of enteritis across Canada were evaluated and characterized in this study (Table 1). These isolates were submitted to the National Laboratory for Enteric Pathogens (NLEP) by Canadian provincial public health laboratories between 1986 and 1997 as part of the normal reference and enteric disease surveillance efforts in Canada. Reference strains ATCC 7966 (A. hydrophila), ATCC 9071 (A. sobria), ATCC 35622 (A. veronii bv. veronii), ATCC 15468 (A. caviae), ATCC 49568 (A. jandaei), and ATCC 43700 (A. schubertii) were also investigated. The hemolytic isolate of A. hydrophila, isolate CA11, was obtained from the Center for Food Safety and Applied Nutrition of the Food and Drug Administration, Cincinnati, Ohio, and was used as a positive control. All 128 isolates were grown on Mueller-Hinton agar (Oxoid Ltd., Nepean, Ontario, Canada) containing 5% defibrinated sheep blood erythrocytes and were identified by standardized biochemical protocols (2). Cultures were stored either on slants of Institut Pasteur (IP) maintenance medium in the dark or in brain heart infusion broth containing 15% glycerol at −80°C. Positive hemolytic activity was defined as a zone of hemolysis around colonies on the blood agar plate after 18 to 24 h of incubation at 37°C.
TABLE 1.
Correlation of hemolytic phenotype with hemolysin gene content as determined by multiplex PCRa
| Organismb | Phenotype | No. of isolates with each hemolysin genotype
|
No. of isolates | ||||
|---|---|---|---|---|---|---|---|
| ahh1 (genotype 1) | asa1 (genotype 2) | ahh1 + asa1 (genotype 3) | ahh1 + A. hydrophila aerA (genotype 4) | No hemolysin gene (genotype 5) | |||
| A. hydrophila (87) | Hemolytic | 35c | 1 | 46 | 82 | ||
| Nonhemolytic | 3d | 2d | 5 | ||||
| A. veronii bv. sobria (21) | Hemolytic | 9 | 4 | 1d | 14 | ||
| Nonhemolytic | 2d | 1d | 4 | 7 | |||
| A. caviae (15) | Hemolytic | 1d | 1 | ||||
| Nonhemolytic | 3d | 11 | 14 | ||||
| A. veronii bv. veronii (1) | Hemolytic | 1 | 1 | ||||
| A. trota (2) | Hemolytic | 2 | 2 | ||||
| A. jandaei (1) | Hemolytic | 1 | 1 | ||||
| A. schubertii (1) | Nonhemolytic | 1d | 1 | ||||
| Total | 46 | 11 | 5 | 48 | 18 | 128 | |
All isolates produced 16S rRNA amplicons.
Numbers in parentheses are the total number of isolates examined within each species.
Includes positive control strain CA11.
DNA hybridization was performed with all of these isolates.
Nucleic acid isolation.
Nucleic acids were isolated by lysis of the bacteria in a solution containing 0.5% sodium dodecyl sulfate and 5 mg of lysozyme per ml, followed by extraction with phenol-chloroform (21). Nucleic acid samples were precipitated with ethanol and dissolved in TE buffer (10 mM Tris chloride, 1 mM EDTA [pH 8.0]). The nucleic acid content was quantified by determining the optical density (OD) at 260 nm (OD260) and was adjusted to give a final concentration of 200 μg/ml in TE buffer. Template DNA for PCR was prepared by further dilution in distilled H2O to a concentration of 2 μg/ml.
Oligonucleotide primers and PCR conditions.
The AHH1 primer set was designed to amplify a 130-bp fragment of A. hydrophila extracellular hemolysin gene ahh1 (20). The AH-aerA primer set amplified a 309-bp fragment of the A. hydrophila aerolysin gene aerA (GenBank accession no. M16495); the nucleotide sequence of this gene had 89% similarity to that of the hemolysin gene of AHH5 (GenBank accession no. X65045). Similarly, the ASA1 primer set was designed to amplify a 249-bp fragment of A. sobria hemolysin gene asa1 (GenBank accession no. X65046). Primers based on the A. hydrophila ATCC 7966 16S rRNA sequence (GenBank accession no. X74677) were designed to amplify a portion of the 16S rRNA gene as an internal control (Table 2). DNA samples (5 ng per reaction mixture) were amplified in a 25-μl reaction mixture consisting of 50 mM potassium chloride; 10 mM Tris chloride (pH 8.3); 1.25 mM magnesium chloride; 200 μM (each) dATP, dCTP, dGTP, and dTTP; 2.0 μM (each) AHH1 primers; 1.5 μM (each AH-aerA and ASA1 primer) 0.05 μM (each) A16S primers (Table 2); and 1.25 U of FastStart Taq DNA polymerase (Roche Diagnostic GmbH, Mannheim, Germany). Amplifications were performed with a model 2400 DNA thermal cycler (Applied Biosystems, Foster City, Calif.). Parameters for the amplification included an initial denaturation at 95°C for 5 min, followed by 50 cycles of denaturation at 95°C for 0.5 min, annealing of the primers at 59°C for 0.5 min, and primer extension at 72°C for 0.5 min. A final extension at 72°C for 7 min was used. Ten microliters of the reaction mixture was then analyzed by submarine gel electrophoresis in 1.5% agarose at 5 V/cm, and the reaction products were visualized with UV light after staining with ethidium bromide. The identities of the amplicons were confirmed by comparison of the amplicon sizes with the predicted sizes, as indicated in Table 2, and by digestion with restriction endonucleases. While the internal control (the amplification product obtained by PCR with the A16S primer set) gave 113- and 243-bp fragments after digestion with NheI, the product obtained by PCR with the AH-aerA primer set yielded 141- and 168-bp fragments after SacI digestion. The amplification product obtained by PCR with the AHH1 primer set showed 55- and 75-bp fragments following HinfI digestion, and the amplicon obtained by PCR with the ASA1 primer set produced 116- and 133-bp fragments after digestion with PuvII.
TABLE 2.
Primer pairs used for PCR amplification
| Primer pair | Sequence (5′ to 3′) | Target gene | Location within gene | Size of PCR amplicon (bp) | Reference or GenBank accession no. |
|---|---|---|---|---|---|
| AHH1F | GCCGAGCGCCCAGAAGGTGAGTT | ahh1a | 961-983 | 130 | 20 |
| AHH1R | GAGCGGCTGGATGCGGTTGT | 1090-1071 | |||
| AH-aerAF | CAAGAACAAGTTCAAGTGGCCA | A. hydrophila aerA | 1323-1344 | 309 | M16495 |
| AH-aerAR | ACGAAGGTGTGGTTCCAGT | 1631-1613 | |||
| ASA1F | TAAAGGGAAATAATGACGGCG | asa1 | 863-883 | 249 | X65046 |
| ASA1R | GGCTGTAGGTATCGGTTTTCG | 1111-1091 | |||
| A16SF | GGGAGTGCCTTCGGGAATCAGA | 16S rRNA | 1020-1041 | 356 | X74677 |
| A16SR | TCACCGCAACATTCTGATTTG | 1375-1355 |
From strain ATCC 7966.
Cell culture cytotoxicity assay.
Vero cell cultures were grown for 48 h to give confluent monolayers of cells in 96-well cell culture plates. The cell culture plates contained minimum essential medium with heat-inactivated 10% fetal bovine serum (Sigma-Aldrich, St. Louis, Mo.), 40 U of penicillin-streptomycin (Invitrogen, Burlington, Ontario, Canada) per ml, 40 μg of gentamicin (Invitrogen) per ml, 0.1 mM nonessential amino acids (Invitrogen), and 2 mM l-glutamine (Invitrogen). The bacterial cultures were grown in 5 ml of brain heart infusion broth at 37°C on a rotating drum for 24 h, and the organisms were removed by centrifugation. Supernatants were filtered through a 0.22-μm-pore size syringe filter (Millipore, Nepean, Ontario, Canada), and 20-μl aliquots of each filtrate were diluted twofold in the cell culture medium and added in duplicate to the Vero cell monolayers for the toxin assay. After incubation at 37°C in a humid atmosphere containing 5% CO2, the cytotoxic activity of the culture supernatants was assessed microscopically after 24 and 48 h of incubation. At the end of the 48-h incubation period, all monolayers were stained with 2% crystal violet (Sigma-Aldrich) diluted 1/80 with buffered formalin (Fisher, Nepean, Ontario, Canada). After 30 min the detached and dead cells in all of the wells were removed by rinsing the wells with distilled H2O. The stained live cells were solubilized in situ in buffer containing 0.3% acetic acid and 0.5% sodium dodecyl sulfate. The OD630s of the plates were read in a Dynatech MRX plate reader (Dynex Technologies, Chantilly, Va.) by using Revelation software, and the cytotoxicities were determined by comparing the OD values for the test wells with those for the control wells to which only phosphate-buffered saline had been added. The percent cytotoxicity was determined as follows: 100 − (TOD/NCOD) × 100, where TOD is the OD630 of a test well, and NCOD is the average OD630 for the negative controls. The inverse of the dilution that resulted in what was equal or closest to 50% cytotoxicity was determined to be the endpoint titer. The data were subjected to statistical evaluation by the t test.
DNA hybridization.
On the basis of the data obtained from the PCR and the hemolysin and cytotoxin tests, a total of 14 isolates were further investigated for the presence of the ahh1, A. hydrophila aerA, and asa1 genes by DNA hybridization with an enhanced chemiluminescence direct nucleic acid labeling and detection system according to the instructions of the manufacturer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) under high-stringency conditions. These samples included 11 isolates of genotypes 1, 2, and 4 that were phenotypically hemolysin negative and 3 isolates of genotype 5 that were positive for cytotoxicity (Table 1; see Table 3). The probes were prepared from the amplification products obtained by PCR with the AHH1, AH-aerA, and ASA1 primer sets, respectively.
TABLE 3.
Evaluation of degree of cytotoxicity expressed in Vero cells compared with genotype results
| Genotype | Hemolysin-positive isolates
|
Hemolysin-negative isolates
|
||||
|---|---|---|---|---|---|---|
| No. of isolates | CTa range | Mean ± SEb CT | No. of isolates | CT range | Mean ± SE CT | |
| 1 (ahh1) | 38 | 2-256 | 32.37 ± 8.97 | 8 | 0-16 | 3.00 ± 2.10 |
| 2 (asa1) | 10 | 2-16 | 9.20 ± 1.96 | 1 | 0 | 0 |
| 3 (ahh1 and asa1) | 5 | 4-32 | 12.0 ± 5.06 | 0 | 0 | 0 |
| 4 (ahh1 and A. hydrophila aerA) | 46 | 0-256 | 79.48 ± 13.38c | 2 | 0-16 | 8.00 ± 8.0 |
| 5 (no hemolysin genes detected) | 4d | 0-16 | 5.0 ± 3.79 | 14e | 0-2 | 0.14 ± 0.14 |
CT in Vero cells. Values are the titers obtained in cell culture assays.
SE, standard error of the mean.
P < 0.001 compared to the mean CTs for the other genotypes.
None of the four hemolysin-positive isolates produced a hemolysin-aerolysin amplicon in the multiplex PCR assay; one isolate each of A. caviae and A. sobria was cytotoxic.
One isolate (A. schubertii) showed a low level of cytotoxicity.
RESULTS
The sizes of the amplification products obtained by the multiplex PCR were identical to those predicted from the design of the primers (Fig. 1; Table 2). Amplicons from the control isolates were subjected to further characterization by digestion with restriction endonucleases with known cleavage sites within the amplified sequence. Restriction fragments of the anticipated sizes were obtained in each case.
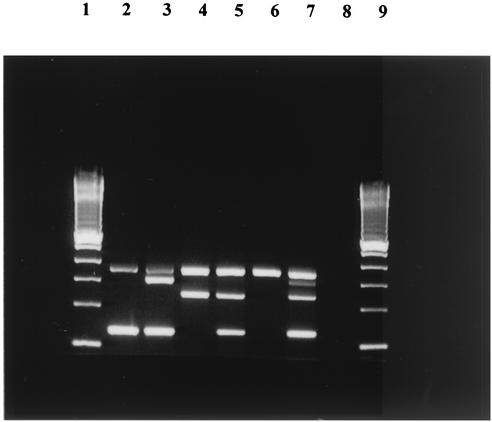
FIG. 1.
Detection and identification of Aeromonas hemolysin and aerolysin genes by amplification of fragments in the multiplex PCR assay. Lanes 1 and 9, 100-bp ladder (Invitrogen); lane 2, A. hydrophila genotype 1 showing the ahh1 and 16S rRNA genes (130- and 356-bp fragments, respectively); lane 3, A. hydrophila genotype 4 showing the ahh1, aerA, and 16S rRNA genes (130-, 309-, and 356-bp fragments, respectively); lane 4, A. sobria gentotype 2 showing the asa1 and 16S rRNA genes (249- and 356-bp fragments, respectively); lane 5, A. sobria genotype 3 showing the ahh1, asa1, and 16S rRNA genes (130-, 249-, and 356-bp fragments, respectively); lane 6, 16S rRNA internal control showing a 356-bp fragment only; lane 7, PCR positive control with a DNA mixture showing PCR amplification fragments for all four genes, ahh1, aerA, asa1, and 16S rRNA (130-, 309-, 249-, and 356-bp fragments, respectively); lane 8, PCR negative control.
Strains of each of the Aeromonas phenotypes carried the hemolysin genes at different frequencies (Table 1). Of the 128 Aeromonas spp. isolates tested, 99 (77%) were ahh1 positive; these included 87 of 87 (100%) of the A. hydrophila isolates, 6 of 21 (29%) of the A. sobria isolates, 3 of 15 (20%) of the A. caviae isolates, 2 of 2 (100%) of the A. trota isolates, and the single A. jandaei isolate examined. By PCR with the AH-aerA primer set, amplicons were produced only by A. hydrophila isolates (48 of 87 [55%]). Sixteen of the 128 Aeromonas isolates (12.5%) were asa1 positive, including 14 of 21 (67%) of the A. sobria isolates, the single A. veronii bv. veronii isolate examined, and 1 of 87 (1%) of the A. hydrophila isolates (NLEP A1607). Hemolysin genes were not detected in 18 of 128 (14%) of the isolates overall. Most A. caviae isolates (12 of 15 [80%]) produced no hemolysin gene amplicons when they were subjected to the multiplex PCR, although one isolate did produce detectable hemolysis on agar containing sheep erythrocytes (Table 1). In addition, hemolysin genes were not detected in 5 of 21 (24%) of the A. sobria isolates or in the single isolate of A. schubertii tested. All Aeromonas isolates were PCR positive for the 16S rRNA internal positive control.
Five genotypes were defined on the basis of the results of the multiplex PCR (Table 1). The reproducibility of the hemolysis data for all isolates was demonstrated in triplicate on agar plate assays containing sheep blood. In addition, the findings for 29 gene-positive isolates were further confirmed by using horse blood agar. Of these 29 isolates, 22 were positive for hemolysis on both sheep blood agar and horse blood agar, while the remaining 7 isolates (2 A. hydrophila, 2 A. caviae, and 3 A. sobria isolates) were negative on both types of blood agar (data not shown). Of the 27 Aeromonas isolates that expressed no hemolytic activity, 8 isolates (3 A. hydrophila, 2 A. sobria, and 3 A. caviae isolates) were genotype 1, 1 isolate (A. sobria) was genotype 2, 2 isolates (both of which were A. hydrophila) were genotype 4, and 16 were negative for hemolysins in the multiplex PCR assay. All 128 isolates were further analyzed for cytotoxins by the Vero cell culture assay (Table 3). A higher proportion of high cytotoxicity titers (CTs) were obtained for isolates of genotype 4 than for isolates of the other genotypes tested, suggesting a possible relationship between the hemolysin genes present and the degree of cytotoxicity produced by each genotype. However, the ranges of CTs were very similar for genotype 1 and 4 isolates, as were the mean titers for each population of bacteria. Cytotoxicity was detected in three isolates that were negative for the hemolysins targeted by the multiplex PCR assay. Two of these isolates (A. caviae A1833 and A. sobria A1432) were positive for both hemolytic and cytotoxic activities, and the third one (A. schubertii) was weakly cytotoxic only. DNA hybridization showed a stronger signal for A. sobria A1432 (with the ASA1 probe) and for both A. caviae A1833 and A. schubertii (with the AH-aerA probe). Among the 11 nonhemolytic isolates, which included 5 A. hydrophila isolates, 3 nonhemolytic A. sobria isolates, and 3 nonhemolytic A. caviae isolates, all of which were positive for one or more hemolysin genes by PCR, consistent positive DNA hybridization results were obtained with the corresponding PCR probe. In addition, two genotype 1 A. hydrophila isolates also showed a stronger signal with the AH-aerA probe; these were isolates A1915 (CT = 16) and A1556 (CT = 0). Hemolysis was always found in genotype 3 isolates, which carried both the ahh1 and the asa1 genes. However, 8 of 46 (17%) genotype 1 isolates, 1 of 11 (9%) genotype 2 isolates, and 2 of 48 (4%) genotype 4 isolates were nonhemolytic, while 2 of 18 (11%) isolates that did not have hemolysin or aerolysin genes detectable by PCR were phenotypically hemolytic (Table 1). There was a statistically significant correlation between the Vero cell CT and the hemolysin genotype (P < 0.001). This was demonstrated by a tendency for isolates carrying both the ahh1 and the A. hydrophila aerA genes to have higher CTs than isolates with other genotypes or no hemolysin genes (Table 3). Indeed, of the 110 isolates with at least one hemolysin gene, 99 of 110 (90%) were cytotoxic in Vero cells and showed hemolytic activity (Table 3).
DISCUSSION
Hemolysin genes did not appear to be distributed randomly among the different Aeromonas species. All the A. hydrophila isolates had the ahh1 toxin gene, while some also carried the aerolysin gene. However, none of the A. hydrophila isolates were positive only for the A. hydrophila aerA gene. A PCR and Southern hybridization survey indicated that all virulent A. hydrophila isolates were both hlyA (AHH1) and aerA (aerolysin) positive. Of the 61 A. hydrophila isolates investigated, only 2 carried the aerolysin gene alone, and both of these were isolated from healthy fish (19). Although a majority of the A. veronii bv. sobria isolates also carried hemolysin genes, the most prevalent gene detected by the multiplex PCR was asa1. In contrast, most A. caviae isolates tested did not produce an amplicon in the multiplex PCR developed in this study, and the few that did carried only the ahh1 gene. Indeed, the most common hemolysin gene carried by Aeromonas isolates overall was ahh1 (99 of 128 [77%] isolates). Johnson and Lior (24) previously reported that 80.7% of A. hydrophila isolates from stools were cytotoxic. Of 445 Aeromonas isolates screened for the presence of aerolysin by PCR in a second study, 79% carried the aerolysin gene and 83% showed cytotoxicity in Vero cell culture assays (33). The investigators found a high frequency of aerolysin gene carriage among all Aeromonas spp. tested, including A. caviae. Results from other toxin surveys (16, 25) supported the distribution of cytotoxic enterotoxins (hemolysins and aerolysins) noted in this study.
Genotype 2 was characteristic of most hemolytic A. sobria isolates and was also found in an atypical A. hydrophila isolate, NLEP A1607. This isolate displayed weak hemolytic activity but was positive for both the asa1 and the ahh1 genes. On the basis of the results of biochemical assays, this isolate was classified as A. hydrophila (esculin test positive, salicin test negative); however, sequence analysis of NLEP A1607 (GenBank accession no. AF410466) confirmed that it had 96% identity with the A. sobria hemolysin gene (asa1; GenBank accession no. X65046). Furthermore, the partial 16S rRNA sequence of A. hydrophila isolate NLEP A1607 (GenBank accession no. AF410780) shared 99% similarity with the 16S rRNA sequence of A. sobria isolate ATCC 9071. It is possible that isolate NLEP A1607 represents a novel variant or a new subspecies of A. hydrophila.
Immunologic studies have demonstrated that the hemolysins produced by A. hydrophila can be divided into two major groups (26). A comparison of the nucleotide sequences of the extracellular hemolysin and aerolysin genes detected only 23.8% nucleotide sequence homology. It is believed that the evolutionary origins of these two genes may differ. Many isolates characterized in this study carried more than one hemolysin: ahh1 and either A. hydrophila aerA or asa1. Previous investigations have also found A. hydrophila isolates that produced more than one hemolytic toxin (19, 44). Inactivation of both hemolytic toxin genes in an isolate of A. hydrophila was necessary to completely attenuate the virulence of the isolate in a mouse model of infection, suggesting that assessment of the toxin gene contents of clinical isolates may be critical for prediction of the virulence potentials of the organisms.
It was difficult to predict the hemolytic or cytotoxic phenotype that a isolate would express solely on the basis of the hemolysin genotype alone. Among the various Aeromonas spp. tested, eight isolates carrying the ahh1 gene, one isolate carrying the asa1 gene, and two isolates carrying both the ahh1 and the A. hydrophila aerA genes were nonhemolytic on sheep blood agar. Both of the last two isolates were A. hydrophila. Although sheep erythrocytes appear to be less sensitive than erythrocytes from other mammals (7, 26), the standardization of the hemolysis assay for all isolates used in the present study suggests that the differences among isolates have a biological basis and are not likely solely due to the sensitivity of the assay. It is possible that the nonhemolytic isolates carried hemolysin genes either that could not be expressed or that had mutations affecting domains responsible for the hemolytic phenotype. Hemolysin production is significantly correlated (P < 0.05) with enterotoxin production, as measured in suckling mouse assays (9, 40). Both phenotypes may be subject to regulation at the phenotypic level, in that all Aeromonas isolates that failed to produce enterotoxin in the initial test (44.2%) showed enterotoxic effects after one to three consecutive passages through rabbit ileal loops, suggesting expression of previously repressed toxin genes (40). Little is known about the regulatory mechanisms responsible for these effects and whether they operate in all Aeromonas isolates. The hemolytic activities expressed by A. caviae and A. veronii bv. sobria isolates lacking detectable amplicons in the multiplex PCR most likely resulted from the expression of alternate cytotoxin and hemolysin gene variants (43). Recently, it has been confirmed that lecithinase (phospholipase C [PLC]) cloned from A. hydrophila was clearly cytotoxic in Vero cells but showed no hemolytic activity (28).
The high degree of correlation between the Vero cell CTs and the hemolysin genotype was striking. The range of cytotoxicity values for each genotype (Table 3) suggested that the cytotoxicity in Aeromonas spp. is multifactorial and may involve the products of a number of different genes acting either alone or in concert. Those described in the literature include α and β hemolysins (40); aerolysin (8, 16); Act, Alt, and Ast (38); PLC (28); cholera toxin-like factor (42); and proteases and RNase (15). The Vero cell CTs for isolates carrying both the ahh1 and the asa1 genes were quite low, while those for those isolates positive for the ahh1 gene together with the A. hydrophila aerA gene were high. One intriguing possibility is that interactions between different hemolysin and aerolysin genes affect the expression of these genes in A. hydrophila isolates. Alternatively, mutagenesis studies have suggested that both the hemolysin and the aerolysin toxins act by inducing pore formation in the membranes of affected cells and that the effect of these together is likely to be synergistic (19, 44). In addition, gene variation, plasmid-mediated gene regulation, quorum sensing-dependent regulation, and toxin protein production as well as secretion might all affect cytotoxin expression levels (8, 10, 15, 17, 41). The present data were in agreement with previous observations demonstrating a very close relationship between hemolytic activity and cytotoxicity for all Aeromonas spp. tested (33, 43). Similarly, purified hemolysin from A. hydrophila was found to cause fluid secretion in infant mouse assays and cytotoxicity in Vero cell assays (3). In the present study, however, Vero cell cytotoxicity did not correlate absolutely with the hemolysin gene content, as determined by PCR. One isolate each of A. caviae, A. sobria, and A. schubertii did not amplify any of the hemolysin genes tested in the multiplex PCR but showed weak hemolytic or cytotoxic phenotypic activity, suggesting that other virulence traits exist. Indeed, factors such as the export of aerolysin and activation of the protoxin may also affect the ability to detect cytotoxicity (8). Furthermore, DNA hybridization data indicated that all three isolates possessed both hemolysin and aerolysin genes; hence, sequence variations in the target DNA may offer one explanation for a lack of PCR amplification products and for the low level of cytotoxicity detected.
Although the production of hemolytic cytotoxins has been regarded as strong evidence of pathogenic potential in Aeromonas spp., nonhemolytic aeromonads have been implicated as human pathogens (31). Cytotonic enterotoxins also contribute to the pathogenesis caused by Aeromonas spp. (38). The presence of these toxins may also need to be assessed, especially in A. caviae isolates with no detectable cytotoxins. It has been shown that many enzymes such as lipase, PLC, protease, and RNase are putative virulence factors for Aeromonas spp. (15, 28) and that cytotoxicity may be affected by a number of environmental factors or growth conditions. Indeed, Granum et al. (16) confirmed that the optimal growth temperature for toxin production by A. caviae was 30°C rather than 37°C, while Mateos et al. (27) reported that Aeromonas spp. isolated from humans showed higher levels of toxin production at 37°C. These findings suggest that as yet undefined factors exist. In the present study, one A. hydrophila isolate, one A. sobria isolate, and eight A. caviae isolates showed no cytotoxicity at either 30 or 37°C, while one A. hydrophila isolate expressed relatively low levels of cytotoxic activity at 30°C compared to that at 37°C and three other isolates showed higher levels of activity at 30°C compared to that at 37°C (data not shown). A recent study on serine protease activity in A. hydrophila confirmed that protease production at 22 and 30°C was under quorum-sensing control but was inhibited at 37°C in a quorum sensing-independent fashion (41). Positive DNA hybridization results for the three genotype 5 isolates (one isolate each of A. sobria, A. caviae, and A. schubertii [Table 1]) suggested that PCR-based detection may be more specific than DNA hybridization, although it may have a lower sensitivity. One genotype 1 isolate (isolate A1556) that was phenotypically both hemolysin and cytotoxin negative tested positive with the AH-aerA probe, further indicating that other factors affect gene expression (8, 10, 15, 17, 41).
Together, these observations suggest that an evaluation of Aeromonas virulence requires the assessment of virulence phenotypes and complete virulence gene sets. Phenotypic methods may not detect the presence of toxins. A. caviae isolates that did not produce detectable cytotoxic or hemolytic activity were found to regain the ability to express toxins after animal passage, only to lose it again upon subsequent subculture (40). Screening for specific cytotoxin and hemolysin genes appears to be the most effective way of detecting and characterizing Aeromonas virulence factors. The multiplex PCR assay described in this study should prove to be a useful method for the identification of hemolysin and aerolysin genotypes when screening for the major species-specific isolates of Aeromonas species.
Acknowledgments
We thank Erin Becker, Roman Benes, and Lorelee Tschetter for valuable technical support. We gratefully acknowledge the provincial laboratories of public health across Canada as well as our national and international collaborators who submitted isolates of Aeromonas to NLEP for characterization.
REFERENCES
- 1.Aggar, W. A., J. D. McCormick, and M. J. Gurwith. 1985. Clinical and microbiological features of Aeromonas hydrophila-associated diarrhea. J. Clin. Microbiol. 21:909-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altwegg, M. 1999. Aeromonas and Plesiomonas, p. 507-516. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical of microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 3.Asao, T., Y. Kinoshita, S. Kozaki, T. Uemura, and G. Sakaguchi. 1984. Purification and some properties of Aeromonas hydrophila hemolysin. Infect. Immun. 46:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, B., and D. A. Austin. 1987. Bacterial fish pathogens: disease in farmed and wild fish. Halsted Press, New York, N.Y.
- 5.Baloda, S. B., K. Krovacek, L. Eriksson, T. Linne, and L. Mansson. 1995. Detection of aerolysin gene in Aeromonas strains isolated from drinking water, fish and foods by the polymerase chain reaction. Comp. Immunol. Microbiol. Infect. Dis. 18:17-26. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanovic, R., M. Cobeljic, M. Markovic, V. Nikolic, M. Ognjanovic, L. Sarjanovic, and D. Makic. 1991. Haemolytic-uraemic syndrome associated with Aeromonas hydrophila enterocolitis. Pediatr. Nephrol. 5:293-295. [DOI] [PubMed] [Google Scholar]
- 7.Brenden, R., and J. M. Janda. 1987. Detection, quantitation and stability of the beta hemolysin of Aeromonas spp. J. Med. Microbiol. 24:247-251. [DOI] [PubMed] [Google Scholar]
- 8.Buckley, J. T. 1991. Secretion and mechanism of action of the hole-forming toxin aerolysin. Experientia 47:418-419. [PubMed] [Google Scholar]
- 9.Burke, V., J. Robinson, J. Beaman, M. Gracey, M. Lesmana, R. Rockhill, P. Echeverria, and J. M. Janda. 1983. Correlation of enterotoxicity with biotype in Aeromonas spp. J. Clin. Microbiol. 18:1196-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopra, A. K., C. W. Houston, and A. Kurosky. 1991. Genetic variation in related cytolytic toxins produced by species of Aeromonas. FEMS Microbiol. Lett. 62:231-237. [DOI] [PubMed] [Google Scholar]
- 11.Chopra, A. K., C. W. Houston, J. W. Peterson, and G. F. Jin. 1993. Cloning, expression, and sequence analysis of a cytolytic enterotoxin gene from Aeromonas hydrophila. Can. J. Microbiol. 39:513-523. [DOI] [PubMed] [Google Scholar]
- 12.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 1:1129-1137. [DOI] [PubMed] [Google Scholar]
- 13.Cumberbatch, N., M. J. Gurwith, C. Langston, R. B. Sack, and J. L. Brunton. 1993. Cytotoxic enterotoxin produced by Aeromonas hydrophila: relationship of toxigenic isolates to diarrheal disease. Infect. Immun. 23:267-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, M. R., X. J. Xu, C. W. Houston, J. W. Peterson, and A. K. Chopra. 1995. Amino-acid residues involved in biological functions of the cytolytic enterotoxin from Aeromonas hydrophila. Gene 156:79-83. [DOI] [PubMed] [Google Scholar]
- 15.Gosling, P. J. 1996. Pathogenic mechanisms, p. 245-265. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley & Sons Ltd., Chichester, England.
- 16.Granum, P. E., K. O'Sullivan, J. M. Tomás, and Ø. Ørmen. 1998. Possible virulence factors of Aeromonas spp. from food and water. FEMS Immunol. Med. Microbiol. 21:131-137. [DOI] [PubMed] [Google Scholar]
- 17.Hanes, D. E., and D. K. Chandler. 1993. The role of a 40-megadalton plasmid in the adherence and hemolytic properties of Aeromonas hydrophila. Microb. Pathog. 15:313-317. [DOI] [PubMed] [Google Scholar]
- 18.Haque, Q. M., A. Sugiyama, Y. Iwade, Y. Midorikawa, and T. Yamauchi. 1996. Diarrheal and environmental isolates of Aeromonas spp. produce a toxin similar to Shiga-like toxin 1. Curr. Microbiol. 32:239-245. [DOI] [PubMed] [Google Scholar]
- 19.Heuzenroeder, M. W., C. Y. Wong, and R. L. Flower. 1999. Distribution of two hemolytic toxin genes in clinical and environmental isolates of Aeromonas spp.: correlation with virulence in a suckling mouse model. FEMS Microbiol. Lett. 174:131-136. [DOI] [PubMed] [Google Scholar]
- 20.Hirono, I., and T. Aoki. 1991. Nucleotide sequence and expression of an extracellular hemolysin gene of Aeromonas hydrophila. Microb. Pathog. 11:189-197. [DOI] [PubMed] [Google Scholar]
- 21.Hirono, I., T. Aoki, T. Asao, and S. Kozaki. 1992. Nucleotide sequences and characterization of hemolysin genes from Aeromonas hydrophila and Aeromonas sobria. Microb. Pathog. 13:433-446. [DOI] [PubMed] [Google Scholar]
- 22.Howard, S. P., W. J. Garland, M. J. Gree, and J. T. Buckley. 1987. Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J. Bacteriol. 169:2869-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda, J. M. 1991. Recent advances in the study of the taxonomy, pathogenicity, and infectious syndromes associated with the genus Aeromonas. Clin. Microbiol. Rev. 4:397-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, W. M., and H. Lior. 1981. Cytotoxicity and suckling mouse reactivity of Aeromonas hydrophila isolated from human sources. Can. J. Microbiol. 27:1019-1027. [DOI] [PubMed] [Google Scholar]
- 25.Kingombe, C. I., G. Huys, M. Tonolla, M. J. Albert, J. Swings, R. Peduzzi, and T. Jemmi. 1999. PCR detection, characterization, and distribution of virulence genes in Aeromonas spp. Appl. Environ. Microbiol. 65:5293-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozaki, S., A. Tsutomu, K. Yoichi, and G. Sakaguchi. 1989. Characterization of Aeromonas sobria hemolysin by use of monoclonal antibodies against Aeromonas hydrophila hemolysins. J. Clin. Microbiol. 27:1782-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos, D., J. Anguita, G. Naharro, and C. J. Paniagua. 1993. Influence of growth temperature on the production of extracellular virulence factors and pathogenicity of environmental and human strains of Aeromonas hydrophila. Appl. Bacteriol. 74:111-118. [DOI] [PubMed] [Google Scholar]
- 28.Merino, S., A. Aguilar, M. M. Nogueras, M. Regue, S. Swift, and J. M. Tomas. 1999. Cloning, sequencing, and role in virulence of two phospholipases (A1 and C) from mesophilic Aeromonas sp. serogroup O:34. Infect. Immun. 67:4008-4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nacescu, N., A. Israil, C. Cedru, and D. Caplan. 1992. Hemolytic properties of some Aeromonas strains. Roum. Arch. Microbiol. Immunol. 51:147-156. [PubMed] [Google Scholar]
- 30.Nakano, H., T. Kameyama, K. Venkateswaran, H. Kawakami, and H. Hashimoto. 1990. Distribution and characterization of hemolytic, and enteropathogenic motile Aeromonas in aquatic environment. Microbiol. Immunol. 34:447-458. [DOI] [PubMed] [Google Scholar]
- 31.Namdari, H., and E. J. Bottone. 1990. Microbiologic evidence supporting the role of Aeromonas caviae as a pediatric enteric pathogen. J. Clin. Microbiol. 28:837-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Namdari, H., and E. J. Bottone. 1990. Cytotoxin and enterotoxin production as factors delineating enteropathogenicity of Aeromonas caviae. J. Clin. Microbiol. 28:1796-1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ørmen, Ø., and Ø. Østensvik. 2001. The occurrence of aerolysin-positive Aeromonas spp. and their cytotoxicity in Norwegian water sources. J. Appl. Microbiol. 90:797-802. [DOI] [PubMed] [Google Scholar]
- 34.Pitarangsi, C., P. Echeverria, R. Whitmore, C. Tirapat, S. Formal, G. J. Dammin, and M. Tingtalapong. 1982. Enteropathogenicity of Aeromonas hydrophila and Plesiomonas shigelloides: prevalence among individuals with and without diarrhea in Thailand. Infect. Immun. 35:666-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poirier, T., E. Laurens, J. Y. Viaud, B. Poujol, and G. Lorre. 1993. Nosocomial Aeromonas hydrophila pneumonia complicating toxic coma. Ann. Fr. Anesth. Reanim. (France) 12:72-74. [DOI] [PubMed] [Google Scholar]
- 36.Pollard, D. R., W. M. Johnson, H. Lior, S. D. Tyler, and K. R. Rozee. 1990. Detection of the aerolysin gene in Aeromonas hydrophila by the polymerase chain reaction. J. Clin. Microbiol. 28:2477-2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robson, W. L., A. K. Leung, and C. L. Trevenen. 1992. Haemolytic-uraemic syndrome associated with Aeromonas hydrophila enterocolitis. Pediatr. Nephrol. 6:221-222. [DOI] [PubMed] [Google Scholar]
- 38.Sha, J., E. V. Kozlova, and A. K. Chopra. 2002. Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibata, M., K. Morita, N. Wanatabe, H. Wada, T. Okitsu, S. Yamai, K. Itoh, T. Shimada, H. Wanatabe, and M. Kanamori. 1996. Rapid detection of the hemolysin genes in Aeromonas sobria by polymerase chain reaction. Kansenshogaku Zasshi. 70:1266-1270. [DOI] [PubMed] [Google Scholar]
- 40.Singh, D. V., and S. C. Sanyal. 1992. Production of hemolysis and its correlation with enterotoxicity in Aeromonas spp. J. Med. Microbiol. 37:262-267. [DOI] [PubMed] [Google Scholar]
- 41.Swift, S., M. J. Lynch, L. Fish, D. F. Kirke, M. J. Tomas, G. S. Stewart, and P. Williams. 1999. Quorum sensing-dependent regulation and blockade of exoprotease production in Aeromonas hydrophila. Infect. Immun. 67:5192-5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vadivelu, J., S. D. Puthucheary, M. Phipps, and Y. W. Chee. 1995. Possible virulence factors involved in bacteraemia caused by Aeromonas hydrophila. J. Med. Microbiol. 42:171-174. [DOI] [PubMed] [Google Scholar]
- 43.Wang, G., K. D. Tyler, C. K. Munro, and W. M. Johnson. 1996. Characterization of cytotoxic, hemolytic Aeromonas caviae clinical isolates and their identification by determining presence of a unique hemolysin gene. J. Clin. Microbiol. 34:3203-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wong, C. Y., M. W. Heuzenroeder, and R. L. Flower. 1998. Inactivation of two haemolytic toxin genes in Aeromonas hydrophila attenuates virulence in a suckling mouse model. Microbiology 144:291-298. [DOI] [PubMed] [Google Scholar]