Abstract
We evaluated the molecular mechanism for resistance of 360 enterococci for which the gentamicin MICs were ≥128 μg/ml. The aac(6′)-Ie-aph(2")-Ia, aph(2")-Ic, and aph(2")-Id genes were identified by PCR in isolates from animals, food, and humans. The aph(2")-Ib gene was not identified in any of the isolates. Two Enterococcus faecalis isolates (MICs > 1,024 μg/ml) from animals failed to generate a PCR product for any of the genes tested and likely contain a new unidentified aminoglycoside resistance gene. Pulsed-field gel electrophoresis (PFGE) analysis showed a diversity of strains. However, 1 human and 18 pork E. faecalis isolates from Michigan with the aac(6′)-Ie-aph(2")-Ia gene had related PFGE patterns and 2 E. faecalis isolates from Oregon (1 human and 1 grocery store chicken isolate) had indistinguishable PFGE patterns. We found that when a gentamicin-resistant gene was present in resistant enterococci from animals, that gene was also present in enterococci isolated from food products of the same animal species. Although these data indicate much diversity among gentamicin-resistant enterococci, the data also suggest similarities in gentamicin resistance among enterococci isolated from humans, retail food, and farm animals from geographically diverse areas and provide evidence of the spread of gentamicin-resistant enterococci from animals to humans through the food supply.
Enterococci are important nosocomial pathogens. Multidrug resistance is common among enterococci and presents a formidable treatment problem. High-level (MIC ≥ 2,000 μg/ml) gentamicin resistance (HLGR) in enterococci, first described in 1979, is a significant therapeutic problem, particularly for patients with endocarditis (21, 23). Until recently, aac(6′)-Ie-aph (2")-Ia was the only reported gene associated with HLGR in enterococci (6, 14). This bifunctional gene confers resistance to essentially all clinically available aminoglycosides except streptomycin, thereby eliminating synergism between aminoglycosides and a cell-wall-active agent such as ampicillin or vancomycin (6, 7). This gene has been detected in various species of enterococci significant to human infection and among enterococci isolated from food-producing animals.
Three recently identified gentamicin-modifying genes are also associated with gentamicin resistance in enterococci and the elimination of synergy between aminoglycosides and cell-wall-active agents (8, 16, 27). The aph(2")-Ib gene is associated with gentamicin (MIC ≥ 500 μg/ml) and other types of aminoglycoside resistance in Enterococcus faecium and Escherichia coli and appears to be linked with the aac(6′)-Im aminoglycoside resistance gene in both enterococci and gram-negative bacilli (9, 16). This gene has been detected in vancomycin-resistant E. faecium isolates from hospitalized patients (16). The aph(2")-Ic gene is associated with gentamicin MICs of 128 to 512 μg/ml and the elimination of ampicillin/gentamicin synergism (8). The aph(2")-Ic gene was first described in 1997 in a veterinary isolate of Enterococcus gallinarum and has also been identified in human E. faecium and Enterococcus faecalis isolates (8). The aph(2")-Id gene, first described in 1998 in a human Enterococcus casseliflavus isolate, confers high-level resistance to gentamicin but not to amikacin (27). This gene has been detected in several vancomycin-resistant E. faecium isolates from hospitalized patients (27).
To further the understanding of the potential spread of gentamicin resistance genes among enterococci, we evaluated the gentamicin-resistant enterococci isolated from humans, food, and farm animals from six states for strain relatedness and molecular mechanisms of resistance.
MATERIALS AND METHODS
Source of isolates.
Forty-one enterococci were isolated from human stool specimens from outpatients submitted for diagnostic purposes to clinical microbiology laboratories. This included isolates from outpatients in Georgia, Maryland, Michigan, Minnesota, and Oregon. Human stool specimens were collected from 1998 to 2000 except specimens from Michigan, which were collected in 1999 and 2000.
One hundred five isolates were obtained from food purchased in grocery stores: 39 isolates were from whole chickens purchased from at least 28 stores in Georgia, Maryland, Michigan, Minnesota, and Oregon in 1998 or 1999, and 66 isolates were from ground pork purchased from 12 stores in Michigan in 1999 and 2000. Two hundred fourteen isolates were isolated from healthy animals on farms: 15 isolates from six chicken farms, 107 isolates from nine dairy cattle farms, 50 isolates from nine swine farms, and 42 isolates from eight turkey farms. Isolates were collected in Michigan between 1995 and 2000 except the chicken isolates, which were collected in Indiana in 2000. Except for 13 turkey isolates that were obtained from cloacal cultures of individual animals in 1995 and 1996, chicken and turkey specimens were collected via drag culture of the poultry litter, which contained fecal waste. Cattle and swine isolates were isolated from stool specimens collected from individual animals.
Isolation and identification.
A total of 5.0 ml of selective broth was added to approximately 0.5 g of human stool or animal feces. Drag swabs were processed by vortexing specimens in 300 ml of sterile saline and inoculating 1 ml of suspension into 10 ml of selective broth. Grocery store chicken specimens were sampled by placing whole chicken carcasses in a sterile bag and adding 400 ml of sterile buffered peptone water and massaging for 30 s to ensure that all surfaces were rinsed. A total of 100 ml was withdrawn and incubated at 37°C for 24 h, and then 0.5 ml was inoculated to 5 ml of selective broth. Pork specimens were sampled by adding 25 g of ground pork to 100 ml of buffered peptone water in a sampling filter bag (Cole-Parmer Instrument Co., Vernon Hills, Ill.) and massaged for 2 min. The filter bag was removed, the rinse water was incubated at 37°C for 24 h, and then 0.5 ml of the ground pork rinse was inoculated to 5 ml of selective broth. Selective broth for all cultures consisted of Enterococcosel (Becton Dickinson, Cockeysville, Md.) broth containing gentamicin at 100 μg/ml. After 72 h of incubation at 37°C, selective broth cultures were subcultured to solid media and incubated at 37°C. Selective broths from human and food samples were cultured on solid media consisting of Ford agar base, pH 7.8 (consisting of Columbia agar base, phenol red broth base, and raffinose), containing 100 μg of gentamicin/ml (15). Selective broths from animal feces and drag samples were cultured on solid media consisting of enterococcal agar containing 100 μg of gentamicin (American Pharmaceutical Partners, Los Angeles, Calif.)/ml. Representative colonies were selected from solid media and identified by conventional biochemical reactions (13). Susceptibility to gentamicin was determined by a standardized microdilution method (22). Strains for which MICs were ≥2,000 μg/ml were considered HLGR.
PFGE.
Genomic DNA was prepared according to previously published methods (12). Sample plugs were digested with 30 U of SmaI (New England Biolabs, Beverly, Mass.), loaded on a 0.8% agarose gel in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA), and electrophoresed on a CHEF-DRIII apparatus (Bio-Rad) with the following parameters: initial switch time, 5 s; final switch time, 35 s; start ratio, 1; voltage, 6 V/cm; run time, 17 h; temperature, 14°C. A Lambda ladder pulsed-field gel electrophoresis (PFGE) marker (New England Biolabs) was used as the size standard. The gel was stained with ethidium bromide and visualized on a UV transilluminator. Strains were considered related by PFGE if their patterns differed by one to three bands (25).
PCR.
DNA was amplified by PCR with a PerkinElmer model 480 thermal cycler. The reaction mixtures contained 1× PCR buffer (GIBCO BRL), 200 μM deoxynucleoside triphosphate, 1.25 U of Taq polymerase (GIBCO BRL), one or two colonies of organism, and 0.1 μg of each primer. The final reaction volume was 50 μl, and samples were covered with 1 drop of mineral oil. PCR was performed with a precycle of 10 min at 94°C and cycles of 1 min at 94°C, 1 min at 55°C, and 3 min at 72°C; these were followed by 10 min at 72°C. The synthetic oligonucleotide primers used to detect the genes are as follows: for the aac(6′)-Ie-aph(2")-Ia gene, 5′-GAGCAATAAGGGCATACCAAAAATC-3′ and 5′-CCGTGCATTTGTCTTAAAAAACTGG-3′; for the aph(2")-Ib gene, 5′-TATGGATTCATGGTTAACTTGGACGCTGAG-3′ and 5′-ATTAAGCTTCCTGCTAAAATATAAACATCTCTGCT-3′; for the aph(2")-Ic gene, 5′-GAAGTGATGGAAATCCCTTCGTG-3′ and 5′-GCTCTAACCCTTCAGAAATCCAGTC-3′; and for the aph(2")-Id gene, 5′-GGTGGTTTTTACAGGAATGCCATC-3′ and 5′-CCCTCTTCATACCAATCCATATAACC-3′.
Filter matings.
Filter matings were performed with E. faecium and E. faecalis isolates by using the plasmid-free E. faecium GE-1 recipient strain and the plasmid-free E. faecalis FA2-2 recipient strain. Filters were incubated for 24 h, and transconjugants were selected on brain-heart infusion agar (EM Science) containing 500 μg of gentamicin (APP)/ml, 50 μg of rifampin (Sigma)/ml, and 25 μg of fusidic acid (Sigma)/ml. Transfer frequencies were expressed as the number of transconjugants divided by the number of recipients.
RESULTS
We evaluated 360 enterococci for which gentamicin MICs were ≥128 μg/ml: 258 isolates with HLGR (MIC ≥ 2,000 μg/ml) and 102 for which the gentamicin MICs were 128 to 1,024 μg/ml. Of the 360 isolates, there were 251 (69.7%) E. faecalis, 104 (28.9%) E. faecium, 3 (0.8%) E. casseliflavus, and 2 (0.6%) E. gallinarum isolates.
Of the 360 gentamicin-resistant enterococci evaluated, 259 (72%) contained the aac(6′)-Ie-aph (2")-Ia gene, 66 (18%) contained the aph(2")-Ic gene, and 33 (9%) contained the aph(2")-Id gene (Table 1). The aph(2")-Ib gene was not detected in any of the isolates tested. Two E. faecalis isolates (MICs > 1,024 μg/ml), one from pork and one from a chicken farm, failed to generate a PCR product for any of the genes tested.
TABLE 1.
Gentamicin resistance gene content of gentamicin-resistant enterococci from humans, food, and food-producing animals
| Gentamicin resistance gene and species | Total | Sources/No. positive
|
||||||
|---|---|---|---|---|---|---|---|---|
| Human | Food
|
Food-producing animal
|
||||||
| Chicken | Pork | Chicken | Turkey | Swine | Dairy | |||
| aac(6′)-Ie-aph(2")-Ia | ||||||||
| E. faecalis | 189 | 28 | 24 | 56 | 6 | 15 | 36 | 24 |
| E. faecium | 65 | 11 | 2 | 0 | 4 | 7 | 3 | 38 |
| E. casseliflavus | 3 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| E. gallinarum | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| Total (%) | 259 (72) | 39 (95) | 27 (69) | 56 (85) | 10 (71) | 26 (62) | 39 (78) | 62 (58) |
| aph(2")-Ic | ||||||||
| E. faecalis | 60 | 0 | 0 | 9 | 0 | 0 | 10 | 41 |
| E. faecium | 6 | 1 | 0 | 0 | 0 | 0 | 1 | 4 |
| Total (%) | 66 (18) | 1 (2.5) | 0 (0) | 9 (14) | 0 (0) | 0 (0) | 11 (22) | 45 (42) |
| aph(2")-Id | ||||||||
| E. faecalis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. faecium | 33 | 1 | 12 | 0 | 4 | 16 | 0 | 0 |
| Total (%) | 33 (9) | 1 (2.5) | 12 (31) | 0 (0) | 4 (27) | 16 (38) | 0 (0) | 0 (0) |
| Total (%) | 360a | 41 | 39 | 66a | 15a | 42 | 50 | 107 |
Two E. faecalis isolates (gentamicin MICs > 1,024 μg/ml) from pork and a chicken farm failed to generate a PCR product for any of the genes tested.
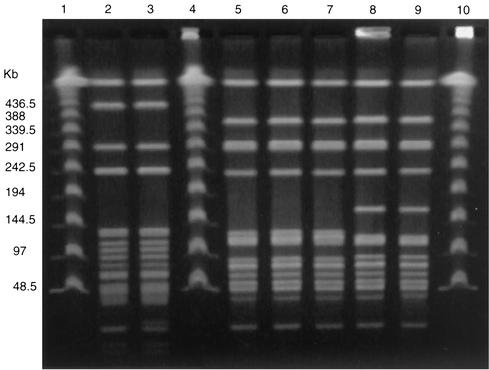
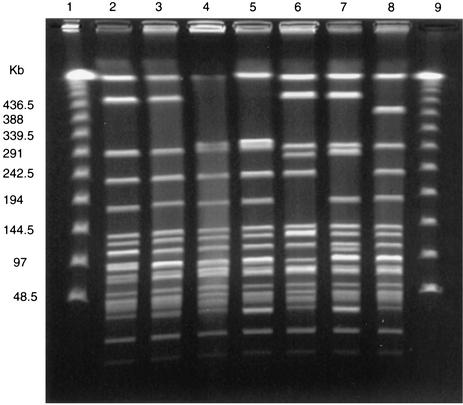
Of the 259 isolates that contained the aac(6′)-Ie-aph(2")-Ia gene, there were 223 that were HLGR and 36 for which gentamicin MICs were 128 to 1,024 μg/ml. The aac(6′)-Ie-aph(2")-Ia gene was detected in 39 (95%) of 41 human stool isolates; 83 (79%) of 105 food isolates, which consisted of 27 (69%) of 39 grocery store chicken isolates and 56 (85%) of 66 ground pork isolates; and 137 (64%) of 214 animal isolates, which consisted of 10 (67%) of 15 chicken isolates, 62 (58%) of 107 dairy cow isolates, 39 (78%) of 50 swine isolates, and 26 (62%) of 42 turkey isolates. Seventy-five percent of E. faecalis isolates, 63% of E. faecium isolates, all three E. casseliflavus isolates, and both of the E. gallinarum isolates tested contained the aac(6′)-Ie-aph(2")-Ia gene. The 259 isolates that contained aac(6′)-Ie-aph(2")-Ia yielded 88 unique PFGE strain types. However, five E. faecalis isolates with aac(6′)-Ie-aph(2")-Ia isolated from grocery store chickens purchased in Georgia, Maryland, and Oregon had related PFGE patterns (Fig. 1). Furthermore, three isolates from Georgia had indistinguishable PFGE patterns and one isolate from Maryland and one from Oregon were also indistinguishable (Fig. 1). Two E. faecalis isolates with aac(6′)-Ie-aph(2")-Ia, isolated from human stools collected in Oregon and a grocery store chicken purchased in Oregon, also were indistinguishable by PFGE (Fig. 1). Furthermore, 19 E. faecalis isolates with the aac(6′)-Ie-aph(2")-Ia gene (isolated from 1 human stool and 18 pork specimens) had related PFGE patterns; the human isolate and 9 of the pork isolates had indistinguishable PFGE patterns, while the other 9 pork isolates differed from the human isolate by one to three bands (Fig. 2). These pork isolates were isolated from ground pork purchased from 12 stores in Michigan over a 12-month period in 2000. The human isolate was isolated during the same period in Michigan. This strain type was not seen in any of the chicken or pork isolates from the other states.
FIG. 1.
PFGE of SmaI-digested genomic DNA from gentamicin-resistant E. faecalis from grocery store chicken isolates and one human isolate. Lanes 1, 4, and 10, Lambda ladder DNA standard; lanes 2 and 3, human and grocery store chicken isolates, respectively, from Oregon with indistinguishable PFGE patterns; lanes 5 through 7, grocery store chicken isolate from Georgia with indistinguishable PFGE patterns; lanes 8 and 9, grocery store chicken isolates from Maryland and Oregon with indistinguishable PFGE patterns.
FIG. 2.
PFGE of SmaI-digested genomic DNA from gentamicin-resistant E. faecalis isolated from human and ground pork samples in Michigan. Lanes 1 and 9, Lambda ladder DNA standard; lane 2, human isolate; lanes 3 through 8, ground pork isolates with related PFGE patterns.
Of the 66 isolates with aph(2")-Ic, 3 were HLGR, while the MICs for the other 63 ranged from 128 to 1,024 μg/ml. One (2.5%) of 41 human, 9 (14%) of 66 ground pork, 45 (42%) of 107 dairy cattle, and 11 (22%) of 50 swine isolates had the aph(2")-Ic gene. aph(2")-Ic was not detected in farm chicken isolates or grocery store chicken isolates. Ninety-one percent of isolates with aph(2")-Ic were E. faecalis, and 9% were E. faecium. Of all the isolates tested, 24% of E. faecalis isolates and 6% of E. faecium isolates contained the aph(2")-Ic gene. The 66 isolates with aph(2")-Ic yielded 17 unique PFGE patterns. However, there were four groups of E. faecalis isolates with aph(2")-Ic that had PFGE patterns that, while distinguishable between groups, were indistinguishable among isolates with each group: three swine isolates, four dairy cattle isolates, six pork isolates, and a group of nine isolates that included eight dairy cattle isolates and one swine isolate. Furthermore, one isolate from a dairy cow and six isolates from swine had related PFGE patterns. Transfer frequencies for E. faecium and E. faecalis isolates containing the aph(2")-Ic gene were 10−9 to 10−2.
Of the 33 isolates with aph(2")-Id, all were HLGR E. faecium. The aph(2")-Id was detected in 1 (2.5%) of 41 human, 12 (31%) of 39 grocery store chicken, 4 (27%) of 15 chicken, and 16 (38%) of 42 turkey isolates. The one isolate from a human was collected in Michigan. The 32 isolates with aph(2")-Id from food or animals were all isolated from poultry (chickens or turkeys): 12 isolates from grocery store chickens from five states, which consisted of 2 from Georgia, 1 from Maryland, 2 from Michigan, 1 from Minnesota, and 6 from Oregon; 4 isolates from two chicken farms in Indiana; and 16 isolates from turkey farms in Michigan. The 33 isolates with aph(2")-Id yielded 30 unique PFGE patterns. However, two E. faecium isolates with aph(2")-Id from grocery store chickens from Oregon were indistinguishable and two E. faecium isolates from chicken farms in Indiana were indistinguishable. Transfer frequencies for these E. faecium isolates ranged from <10−9 to 10−3.
DISCUSSION
The reservoirs and sources of gentamicin resistance in enterococci have not been fully defined, particularly for the three most recently identified genes. The purpose of this study was to compare PFGE strain types and the aminoglycoside resistance gene content of enterococci among different sources (humans, animals, and retail food) rather than determine the prevalence of gentamicin resistance among enterococci from these sources. Although, earlier studies have reported the frequent isolation of gentamicin-resistant enterococci in food-producing animals (S. M. Donabedian, P. S. Bozigar, M. B. Perri, E. Hershberger, L. A. Thal, P. Bartlett, R. Jones, and M. J. Zervos, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1873, 2001), little information is available on the gentamicin-resistant enterococci isolated from retail food or from humans in the community. In our study, we confirmed gentamicin resistance from human stool specimens and from chicken and pork purchased in grocery stores. We also found evidence of dissemination across a broad geographical area of the United States.
The aac(6′)-Ie-aph(2")-Ia gene was the most common gene among the gentamicin-resistant isolates evaluated in this study. This gene was detected in various enterococcal species, including the E. faecalis, E. faecium, E. gallinarum, and E. casseliflavus isolates collected from human stool, chicken, and pork purchased in grocery stores and chickens, dairy cattle, swine, and turkeys on farms. These observations provide evidence of a large reservoir for this resistance gene in humans, food, and food-producing animals, indicating widespread dissemination of this resistance determinant. E. faecalis isolates with indistinguishable PFGE patterns that contained the aac(6′)-Ie-aph(2")-Ia gene were isolated from a human stool specimen and pork samples from different grocery stores in the same state. Similarly, E. faecalis isolates with indistinguishable PFGE patterns that contained the aac(6′)-Ie-aph(2")-Ia gene were isolated from a human stool specimen and chicken purchased from a grocery store in the same state.
In a previous study, the aph(2")-Ic gene was found infrequently and was limited to human E. faecalis and E. faecium, equine E. gallinarum, and goose E. faecium (8). The human isolates were from clinically ill persons. Our report is the first description of the aph(2")-Ic gene in E. casseliflavis and the first identification of the gene in enterococci isolated from food (pork) purchased from a grocery store. In addition, we detected the aph(2")-Ic gene in gentamicin-resistant enterococci isolated from dairy cattle and swine farms. It is notable that we detected the aph(2")-Ic gene in 14% of gentamicin-resistant enterococci from pork purchased from grocery stores and 22% of gentamicin-resistant enterococci from pigs on the farm but did not detect the gene in gentamicin-resistant enterococci isolated from the chicken from grocery stores or on farms. Further sampling is necessary to confirm this specificity.
The aph(2")-Id gene has previously been described in only a few human strains of vancomycin-resistant enterococci: a vancomycin-resistant E. casseliflavus isolate and vancomycin-resistant E. faecium isolates from an outbreak in four hospitals (27). Our report is the first identification of the aph(2")-Id gene in enterococci isolated from food purchased from grocery stores and food-producing animals on farms. It is notable that we detected the aph(2")-Id gene only in E. faecium isolates and only in poultry. We detected the gene from 31% of gentamicin-resistant enterococci from chicken purchased from grocery stores and 29% of gentamicin-resistant enterococci from chickens on farms. We also detected the gene in 38% of gentamicin-resistant isolates from turkey farms. Again, further sampling is necessary to confirm this specificity, but these data suggest that poultry may be a specific reservoir for this gene, which has been associated with clinical illness in humans. It is also noteworthy that two HLGR isolates, one from pork purchased from a grocery store and one from a chicken farm, failed to generate a PCR product for any of the genes tested and therefore may contain a new gentamicin resistance gene; further studies are ongoing.
The results of this study show a commonality of gentamicin-resistant determinants and gentamicin-resistant enterococcal isolates among humans, food, and food-producing animals over a broad geographical area. Because we have shown that enterococci isolated from animals and humans possess the same aminoglycoside resistance gene content, it will be important to determine the transferability of these resistance genes since the dissemination of genes can occur by horizontal transfer. Our additional findings of a high prevalence of gentamicin-resistant enterococci in the feces of food-producing animals on farms also suggest that the occurrence of gentamicin-resistant enterococci in food can be attributed to the presence of the organism in food-producing animals. Furthermore, it is likely that the occurrence of gentamicin-resistant enterococci in food-producing animals is a consequence of gentamicin use in these animals. There is clear evidence that, with an increase in the consumption of antimicrobial agents by humans or animals, there is a resultant increase in antimicrobial resistance (1-5, 10, 11, 18-20, 26, 28-34). Gentamicin is commonly used in swine and widely used in chickens and turkeys. For example, it is estimated that 80% of the 8 billion chickens raised for meat (broilers) each year in the United States are treated with gentamicin in the absence of disease for the prevention of early mortality (20).
Importantly, our findings demonstrate that food-producing animals are an important reservoir for gentamicin-resistant enterococci and are a source of such bacteria for humans. We found that when gentamicin-resistant genes are present in the resistant enterococci isolated from animals, they are also present in the enterococci isolated from food products of the same animal species. Similarly, when they are absent in the animals, they are absent in the corresponding food product. We also found that two genes that are found rarely in human isolates, aph(2")-Ic and aph(2")-Id, were more prevalent in several food-producing animals. Finally, we found indistinguishable gentamicin-resistant enterococcal isolates with common resistant determinants from humans and food. Taken together, these findings suggest that gentamicin-resistant enterococci, which are clinically important pathogens, are transmitted from food-producing animals to humans through the food supply. Since the use of gentamicin in food-producing animals will create selective pressure to increase the emergence and dissemination of gentamicin-resistant enterococci, there is a need to prevent the misuse and overuse of gentamicin in food-producing animals.
Acknowledgments
This work was supported in part by Food and Drug Administration-Center for Veterinary Medicine Department of Health and Human Services Cooperative Agreement FD-U-001577-01.
REFERENCES
- 1.Aarestrup, F. M. 1995. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb. Drug. Resist. 1:255-257. [DOI] [PubMed] [Google Scholar]
- 2.Aarestrup, F. M., P. Ahrens, M. Madsen, L. V. Pallesen, R. L. Poulsen, and H. Westh. 1996. Glycopeptide susceptibility among Danish Enterococcus faecium and Enterococcus faecalis isolates of animal and human origin and PCR identification of genes within the VanA cluster. Antimicrob. Agents Chemother. 40:1938-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, J., J. Z. Jordens, and D. T. Griffiths. 1994. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J. Antimicrob. Chemother. 34:507-514. [DOI] [PubMed] [Google Scholar]
- 4.Bates, J., J. Z. Jordens, and J. B. Selkon. 1993. Evidence for an animal origin of vancomycin-resistant enterococci. Lancet 342:490-491. [DOI] [PubMed] [Google Scholar]
- 5.Chaslus-Dancla, E., P. Pohl, M. Meurisse, M. Marin, and J. P. Lafont. 1991. High genetic homology between plasmids of human and animal origins conferring resistance to the aminoglycosides gentamicin and apramycin. Antimicrob. Agents Chemother. 35:590-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, J. W. 2000. Aminoglycoside resistance in enterococci. Clin. Infect. Dis. 31:586-589. [DOI] [PubMed] [Google Scholar]
- 7.Chow, J. W., S. M. Donabedian, S. M. Clewell, D. B. Sahm, and M. J. Zervos. 1998. In vitro susceptibility and molecular analysis of gentamicin-resistant enterococci. Diagn. Microbiol. Infect. Dis. 32:141-146. [DOI] [PubMed] [Google Scholar]
- 8.Chow, J. W., M. J. Zervos, S. A. Lerner, L. A. Thal, S. M. Donabedian, D. D. Jaworski, S. Tsai, K. J. Shaw, and D. B. Cewell. 1997. A novel gentamicin resistance gene in Enterococcus. Antimicrob. Agents Chemother. 41:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow, J. W., V. Kak, I. You, S. J. Kao, J. Petrin, D. B. Clewell, S. A. Lerner, G. H. Miller, and K. J. Shaw. 2001. Aminoglycoside resistance genes aph(2")-Ib and aac(6′)-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob. Agents Chemother. 45:2691-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coque, T. M., R. C. Arduino, and B. E. Murray. 1995. High-level resistance to aminoglycosides: comparison of community and nosocomial fecal isolates of enterococci. Clin. Infect. Dis. 20:1048-1051. [DOI] [PubMed] [Google Scholar]
- 11.Coque, T. M., J. F. Tomayko, S. C. Ricke, P. C. Okhuysen, and B. E. Murray. 1996. Vancomycin-resistant enterococci from nosocomial, community, and animal sources in the United States. Antimicrob. Agents Chemother. 40:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donabedian, S. M., J. W. Chow, D. M. Shlaes, M. Green, and M. J. Zervos. 1995. DNA probes and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J. Clin. Microbiol. 33:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., K. S. Gilmore, and P. Courvalin. 1986. Nucleotide sequence analysis of the gene specifying the bifunctional 6′-aminoglycoside acetyltransferase enzyme in Streptococcus faecalis and identification and cloning of gene regions specifying the two activities. J. Bacteriol. 167:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford, M., J. D. Perry, and F. K. Gould. 1994. Use of cephalexin-aztreonam-arabinose agar for selective isolation of Enterococcus faecium. J. Clin. Microbiol. 32:2999-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2")-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kariyama, R., H. Kumon, L. Chow, M. J. Zervos, T. Takata, M. Tabata, and J. W. Chow. 1998. In-vitro activity of the combination of ampicillin and arbekacin against high-level gentamicin-resistant enterococci. J. Antimicrob. Chemother. 42:836-838. [DOI] [PubMed] [Google Scholar]
- 18.Klare, I., H. Heier, H. Claus, G. Böhme, S. Marin, G. Seltmann, et al. 1995. Enterococcus faecium strains with vanA-mediated high-level glycopeptide resistance isolated from animal foodstuffs and fecal samples of humans in the community. Microb. Drug Resist. 1:265-272. [DOI] [PubMed] [Google Scholar]
- 19.Levy, S. B., G. B. Fitzgerald, and A. B. Macone. 1976. Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 260:40-42. [DOI] [PubMed] [Google Scholar]
- 20.Mellon, M., C. Benbrook, and K. L. Benbrook. Hogging it! Union of concerned scientists. UCS Publishing, Cambridge, Mass.
- 21.Murray, B. E. 1990. The life and times of the enterococcus. Clin. Microbiol. Rev. 3:46-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 1997. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.Patterson, J. E., and M. J. Zervos. 1990. High-level gentamicin resistance in Enterococcus: microbiology, genetic basis, and epidemiology. Rev. Infect. Dis. 12:644-652. [DOI] [PubMed] [Google Scholar]
- 24.Silverman, J., L. A. Thal, M. B. Perri, G. Bostic, and M. J. Zervos. 1998. Epidemiologic evaluation of antimicrobial resistance in community-acquired enterococci. J. Clin. Microbiol. 36:830-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thal, L. A., J. W. Chow, R. Mahayni, H. Bonilla, M. B. Perri, S. M. Donabedian, J. Silverman, S. Taber, and M. J. Zervos. 1995. Characterization of antimicrobial resistance in enterococci of animal origin. Antimicrob. Agents Chemother. 39:2112-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai, S. F., M. J. Zervos, D. B. Clewell, S. M. Donabedian, D. F. Sahm, and J. W. Chow. 1998. A new high-level gentamicin resistance gene, aph(2")-Id, in Enterococcus spp. Antimicrob. Agents Chemother. 42:1229-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Auwera, P., N. Pensart, V. Korten, B. E. Murray, and R. Leclercq. 1996. Influence of oral glycopeptides on the fecal flora of human volunteers: selection of highly glycopeptide-resistant enterococci. J. Infect. Dis. 173:1129-1136. [DOI] [PubMed] [Google Scholar]
- 29.Van Bogaard, A. E., L. B. Jensen, and E. E. Stobberingh. 1997. Vancomycin-resistant enterococci in turkeys and farmers. New Engl. J. Med. 337:1558-1559. [DOI] [PubMed] [Google Scholar]
- 30.Vergis, E. N., M. K. Hayden, J. W. Chow, D. R. Snydman, M. J. Zervos, P. K. Linden, M. M. Wagener, B. Schmitt, and R. R. Muder. 2001. Determinants of vancomycin resistance and mortality rates in enterococcal bacteremia. Ann. Intern. Med. 1:484-492. [DOI] [PubMed] [Google Scholar]
- 31.Welton, L. A., L. A. Thal, M. B. Perri, S. M. Donabedian, J. McMahon, J. W. Chow, and M. J. Zervos. 1998. Antimicrobial resistance in enterococci isolated from turkey flocks fed virginiamycin. Antimicrob. Agents Chemother. 42:705-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witte, W., and I. Klare. 1995. Glycopeptide-resistant Enterococcus faecium outside hospitals: a commentary. Microb. Drug Resist. 1:259-263. [DOI] [PubMed] [Google Scholar]
- 33.Woodford, N., M. J. Palepou, A. P. Johnson, P. R. Chadwick, and J. Bates. 1997. Methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci. Lancet 350:738.. [DOI] [PubMed] [Google Scholar]
- 34.Zervos, M. J., C. A. Kauffman, P. M. Therasse, A. G. Bergman, T. S. Mikesell, and D. R. Schaberg. 1987. Nosocomial infection by gentamicin-resistant Streptococcus faecalis: an epidemiologic study. Ann. Intern. Med. 106:687-691. [DOI] [PubMed] [Google Scholar]