Abstract
Sequences encompassing cleavage sites of fusion protein genes were obtained for avian paramyxovirus 1 isolates from cormorants in Canada. All isolates have the virulent cleavage site SRGRRQKR*FVG. They form a distinct cluster within isolates obtained around the world and may represent a novel genotype closely related to genotype V.
Avian paramyxovirus 1 (APMV1) has been isolated from a variety of species of free-living and domestic birds worldwide. The enveloped virus, belonging to the family Paramyxoviridae, has an over 15,000-nucleotide-long, negative-sense single-stranded RNA genome (7). Virus infections range from inapparent to severe (e.g., Newcastle disease in poultry), depending on the number of viral and host factors (2). The amino acid composition of the fusion protein cleavage site is the major determinant in the virulence of the virus (22). Wild bird species, especially aquatic birds, are considered to be more resistant to the disease and could serve as a virus reservoir (9, 14, 16, 17). However, high mortalities from APMV1 occurred in double-crested cormorants in Canada and the United States in 1990 and 1992 (8, 10, 21, 28). Based on fusion and matrix protein sequence data, the viruses from cormorants were related to viruses of psittacine origin and to viruses isolated during the 1970s California outbreak (24, 25, 26).
Canada has experienced several smaller outbreaks in cormorants since 1992. Relatively large outbreaks occurred in 1995 in Saskatchewan (18, 19), in 1996 in Ontario, and in 1999 in Alberta (6). Isolated submissions were also received from different parts of Canada throughout the years 1995 to 2000.
The purpose of this work was to characterize on the molecular level cormorant isolates submitted from 1995 to 2000, confirm their in vivo pathotype by using predicted amino acid composition of the fusion protein cleavage site, and determine their genotype.
Archived cormorant APMV1 isolates (allantoic fluid [AF] from embryonated chicken eggs inoculated with 0.1 ml of 10% homogenate from pooled cormorant tissues), stored at the National Centre for Foreign Animal Disease, Winnipeg, Manitoba, Canada, were used in the molecular analysis. AF was tested for the presence of APMV1 by the hemagglutination-inhibition test (5). AFs from dead embryonated chicken eggs yielding negative or weak hemagglutinating activity were retested on Vero cells by the indirect immunoperoxidase assay adapted from the work of Afshar et al. (1). The intracerebral pathogenicity index (ICPI) and the intravenous pathogenicity index (IVPI) were determined as described in the Office International des Epizooties manual of standards (29). Viral RNA was extracted from AF by using TriPure isolation reagent (Boehringer Mannheim). One-step reverse transcription-PCR (RT-PCR) was performed with the Qiagen OneStep RT-PCR kit and the following RT-PCR program: 50°C for 30 min; holding at 95°C for 5 min; and 30 cycles of 95°C for 60 s, 56°C for 30 s, and 72°C for 30 s. The forward (5′-TGTCGCAGTGACTGCTGACC) and reverse (5′-GTCAGTGACCTCGTGCACAG) primers yielded an amplicon of approximately 750 nucleotides, which was sequenced with the RT-PCR primers. The ClustalW multiple alignment algorithm (European Bioinformatics Institute) was used to align the sequences, and the obtained PHYLIP output file (J. Felsenstein, Department of Genetics, University of Washington) was applied in TREECON, version 1.3b (27) to construct a dendrogram, with the neighbor-joining algorithm (23), based on a distance estimation model of Jukes and Cantor (15).
The 1995 to 2000 AMPV1 isolates from cormorants appear to form a separate cluster from the viruses isolated from cormorants in 1990 and 1992, based on the analysis of the 643-nucleotide sequence spanning from the 5′-terminal nucleotides of the matrix protein gene past the fusion protein cleavage site coding sequence and supported by a decrease or lack of hemagglutinating activity not observed for the earlier isolates (Table 1) (11). The phylogenetic analysis is graphically presented in Fig. 1. The molecular pathotyping (amino acid sequence of the fusion protein cleavage sites) corresponded with the in vivo pathotyping (Table 1). Compared to over 400 fusion protein cleavage site sequences published in the GenBank database, this type of cleavage site was identified only in isolates from cormorants, two isolates from exotic birds, and two isolates from pigeons in the United States.
TABLE 1.
Elucidated fusion protein amino acid cleavage site sequences of the APMV1 cormorant isolates, with the GenBank accession numbersa
| Isolate identification (reference) | ICPI (8 days) | IVPI (10 days) | Fusion protein cleavage site (molecular pathotyping) | GenBank accession no. |
|---|---|---|---|---|
| APMV1/CT/Sask/1479/90 (8) | 1.55 | 2.08 | SGGRRQKR*FVG | AF448223 |
| APMV1/CT/Sask/2585/92 (8) | 1.6 | 1 | SRGRRQKR*FVG | AF448844 |
| APMV1/CT/Man/3298/92 | 1.3 | 0.8 | SRGRRQKR*FVG | AF448843 |
| APMV1/CT/Ont/2150/95 | 1.6 | 1.2 | SRGRRQKR*FVG | AF448486 |
| APMV1/CT/Ont/2344/95 | 1.71 | 0.89 | SRGRRQKR*FVG | AF448486 |
| APMV1/CT/Ont/2575/95 | 1.88 | 0.12 | SRGRRQKR*FVG | AY063493 |
| APMV1/CT/Sask/2035/95 (12) | 1.61 | 1.23 | SRGRRQKR*FVG, nonhemagglutinating | AF448845 |
| APMV1/CT/Ont/39/96 | 1.15 | 0.86 | SRGRRQKR*FVG | AF448842 |
| APMV1/CT/Ont/48/96 | 1.66 | 1.41 | SRGRRQKR*FVG | AY063492 |
| APMV1/CT/Sask/3-1125/98 | 1.56 | 1.09 | SRGRRQKR*FVG, nonhemagglutinating | AY063494 |
| APMV1/CT/Alb/35/99 (4) | 1.07 | 0.41 | SRGRRQKR*FVG, nonhemagglutinating | AF448841 |
| APMV1/CT/Sask/45/99 | SRGRRQKR*FVG | AF263615 | ||
| APMV1/CT/Ont/378/00 | 1.3 | 1.64 | SRGRRQKR*FVG, nonhemagglutinating | AY063123 |
| APMV1/CT/Ont/429/00 | ND | ND | SRGRRQKR*FVG | AY063123 |
| APMV1/CT/Ont/410/00 | ND | ND | SRGRRQKR*FVG, nonhemagglutinating | AY063123 |
CT, cormorant. In ICPI the most virulent APMV1 isolates give indices close to 2, in IVPI the most virulent APMV1 isolates give indices close to 3, and in both ICPI and IVPI the avirulent viruses give values close to 0. ND, not done.
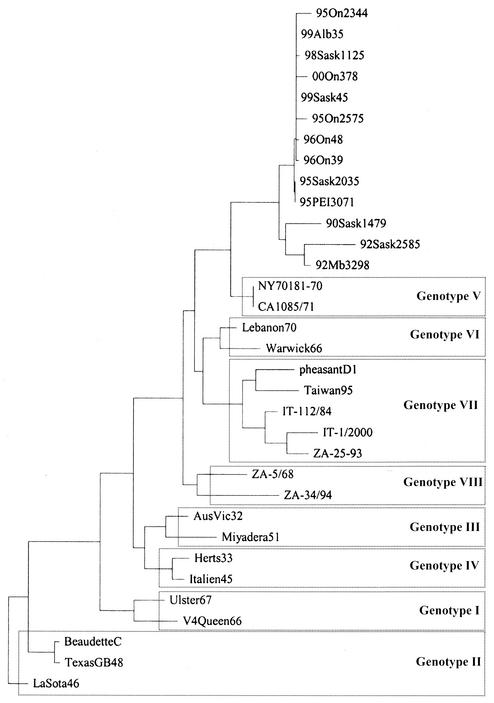
FIG. 1.
Phylogenetic relationships among APMV1 isolates representing individual genotypes and the cormorant isolates from Canada based on a 374-nucleotide sequence including the fusion protein cleavage site. Sequences previously published in GenBank are as follows: NY70181/70, parrot, United States, AF001105; CA1085/71, fowl, United States, AF001106; Lebanon70, AF001110; Warwick66, Z12111; pheasantD1, D-16/93, pheasant, Germany, AF001113; Taiwan95, U62620; IT-112/84, turkey, Bergamo, Italy, AF218127; IT-1/2000, chicken, Padua, Italy, AF293350; ZA-25-93, South AFrica, AF13676; ZA-5/68, ZA 5/(South Africa)/68, AF136762; ZA-34/94, ZA-34/(South Africa)/94, AF136773; AusVic32, Australia, Victoria/32, M21881; Miyadera51, M18456; Herts33, M24702; Italien45, Italien/45, M17710; Ulster67, Ulster 2C/67, D00243; V4Queen66, V4 Queensland/66, M24693; BeaudetteC, X04719; TexasGB48, Texas GB/48, M24692; LaSota46, LaSota/46, U22292.
Genetic characterization of the cormorant isolates was performed by evaluation of 374-nucleotide partial sequences of the variable region of the F gene (Fig. 1), by using corresponding sequences previously published in GenBank. The cormorant isolates appear to form a group related to genotype V in the system proposed by Ballagi- Pordany et al. (3), Lomniczi et al. (20), and Herczeg et al. (12). The phylogenetic distance from the “classical” genotype V ranges from 6.15 to 10.2% (distance matrix not shown).
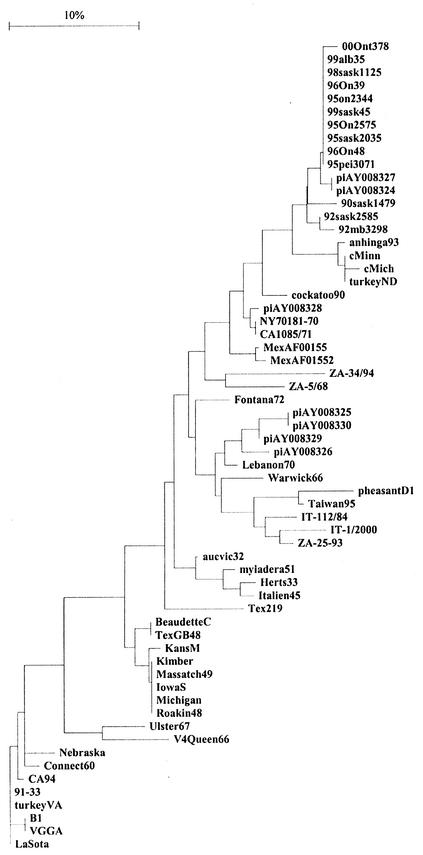
Despite the short sequence used in the analysis, the grouping of the viruses illustrated in Fig. 2 remained close to the grouping in Fig. 1. (The longest published common nucleotide sequence for the representative viruses of the individual genotypes and the North American isolates of AMPV1 is only 93 nucleotides.) Again all cormorant isolates fell into one subgroup, distinct from the remaining viruses in genotype V. Only the anhinga and two pigeon isolates clustered together with cormorant sequences.
FIG. 2.
Phylogenetic relationships among the North American isolates of APMV1 and the representative isolates for each genotype based on a 93-nucleotide sequence including the fusion protein cleavage site. Sequences previously published in GenBank are as follows: cMich 92, cormorant, Michigan, 1992, U22270; cMinn 92, cormorant, Minnesota, 1992, U22269; turkeyND 92, turkey, North Dakota, 1992, U22289; anhinga93, anhinga (exotic—South America)/USA/93, U22265; cockatoo90, cockatoo (Indonesia)/USA/90, AF015508; piAY008326, pigeon/USA/1984, AY008326; c99Sask45, cormorant/Saskatchewan (Canada)/45/1999, AF263615; piAY008329, pigeon/USA/1984, AY008329; piAY008324, pigeon/USA/1975, AY008324; piAY008330, pigeon/USA/1998, AY008330; piAY008327, pigeon/USA/1984, AY008327; piAY008327, pigeon/USA/1984, AY008327; piAY008328, pigeon/USA/1975, AY008328; Fontana, Fontana 72 (CA1083), U22274; Tex219, Texas 219, U22286; Connect60, Connecticut 60, AF206617; CA94, California/1994, U22267; VGGA, VGGA/turkey/USA/1989, U22273; 91-33, isolate 9133, U22288; turkeyVA, turkey, Virginia, U22285; Nebraska, Nebraska, U22282; IowaS, Iowa/Salsbury/1949, U22276; Kimber, Kimber/1947, U22278; Michigan, Michigan, U22281; Massatch49, Massachusetts/MK/1949, U22280; Roakin, Roakin 48 (United States), U22284; KansasM, Kansas/Manhattan/1948, U22277; MexAF015520, Mexico/1996, AF015520; MexAF015518, Mexico/1996, AF015518; B1, B1, U22266.
The finding is supported by previous work of other authors. Seal et al. (24, 25, 26) grouped the APMV1 isolated from parrots imported from South America and viruses isolated in the 1971 California epizootic in a way which in essence agrees with genotype V within the system used by European groups (3, 12, 13, 20). Considering the 6 to 10% phylogenetic distance from the “classical” genotype V, the cormorant isolates may represent a new genotype or a subtype within genotype V (Fig. 1). Clinical signs in cormorants reported for all the outbreaks indicated involvement of the central nervous system, and the isolates were considered neurotropic, velogenic, or mesogenic (4, 6, 10, 11, 18, 19, 21). Regretfully, Canada does not employ an in vivo pathogenicity test for chickens which allows distinction between viscerotropic and neurotropic isolates.
This work suggests that the 1995 to 2000 APMV1 isolates from cormorants may represent an indigenous APMV1 genotype present in the free-living wild bird population in North America, possibly waterfowl (14, 17), which intermittently causes high mortality in species such as cormorants or pigeons. Despite the rigorous biosecurity measures within the poultry industry, there is always the possibility of further spillover into domestic poultry as suggested during the 1992 cormorant outbreak, when a virus simultaneously isolated from a Newcastle disease virus outbreak in turkeys was closely related to the cormorant isolate from Minnesota (24).
REFERENCES
- 1.Afshar, A., G. C. Dulac, and A. Bouffard. 1989. Application of peroxidase labelled antibody assays for the detection of porcine IgG antibodies to hog cholera and bovine viral diarrhea viruses. J. Virol. Methods 23:253-262. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J. 1997. Newcastle disease and other avian Paramyxoviridae infections, p. 541-569. In B. W. Calnek (ed.), Diseases of poultry, 10th ed. Iowa State University Press, Ames.
- 3.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, M., W. M. Reed, S. D. Fitzgerald, and B. Panigraphy. 1994. Neurotropic velogenic Newcastle disease in cormorants in Michigan: pathology and virus characterization. Avian Dis. 38:873-878. [PubMed] [Google Scholar]
- 5.Beard, C. W. 1989. Serologic procedures, p. 192-200. In H. G. Purchase, L. H. Arp, C. H. Domermuth, and J. E. Pearson (ed.), A laboratory manual for the isolation and identification of avian pathogens, 3rd ed. Kendall/Hunt Publishing Company, Dubuque, Iowa.
- 6.Clavijo, A., Y. Robinson, and J. López. 2001. Isolation of Newcastle disease virus and Salmonella typhimurium from the brain of double-crested cormorants (Phalacrocorax auritus). Avian Dis. 45:245-250. [PubMed] [Google Scholar]
- 7.de Leeuw, O., and B. Peeters. 1999. Complete nucleotide sequence of Newcastle disease virus: evidence for the existence of a new genus within the subfamily Paramyxovirinae. J. Gen. Virol. 80:131-136. [DOI] [PubMed] [Google Scholar]
- 8.Docherty, D. E., and M. Friend. 1999. Newcastle disease, p. 175-179. In M. Friend, J. C. Field, and J. C. Franson (ed.), Manual of wildlife diseases: general field procedures and diseases of birds. Biological Resources Division, U.S. Geological Survey, Washington, D.C.
- 9.Erickson, G. A., C. J. Maré, G. A. Gustafson, L. D. Miller, and E. A. Carbrey. 1977. Interactions between viscerotropic velogenic Newcastle disease virus and pet birds of six species. II. Viral evolution through bird passage. Avian Dis. 21:655-669. [PubMed] [Google Scholar]
- 10.Glaser, L., I. K. Barker, D. V. C. Weseloh, J. Ludwig, R. M. Windingstad, D. W. Key, and T. K. Bollinger. 1999. The 1992 epizootic of Newcastle disease in double-crested cormorants in North America. J. Wild. Dis. 35:319-330. [DOI] [PubMed] [Google Scholar]
- 11.Heckert, R. A., M. S. Collins, R. J. Manvell, I. Strong, J. E. Pearson, and D. J. Alexander. 1996. Comparison of Newcastle disease viruses isolated from cormorants in canada and the USA in 1975, 1990 and 1992. Can. J. Vet. Res. 60:50-54. [PMC free article] [PubMed]
- 12.Herczeg, J., E. Wehmann, R. R. Bragg, P. M. Travassos Dias, G. Hadjiev, O. Werner, and B. Lomniczi. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in Southern Africa, one (VIIb) of which reached Southern Europe. Arch. Virol. 144:2087-2099. [DOI] [PubMed] [Google Scholar]
- 13.Herczeg, J., S. Pascucci, P. Massi, M. Luini, L. Selli, I. Capua, and B. Lomniczi. 2001. A longitudinal study of velogenic Newcastle disease virus genotypes isolated in Italy between 1960 and 2000. Avian Pathol. 30:163-168. [DOI] [PubMed] [Google Scholar]
- 14.Hinshaw, V. S., R. G. Webster, and B. Turner. 1980. The perpetuation of orthomyxoviruses and paramyxoviruses in Canadian waterfowl. Can. J. Microbiol. 26:622-629. [DOI] [PubMed] [Google Scholar]
- 15.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. H. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 16.Kaleta, E. F., and C. Baldauf. 1988. Newcastle disease in free-living and pet birds, p. 197-246. In D. J. Alexander (ed.), Newcastle disease. Kluwer Academic Publishers, Boston, Mass.
- 17.Kelleher, C. J., D. A. Halvorson, J. A. Newman, and D. A. Senne. 1985. Isolation of avian paramyxoviruses from sentinel ducks and turkeys in Minnesota. Avian Dis. 29:400-407. [PubMed] [Google Scholar]
- 18.Kuiken, T., F. A. Leighton, G. Wobeser, K. L. Danesik, J. Riva, and R. A. Heckert. 1998. An epidemic of Newcastle disease in double-crested cormorants from Saskatchewan. J. Wild. Dis. 34:457-471. [DOI] [PubMed] [Google Scholar]
- 19.Kuiken, T., G. Wobeser, F. A. Leighton, D. M. Haines, B. Chelack, J. Bogdan, L. Hassard, R. Heckert, and J. Riva. 1999. Pathology of Newcastle disease in double-crested cormorants from Saskatchewan, with comparison of diagnostic methods. J. Wild. Dis. 35:8-23. [DOI] [PubMed] [Google Scholar]
- 20.Lomniczi, B., E. Wehmann, J. Herczeg, A. Ballagi-Pordány, E. F. Kaleta, O. Werner, G. Meulemans, P. H. Jorgensen, A. P. Manté, A. L. J. Gielkens, I. Capua, and J. Damoser. 1998. Newcastle disease outbreaks in recent years in Western Europe were caused by an old (VI) and a novel genotype (VII). Arch. Virol. 143:49-64. [DOI] [PubMed] [Google Scholar]
- 21.Meteyer, C. U., D. E. Docherty, L. C. Glaser, J. C. Franson, D. A. Senne, and R. Duncan. 1997. Diagnostic findings in the 1992 epornitic of neurotropic velogenic Newcastle disease in double-crested cormorants from the upper Midwestern United States. Avian Dis. 41:171-180. [PubMed] [Google Scholar]
- 22.Peeters, B. P. H., O. S. de Leeuw, G. Koch, and A. L. J. Gielkens. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 73:5001-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 24.Seal, B. S., D. J. King, and J. D. Bennet. 1995. Characterization of Newcastle disease virus isolates by reverse transcription-PCR coupled to sequence database for pathotype prediction and molecular epidemiological analysis. J. Clin. Microbiol. 33:2624-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seal, B. S. 1996. Analysis of matrix protein gene nucleotide sequence diversity among Newcastle disease virus isolates demonstrates that recent disease outbreaks are caused by viruses of psittacine origin. Virus Genes 11:217-224. [DOI] [PubMed] [Google Scholar]
- 26.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 28.Wobeser, G., F. A. Leighton, R. Norman, D. J. Myers, D. Onderka, M. J. Pybus, J. L. Neufeld, G. A. Fox, and D. J. Alexander. 1993. Newcastle disease in wild water birds in western Canada, 1990. Can. Vet. J. 34:353-359. [PMC free article] [PubMed] [Google Scholar]
- 29.World Organization for Animal Health. 2000. Newcastle disease, p. 221-232. In Manual of standards for diagnostic tests and vaccines, 4th ed. Office International des Epizooties, Paris, France.