Abstract
To evaluate the prevalence of sulfate-reducing bacteria in septic processes, we searched for these bacteria by culture in 100 consecutive abdominal and pleural pus specimens. Twelve isolates were obtained from abdominal samples and were identified by a multiplex PCR as Desulfovibrio piger (formerly Desulfomonas pigra) (seven strains), Desulfovibrio fairfieldensis (four strains), and Desulfovibrio desulfuricans (one strain).
Sulfate-reducing bacteria (SRB) represent a class of anaerobic microorganisms that conduct dissimilatory sulfate reduction to obtain energy. In this process, sulfate reduction permits the dissimilatory oxidation of organic matter with the release of hydrogen sulfide, a corrosive and cytotoxic compound. SRB are present in the digestive tract (mouth and gut) of animals and humans (3, 17, 18). Human isolates consist of Desulfovibrio species, as Desulfomonas pigra has been recently reclassified as Desulfovibrio piger (2, 9, 19). So far, three species of Desulfovibrio have been isolated from human specimens: Desulfovibrio desulfuricans, Desulfovibrio fairfieldensis, and D. piger (7, 8, 10). These bacteria may play a role in the onset or the perpetuation of inflammatory chronic diseases such as inflammatory bowel disease (4, 8, 14) and periodontitis (10). Desulfovibrio spp. have also been isolated from abdominal and brain abscesses, blood, and urine (1, 5, 6, 7, 11, 12, 16). Most of these infectious processes were consecutive to digestive surgery. Whether SRB are etiologic agents of surgical abdominal infections is not known. Their prevalence in clinical samples is underestimated, as most medical laboratories do not specifically search for SRB. Additionally, these bacteria are seldom isolated because of their slow growth. Colonies appear after more than 3 days of incubation and are generally not noticed, being overgrown by the accompanying flora. Thus, their isolation requires a specific or selective growth medium. Once isolated, identification at the species level may be difficult. For example, it is not possible to differentiate D. desulfuricans and D. fairfieldensis by phenotypic tests, necessitating the use of genotypic tests to differentiate the two species. The aim of this study was to determine the prevalence of SRB in abdominal and pleural pus by using a specific liquid growth medium (Test-kit Labège; Compagnie Française de Géothermie, Orléans, France). A multiplex PCR was devised for the identification of the isolates at the species level.
Over a 6-month period, the consecutive purulent collections of 100 patients (55 women and 45 men) from different surgery units of the University Hospital of Strasbourg, Strasbourg, France, were studied. They consisted of abdominal (n = 88) and pleural (n = 12) samples. The mean age of the patients was 57 years (range, 2 to 95 years). Aerobic and anaerobic bacterial cultures were performed on all samples at the Laboratory of Bacteriology of the University Hospital of Strasbourg. For the specific culture of SRB, 200 μl of each sample was inoculated through the rubber cap of a ready-to-use specific liquid growth medium (Test-kit Labège). This medium was chosen, as it is more sensitive than the commonly used Postgate medium for isolating SRB from clinical samples (8, 10, 15). The inoculated media were incubated at 37°C for 1 month in an anaerobic chamber. SRB were detected by the formation of a black precipitate (ferrous sulfide) and were subsequently identified to species level by a multiplex PCR (8).
DNA extracts were obtained from 500 μl of the SRB-positive culture media (Test-kit Labège). After centrifugation and resuspension in 500 μl of Tris-EDTA buffer (10 mM Tris HCl, 1 mM EDTA, pH 8), bacterial cells were lysed by successively using lysozyme (3 mg ml−1), sodium dodecyl sulfate (1% [wt/vol), and proteinase K (0.25 mg ml−1). After an overnight incubation at 37°C, DNA was then extracted by the standard phenol-chloroform-isoamyl alcohol method. Each 50-μl PCR mixture contained 5 μl of DNA extract (approximately 50 ng of DNA) and final 0.4 μM concentrations of each of the six primers listed below, as well as a 0.8 mM concentration of each deoxynucleoside triphosphate (Boehringer Mannheim Biochemicals, Mannheim, Germany), 20 mM Tris HCl buffer (pH 8.4), 1.5 mM MgCl2, and 1.5 U of Taq DNA polymerase (Gibco-BRL Life Technologies, Paisley, United Kingdom). The primers, designed previously from 16S rRNA gene sequences (8), were Pig-F (5′-CTA GGG TGT TCT AAT CAT CAT CCT AC-3′), P687-R (5′-GAT ATC TAC GGA TTT CAC TCC TAC ACC-3′), Essex-F (5′-CTA CGT TGT GCT AAT CAG CAG CGT AC-3′), 27K-F (5′-CTG CCT TTG ATA CTG CTT AG-3′), 27K-R (5′-GGG CAC CCT CTC GTT TCG GAG A-3′), and Fair-F (5′-TGA ATG AAC TTT TAG GGG AAA GAC-3′) (Table 1). All reactions were carried out by using the GeneAmp PCR System 2400 (Applied Biosystems, Norwalk, Conn.). An initial denaturation step of 94°C for 4 min was followed by 30 cycles of denaturation (94°C, 1 min), annealing (55°C, 1 min), and extension (72°C, 2 min), with a final extension (72°C, 5 min). Amplified products were resolved by electrophoresis in 1.5% (wt/vol) agarose gels containing ethidium bromide (1.6 mg ml−1). A 100-bp DNA ladder was used as a size marker (Gibco-BRL Life Technologies). D. piger, D. desulfuricans strain Essex 6, D. desulfuricans strain MB, and D. fairfieldensis were identified by 255-, 255-, 396-, and 534-bp bands, respectively (Table 1). D. piger and D. desulfuricans Essex 6 were further differentiated by separate PCR assays with their respective specific primers.
TABLE 1.
Primers for the identification of Desulfovibrio strains
| Strain | Primers | Target site on 16S ribosomal DNAa | Length of PCR product (bp) |
|---|---|---|---|
| D. pigerb | Pig-F, P687-R | 453-708 | 255 |
| D. desulfuricans Essex 6 | Essex-F, P687-R | 453-708 | 255 |
| D. desulfuricans MB | 27K-F, 27K-R | 630-1026 | 396 |
| D. fairfieldensis | Fair-F, P687-R | 174-708 | 534 |
Escherichia coli numbering on 16S ribosomal RNA gene.
Formerly D. pigra.
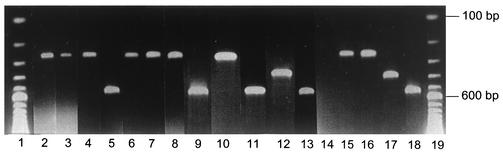
SRB were isolated from 12 samples, all of abdominal origin (10 peritoneal fluids and 2 intra-abdominal collections). The SRB-positive samples were from patients presenting with peritonitis, appendicitis, or abscess after surgery for rectal cancer (Table 2). SRB were identified by PCR as D. piger (seven strains), D. fairfieldensis (four strains), and D. desulfuricans MB (one strain) (Fig. 1). In all cases, SRB isolates were part of mixed aerobic and anaerobic flora. No SRB were detected in the 12 purulent pleural fluids studied.
TABLE 2.
Characteristics of patients from whom SRB were isolated
| Patient no. | Sex | Age (yr) | Sample | Clinical findings | SRB speciesa |
|---|---|---|---|---|---|
| 1 | Male | 64 | Peritoneal fluid | Peritonitis | D. pigerb |
| 2 | Female | 83 | Peritoneal fluid | Peritonitis | D. piger |
| 3 | Female | 81 | Peritoneal fluid | Rectal cancer | D. piger |
| 4 | Male | 32 | Peritoneal fluid | Appendicitis, peritonitis | D. fairfieldensis |
| 5 | Female | 88 | Peritoneal fluid | Peritonitis | D. piger |
| 6 | Male | 14 | Intra-abdominal collection | Appendicitis | D. piger |
| 7 | Male | 18 | Peritoneal fluid | Peritonitis | D. piger |
| 8 | Female | 29 | Peritoneal fluid | Appendicitis, peritonitis | D. fairfieldensis |
| 9 | Male | 9 | Peritoneal fluid | Appendicitis, peritonitis | D. piger |
| 10 | Female | 53 | Peritoneal fluid | Peritonitis | D. fairfieldensis |
| 11 | Male | 80 | Peritoneal fluid | Peritonitis | D. desulfuricans |
| 12 | Male | 21 | Intra-abdominal collection | Appendicitis | D. fairfieldensis |
SRB were identified by PCR.
Formerly D. pigra.
FIG. 1.
Multiplex PCR products obtained with 12 clinical samples and 4 collection strains of Desulfovibrio. Lanes: 1 and 19, 100-bp DNA ladder; 2 to 13, patients 1 to 12; 14, negative control (water); 15, D. piger (formerly D. pigra) ATCC 29098T; 16, D. desulfuricans strain Essex 6 (ATCC 29577T); 17, D. desulfuricans strain MB (ATCC 27774); and 18, D. fairfieldensis ATCC 700045.
SRB have been described in the digestive tract of animals and humans for many years. In humans, neither prevalence nor identification as to predominant species has been determined. Recently, the study of feces from healthy individuals and patients has shown that D. piger is the predominant species in humans (8). Until that study, D. piger had been reported only once in human feces (13). Our results also suggest that it is the most common species of SRB in human abdominal pus, which is a logical finding since the flora of abdominal pus is of enteric origin. In contrast, D. fairfieldensis occurs less commonly in the gut and in purulent abdominal specimens but has been isolated from blood (7, 12) and periodontal pockets (10). D. fairfieldensis is the only species of SRB found in pure culture in septic processes outside the abdomen.
The type species of the genus Desulfovibrio, D. desulfuricans, is commonly isolated from the environment. It has also been considered the most prevalent species of Desulfovibrio in humans (4, 19). However, this species appears to be an uncommon organism in the human intestinal tract, as evidenced by the fact that there was only one isolate obtained from 88 abdominal specimens in this work and that only one isolate was obtained from 151 human feces samples in a previous study (8). These results suggest that D. desulfuricans may occasionally be present in humans as a result of a contamination from the environment but that it is not a common commensal or infectious agent.
The involvement of SRB in pathological processes has recently been proposed. D. fairfieldensis may present with invasive properties that explain its presence in the bloodstream and abscesses (7, 12, 16). Its association with periodontitis deserves further investigation. While D. piger has never been isolated from extra-abdominal samples, recent findings suggest a relationship between this bacterium and inflammatory bowel diseases (8). However, the role of SRB in the digestive tract is still not understood. D. piger and D. fairfieldensis have never been isolated outside human samples, indicating that they may be natural residents of the human digestive tract. The search for these bacteria in various ecological niches should be undertaken in an attempt to further elucidate their role in the human host.
Acknowledgments
We are indebted to the late Wee Tee (University of Melbourne, Melbourne, Australia) for kindly providing four strains of D. fairfieldensis. We thank Muriel Thirion (Laboratoire de Bactériologie, CHU de Strasbourg) for her excellent technical assistance.
REFERENCES
- 1.Baron, E. J., R. Bennion, J. Thompson, C. Strong, P. Summanen, M. McTeague, and S. M. Finegold. 1992. A microbiological comparison between acute and complicated appendicitis. Clin. Infect. Dis. 14:227-231. [DOI] [PubMed] [Google Scholar]
- 2.Gibson, G. R., G. T. McFarlane, and J. H. Cummings. 1988. Occurrence of sulphate-reducing bacteria in human faeces and the relationship of dissimilatory sulphate reduction to methanogenesis in the large gut. J. Appl. Bacteriol. 65:103-111. [DOI] [PubMed] [Google Scholar]
- 3.Gibson, G. R. 1990. Physiology and ecology of sulphate-reducing bacteria. J. Appl. Bacteriol. 69:769-797. [DOI] [PubMed] [Google Scholar]
- 4.Gibson, G. R., J. H. Cummings, and G. T. McFarlane. 1991. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol. Ecol. 86:103-112. [Google Scholar]
- 5.Johnson, C. C., and S. M. Finegold. 1987. Uncommonly encountered, motile, anaerobic gram-negative bacilli associated with infection. Rev. Infect. Dis. 9:1150-1162. [DOI] [PubMed] [Google Scholar]
- 6.La Scola, B., and D. Raoult. 1999. Third human isolate of a Desulfovibrio sp. identical to the provisionally named Desulfovibrio fairfieldensis.J. Clin. Microbiol. 37:3076-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loubinoux, J., F. Mory, I. A. C. Pereira, and A. E. Le Faou. 2000. Bacteremia caused by a strain of Desulfovibrio related to the provisionally named Desulfovibrio fairfieldensis. J. Clin. Microbiol. 38:931-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loubinoux, J., J.-P. Bronowicki, I. A. C. Pereira, J.-L. Mougenel, and A. E. Le Faou. 2002. Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40:107-112. [DOI] [PubMed] [Google Scholar]
- 9.Loubinoux, J., F. M. A. Valente, I. A. C. Pereira, A. Costa, P. A. D. Grimont, and A. E. Le Faou. 2002. Reclassification of the only species of the genus Desulfomonas, Desulfomonas pigra, as Desulfovibrio piger comb. nov. Int. J. Syst. E vol. Microbiol. 52:1305-1308. [DOI] [PubMed] [Google Scholar]
- 10.Loubinoux, J., C. Bisson-Boutelliez, N. Miller, and A. E. Le Faou. 2002. Isolation of the provisionally named Desulfovibrio fairfieldensis from human periodontal pockets. Oral Microbiol. Immunol. 17:321-323. [DOI] [PubMed] [Google Scholar]
- 11.Lozniewski, A., P. Maurer, H. Schuhmacher, J. P. Carlier, and F. Mory. 1999. First isolation of Desulfovibrio sp. as part of a polymicrobial infection from a brain abscess. Eur. J. Clin. Microbiol. Infect. Dis. 18:602-603. [DOI] [PubMed] [Google Scholar]
- 12.MacDougall, R., J. Robson, D. Paterson, and W. Tee. 1997. Bacteremia caused by a recently described novel Desulfovibrio species. J. Clin. Microbiol. 35:1805-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore, W. E. C., J. L. Johnson, and L. V. Holdeman. 1976. Emendation of Bacteroidaceae and Butyrivibrio and descriptions of Desulfomonas gen. nov. and ten new species in the genera Desulfomonas, Butyrivibrio, Eubacterium, Clostridium, and Ruminococcus. Int. J. Syst. Bacteriol. 26:238-252. [Google Scholar]
- 14.Pitcher, M. C. L., and J. H. Cummings. 1996. Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut 39:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 16.Tee, W., M. Dyall-Smith, W. Woods, and D. Eisen. 1996. Probable new species of Desulfovibrio isolated from a pyogenic liver abscess. J. Clin. Microbiol. 34:1760-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van der Hoeven, J. S., C. W. A. Van den Kieboom, and M. J. M. Schaeken. 1995. Sulfate-reducing bacteria in the periodontal pocket. Oral Microbiol. Immunol. 10:288-290. [DOI] [PubMed] [Google Scholar]
- 18.Willis, C. L., G. R. Gibson, C. Allison, S. MacFarlane, and J. S. Holt. 1995. Growth, incidence and activities of dissimilatory sulfate-reducing bacteria in the human oral cavity. FEMS Microbiol. Lett. 129:267-272. [DOI] [PubMed] [Google Scholar]
- 19.Willis, C. L., J. H. Cummings, G. Neale, and G. R. Gibson. 1997. Nutritional aspects of dissimilatory sulfate reduction in the human large intestine. Curr. Microbiol. 35:294-298. [DOI] [PubMed] [Google Scholar]