Abstract
An internal control DNA (ICD) with the same primer binding sequences as the target Chlamydia trachomatis DNA was constructed and evaluated in a PCR assay with immunoenzymatic detection. One hundred urine specimens were tested, and 23 were found to contain inhibitors of the PCR, if not subjected to DNA extraction prior to amplification. Coamplification and detection of the ICD appeared to be a useful method for estimating the effects of inhibitors on C. trachomatis DNA amplification.
Chlamydia trachomatis is a major cause of sexually transmitted disease worldwide (2). In women, infection is frequently asymptomatic, and untreated infections may progress to endometritis, salpingitis, and infertility (1). Thus, the early and rapid detection of these infections is essential. C. trachomatis DNA may be detected by PCR in urine specimens from infected patients (3). However, careful attention must be paid to the possibility of false-negative results in the PCR test due to the presence of compounds that inhibit Taq polymerase, such as hemoglobin and urea in urine (4). Feminine sprays and talcum powder may also interfere with the results of amplification assays (GenProbe BioMérieux technical note). The inclusion of an appropriate internal control DNA (ICD) for coamplification makes it possible to distinguish between PCR failure and truly negative results. A positive result from the ICD target indicates that the amplification reaction was not inhibited, thereby validating a negative result for the primary target.
The overlap extension technique can be used to construct an ICD with the same primer binding sequences as the target DNA (6). However, there is a difference in size between the ICD and the natural amplification products. Using the same primers is an advantage, because multiple sets of primers might interfere with the amplification of one or both of the target genes, due to differences in the primer sequences, sizes and internal sequences of the amplified products, and the relative amounts of the two targets (7).
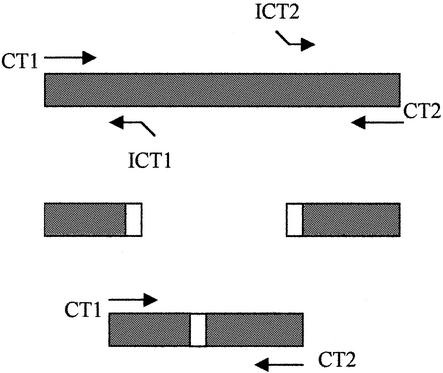
The strategy for constructing ICDs is illustrated in Fig. 1. Primer CT1 (5′-TAGTAACTGCCACTTCATCA-3′) and primer CT2 (5′-biotin-TTCCCCTTGTAATTCGTTGC-3′) are flanking primers that amplify a 201-bp PCR product from C. trachomatis plasmid orf2 DNA (5). The primers ICT1 (5′-AATGCGCAAGCCTGATGTCAGTCAACTTCTGATTTTCAAG-3′) and ICT2 (5′-ATCAGGCTTGCGCATTGCTAGCTACATTACCATGCATTAG-3′) are internal primers. The nucleotides shown in italics are derived from the C. trachomatis sequence. The underlined nucleotides at the 5′ ends are complementary to each other. The 3′ ends of the ICT1 and ICT2 primers bind close to the two ends of the C. trachomatis amplicon. PCR on C. trachomatis genomic DNA was used to generate two smaller DNA products. A 50-bp DNA fragment and a 59-bp DNA fragment were produced with primers CT1 and ICT1 and primers ICT2 and CT2, respectively. The two DNA fragments have overlapping ends due to the complementary sequences at the 5′ ends of primers ICT1 and ICT2 (Fig. 1). These two DNA fragments were used in an additional PCR with the CT1 and CT2 primers. In this reaction, one strand from each fragment contained the overlap sequence and could therefore serve as a primer for the other. Extension of the overlap by Taq polymerase yielded a new 125-bp PCR product, 76 bp shorter than the 201-bp C. trachomatis amplicon produced directly with CT1 and CT2, as visualized on an ethidium bromide-stained agarose gel (data not shown).
FIG. 1.
Construction of internal control DNA by overlap extension. CT1 and CT2 are the primers used for C. trachomatis PCR amplification. ICT1 and ICT2 are designed to bind close to CT1 and CT2. The open boxes at the 5′ ends of the primers indicate complementary sequences.
The 125-bp ICD was purified with the Wizard PCR Preps purification system (Promega Corporation, Madison, Wis.) and inserted into the pGEM T Easy cloning vector (Promega). The recombinant DNA was used to transform competent Escherichia coli JM109 cells, and the ICD was prepared with the Wizard Plus Minipreps DNA purification system (Promega). We used 1 μl of the preparation, containing about 102 copies of the ICD, in subsequent PCR for evaluation of the use of the ICD for urine samples. Thirty-five amplification cycles of 59°C for 1 min, 72°C for 45 s, and 93°C for 30 s were performed.
We designed a C. trachomatis-specific 5′-digoxigenin (DIG)-labeled probe (5′-GCTCAAAATGGGATGG-3′), corresponding to the central region of the C. trachomatis amplicon, which is not contained in the ICD. An ICD-specific 5′-DIG-labeled internal control (IC) probe (5′ ATCAGGCTTGCGCATT 3′), corresponding to the complementary sequences of the ICT1 and ICT2 primers not present in the C. trachomatis target sequence, was also designed. Unique probe binding regions differentiate amplified ICD from target DNA. The ICD described in this study has the same primer binding sequences and the same internal sequence base composition as the C. trachomatis target DNA. Therefore, its amplification should not affect the efficiency of target DNA amplification. After each amplification reaction, the CT probe and the IC probe were added to separate microplate wells to detect amplified target DNA and amplified ICD, respectively, in a colorimetric enzyme immunoassay. Each PCR product was diluted 1/10 in 1× SSC (0.15 M NaCl plus 0.15 M sodium citrate)-0.5% Tween 20 in streptavidin-coated microplate wells, washed with 100 mM Tris-HCl (pH 7.5)-150 mM NaCl, denatured with 0.1 N NaOH, hybridized to appropriate DIG-labeled probes (CT probe and IC probe) at a concentration of 10 pmol/ml in Tris-buffered saline buffer, and detected with an alkaline phosphatase-conjugated anti-DIG antibody/pNPP (paranitrophenyl phosphate) substrate. Samples were considered positive if they gave an A405 of ≥0.05.
As little as one copy of ICD could be detected in a PCR. Various amounts of ICD were incorporated in individual PCR mixtures containing 5 pg of C. trachomatis DNA (equivalent of three elementary bodies [EB]). The addition of 1, 10, or 100 ICD copies did not affect the target DNA signal. Competition with C. trachomatis DNA amplification was observed only at higher ICD copy concentrations. However, detection of the ICD was negatively correlated with C. trachomatis EB copy number in urine, suggesting competitive inhibition of ICD amplification in the presence of large numbers of EB (Table 1). Therefore, competition cannot cause a false-negative result, because an excess of EB will result in a positive signal for the primary C. trachomatis target, even in the absence of a positive signal for the ICD target. False-positive results due to reagent contamination were ruled out by the systematic use of two negative controls: one “no DNA” negative control, which reflects the total reagent handled, and one negative sample control, which had gone through all the sample preparation steps.
TABLE 1.
A405 of PCR amplification products in the colorimetric enzyme immunoassaya
| Amplification products |
A405 with:
|
|
|---|---|---|
| CT probe | IC probe | |
| 30 C. trachomatis EB | 1.5 | <0.05 |
| 102 ICD copies | <0.05 | 0.75 |
| 3 C. trachomatis EB and 1 ICD copy | 1.3 | <0.05 |
| 3 C. trachomatis EB and 10 ICD copies | 1.3 | 0.21 |
| 3 C. trachomatis EB and 102 ICD copies | 1.1 | 0.54 |
| 30 C. trachomatis EB and 102 ICD copies | 1.4 | 0.19 |
| 3 × 102C. trachomatis EB and 102 ICD copies | 1.6 | 0.12 |
| 3 × 103C. trachomatis EB and 102 ICD copies | 2 | <0.05 |
| 3 C. trachomatis EB and 102 ICD copies in urine without DNA extraction | 0.16 | 0.67 |
| 3 C. trachomatis EB and 102 ICD copies in urine with DNA extraction | 0.32 | 0.82 |
Values are means of three independent assays.
We evaluated ICD in tests with 100 clinical urine samples. Urine samples were obtained from 100 patients attending the Center for Sexually Transmitted Diseases (Amiens, France). These samples were tested for C. trachomatis by PCR and then seeded with 10 C. trachomatis EB and treated with DNA extraction (MasterPure DNA purification kit; Epicentre Technologies) and without DNA extraction (the urine centrifugation pellet was dissolved in 10 mM Tris-HCl [pH 7.5]-2.5 mM MgCl2-0.45% Triton X-100-0.45% Tween 20-200 μg of proteinase K/ml and incubated for 1 h at 60°C and for 10 min at 100°C). When the urine samples were initially tested, 2% were positive for C. trachomatis. Several urine samples, containing 300, 30, or 3 C. trachomatis EB, were tested. If urine samples were subjected to DNA extraction before the procedure for amplification and detection, ICD amplification gave a positive result for 100% of the samples (mean A405 = 1.06). In contrast, if urine samples were processed without DNA extraction, ICD amplification gave a positive result for only 77% of the samples (mean A405 = 0.76).
Our results suggest that the use of an ICD is absolutely necessary if urine samples are not subjected to a DNA extraction procedure before amplification, as 23% of urine samples prepared in this way contained substances that inhibited the PCR. However, inhibitory substances are efficiently eliminated by DNA extraction (0% PCR inhibition).
Furthermore, very small amounts of C. trachomatis target DNA (3 to 30 EB) were efficiently amplified only after DNA extraction (Table 1). Such small amounts of C. trachomatis target DNA were generally not amplified in the absence of DNA extraction, even if the ICD was efficiently amplified. PCR inhibitors, probably still present after detergent lysis treatment, decrease amplification efficiency, thereby reducing the amount of PCR products generated and decreasing the signal generated from each amplification product. Once the urine sample had been subjected to DNA extraction, the inclusion of 102 copies of ICD in the PCR mixture allowed the efficient amplification of as little DNA as the equivalent of three C. trachomatis EB (Table 1). Our results suggest that the sensitivity of the PCR assay depends on the sample preparation procedure; the inclusion of the ICD provides further assurance that clinical specimens are successfully amplified and detected. We used this ICD to demonstrate that the frequency of inhibition in the PCR test ranged from 0% if DNA extraction was performed before amplification to 23% if it was not performed before amplification. Thus, the incorporation of an ICD into PCR-based tests may be superfluous or necessary depending on the sample preparation procedure used.
Acknowledgments
This work was supported by funds from the Conseil Régional de la Picardie.
REFERENCES
- 1.Brunham, R. C., I. W. Maclean, B. Binns, and R. Peeling. 1985. Chlamydia trachomatis: its role in tubal infertility. J. Infect. Dis. 152:1275-1282. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control. 1985. Chlamydia trachomatis infection. Policy guidelines for prevention and control. Morb. Mortal. Wkly. Rep. 35:535-574. [Google Scholar]
- 3.DiDomenico, N., H. Link, R. Knobel, T. Caratsch, W. Weschler, Z. G. Loewy, and M. Rosenstraus. 1996. COBAS AMPLICOR: a fully automated RNA and DNA amplification and detection system for routine diagnostic PCR. Clin. Chem. 42:1915-1923. [PubMed] [Google Scholar]
- 4.Gerritsen, M. J., T. Olyhoek, M. A. Smits, and B. A. Bokhout. 1991. Sample preparation method for polymerase chain reaction-based semiquantitative detection of Leptospira interrogans serovar hardjo subtype hardjobovis in bovine urine. J. Clin. Microbiol. 29:2805-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffais, R., and M. Thibon. 1989. Detection of Chlamydia trachomatis by the polymerase chain reaction. Res. Microbiol. 140:139-141. [DOI] [PubMed] [Google Scholar]
- 6.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes; gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 7.Siebert, P. D., and J. W. Larrick. 1992. Competitive PCR. Nature 359:557-558. [DOI] [PubMed] [Google Scholar]