Abstract
A comprehensive assay for the identification of all eight human herpesviruses has been previously reported. This assay was extended to the detection and species-level identification of herpes B virus (Cercopithecine herpesvirus 1) and African green monkey cytomegalovirus (Cercopithecine herpesvirus 5), two herpesviruses of relevance to the clinical virology laboratory.
A previous report from our laboratory described a comprehensive PCR-based method for the detection and species-level identification of all known human herpesviruses (1). In this study we report on the application of this protocol to the detection and species-level identification of two primate herpesviruses.
Herpes B virus (Herpesvirus simiae, Cercopithecine herpesvirus 1 [CeHV-1]) is an alphaherpesvirus commonly found in macaques, such as the rhesus monkey (Macaca mulatta), and is highly pathogenic for humans. Infection can be acquired through direct contact of mucosa or broken skin with saliva or secretions from an infected shedding macaque, by a monkey bite (2, 9), or from infected primary rhesus monkey cells in tissue culture (8) used in clinical and research laboratories. As the infection is lethal in 80% of untreated cases (9), rapid diagnosis and instigation of antiviral therapy are essential. Culture of herpes B virus requires level 3 containment facilities for initial isolation followed by level 4 for propagation (6). Therefore, detection through molecular techniques would be safer as the infectivity of the virus is destroyed in the DNA extraction process, and PCR techniques are usually more sensitive than traditional culture (9).
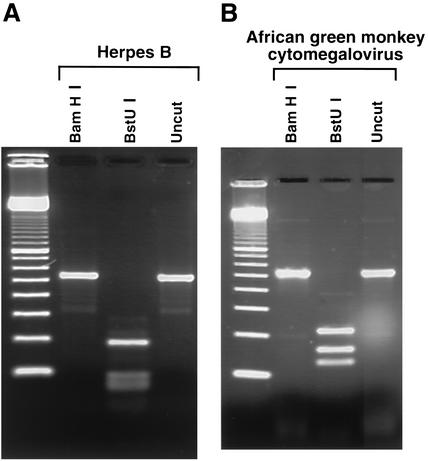
In this study we used the herpes B virus strain E2490 from M. mulatta. Viruses were propagated in Vero cells at the biosafety level 4 laboratory of the Special Pathogens Program, Canadian Science Centre for Human and Animal Health, Winnipeg, Manitoba, Canada. The DNA from cell supernatant was extracted with the QIAamp DNA minikit (catalog no. 51304). The DNA suspension, at a concentration of approximately 20 μg/ml, was received by the Molecular Microbiology Laboratory at the Hospital for Sick Children, Toronto, Ontario, Canada, and was reextracted with guanidium thiocyanate, as described previously (1), to rule out any possibility of working with infective virions of this restricted agent under the containment conditions of a clinical virology laboratory. The resulting DNA suspension was serially diluted for determination of the PCR sensitivity. PCR was performed as described previously (1). In the PCR assay with the primer pair HSV-P1 and HSV-P2 (1), an amplicon of a length comparable to that from human herpesviruses (1) was obtained, from as little as 10−4 ng of template DNA. The patterns obtained by digestion with BamHI and BstUI shown in Fig. 1A were distinct from the patterns previously obtained from human herpesviruses (Fig. 2 in reference 1). Direct sequencing of the amplicon by using the PCR primers as sequencing primers proved to be difficult, possibly because of its high GC content. Therefore, the amplicon was cloned in a plasmid, by using the TA cloning kit (Invitrogen), and sequenced with the M13 forward and reverse primers. The sequencing was performed at the DNA Sequencing Facility, Centre for Applied Genomics, Hospital for Sick Children.
FIG. 1.
Restriction enzyme digestion patterns for herpes B virus (A) and AGM CMV (B). For herpes B virus, BstUI cuts the DNA at eight positions and only the larger fragment is clearly distinguishable.
Relevant properties of the amplicon are summarized in Table 1. The estimated sensitivity of our assay, 10−4 ng, would correspond to approximately 600 genome copies. This is probably an underestimate of the sensitivity, since it relied on the measurement of the DNA recovered from culture supernatant (which probably contained cellular DNA), since it assumes a 100% recovery of the DNA during the second extraction, and since the molecular size of 162 kb for the herpes B virus genome used in the calculation is only an estimate (9). Nonetheless, the sensitivity is comparable to that of other herpes B virus-specific PCR assays, i.e., 100 or 700 genome copies, respectively (3, 4).
TABLE 1.
Characteristics of the amplicons
| Virus | Length (bp) | % G+C content | BamHI fragmenta | BstUI fragments (bp) |
|---|---|---|---|---|
| Herpes B virus (CeHV-1) | 532 | 68.4 | 532 | 191, 91, 76, 62, 56, 25, 19, 10, 2 |
| AGM CMV (CeHV-5) | 523 | 56.0 | 523 | 218, 162, 130, 13 |
Neither amplicon contained a BamHI site.
African green monkey (AGM) cytomegalovirus (CMV) (Cercopithecine herpesvirus 5, CeHV-5 [2]) has long been recognized as an adventitious virus in primary AGM cell cultures (5), including commercial preparations of primary AGM kidney cells. The virus causes a distinct cytopathic effect, and examination of the cell lysate by electron microscopy reveals particles with the typical herpesvirus morphology (5). It is important to accurately identify this agent, since the recovery of a herpesvirus in a cell culture inoculated with a clinical sample can lead to an erroneous diagnosis. In addition, AGM CMV can interfere with the replication of other clinically relevant viruses, such as enteroviruses, myxoviruses, arboviruses, and other herpesviruses (5). In this study AGM primary kidney cells purchased from Bio Whittaker (Walkersville, Md.) were used. In one tube that had been inoculated with a cerebrospinal fluid specimen, a cytopathic effect similar to that of AGM CMV was observed. Extraction of the DNA from the supernatant and PCR were performed as described previously (1). With the primer pair HSV-P1 and HSV-P2 an amplicon of a length comparable to that of other herpesviruses (1) was obtained. In contrast, DNA extracted from the original cerebrospinal fluid specimen tested negative. The patterns obtained by restriction enzyme digestion, shown in Fig. 1B, were distinct from previous patterns (Fig. 2 in reference 1). Both strands of the amplicon were sequenced by using the PCR primers as sequencing primers.
Relevant properties of the amplicon are summarized in Table 1. The sequence was compared with a previously published sequence of a 171-bp segment of the DNA polymerase gene of the AGM CMV (7) (accession number U63460), and an agreement of 166 of 171 bp (97%) was observed, confirming the identity of the virus.
In summary we have shown that the comprehensive PCR for human herpesviruses, which was previously reported (1), can detect and identify two primate herpesviruses that are relevant to the clinical virology laboratory. The assay can be used for quality control of primary cell lines, and in the case of herpes B virus it may be ideally suited for analyzing clinical samples to provide a faster, safer, and more sensitive laboratory diagnostic method than viral isolation.
Nucleotide sequence accession number.
The sequence of the 476-bp segment of the herpes B amplicon internal to the primers was deposited in GenBank (accession number AY117753). The sequence of the 467-bp segment of the AGM CMV amplicon internal to the primers was deposited in GenBank (accession number AY117754).
Acknowledgments
This work was supported by the Department of Paediatric Laboratory Medicine, and the Research Institute, Hospital for Sick Children, Toronto.
REFERENCES
- 1.Johnson, G., S. Nelson, M. Petric, and R. Tellier. 2000. Comprehensive PCR-based assay for detection and species identification of human herpesviruses. J. Clin. Microbiol. 38:3274-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roizman, B., and P. E. Pellett. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 3.Scinicariello, F., R. Eberle, and J. K. Hilliard. 1993. Rapid detection of B virus (herpesvirus simiae) DNA by polymerase chain reaction. J. Infect. Dis. 168:747-750. [DOI] [PubMed] [Google Scholar]
- 4.Slomka, M. J., D. W. G. Brown, J. P. Clewley, A. M. Bennett, L. Harrington, and D. C. Kelly. 1993. Polymerase chain reaction for detection of herpesvirus simiae (B virus) in clinical specimens. Arch. Virol. 131:89-99. [DOI] [PubMed] [Google Scholar]
- 5.Smith, K. O., J. F. Thiel, J. T. Newman, E. Harvey, M. D. Trousdale, W. D. Gehle, and G. Clark. 1969. Cytomegalovirus as common adventitious contaminants in primary African green monkey kidney cell cultures. J. Natl. Cancer Inst. 42:489-497. [PubMed] [Google Scholar]
- 6.U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. 1999. Biosafety in microbiological and biomedical laboratories. U.S. Government Printing Office, Washington, D.C.
- 7.VanDevanter, D. R., P. Warrener, L. Bennett, E. R. Schultz, S. Coulter, R. L. Garber, and T. M. Rose. 1996. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 34:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wells, D. L., S. L. Lipper, J. K. Hilliard, J. A. Stewart, G. P. Holmes, K. L. Herrmann, M. P. Kiley, and L. P. Schonberger. 1989. Herpesvirus simiae contamination of primary rhesus monkey kidney cell cultures. CDC recommendations to minimize risks to laboratory personnel. Diagn. Microbiol. Infect. Dis. 12:333-336. [DOI] [PubMed] [Google Scholar]
- 9.Whitley, R. J., and J. K. Hilliard. 2001. Cercopithecine herpesvirus (B virus), p. 2835-2848. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.