Abstract
A second form of the enzyme acyl-CoA:cholesterol acyltransferase, ACAT2, has been identified. To explore the hypothesis that the two ACAT enzymes have separate functions, the membrane topologies of ACAT1 and ACAT2 were examined. A glycosylation reporter and FLAG epitope tag sequence was appended to a series of ACAT cDNAs truncated after each predicted transmembrane domain. Fusion constructs were assembled into microsomal membranes, in vitro, and topologies were determined based on glycosylation site use and accessibility to exogenous protease. The accessibility of the C-terminal FLAG epitope in constructs was determined by immunofluorescence microscopy of permeabilized transfected cells. Both ACAT1 and ACAT2 span the membrane five times with their N termini in the cytosol and C termini in the ER lumen. The fourth transmembrane domain is located in a different region for each protein, placing the putative active site ACAT1 serine (Ser269) in the cytosol and the analogous residue in ACAT2 (Ser249) in the ER lumen. Mutation of these serines inactivated the ACAT enzymes. The outcome is consistent with the hypothesis that cholesterol ester formation by ACAT2 may be coupled to lipoprotein particle assembly and secretion, whereas ACAT1 may function primarily to maintain the balance of free and esterified cholesterol intracellularly.
INTRODUCTION
In nonhuman primates, a striking association between the amount of coronary artery atherosclerosis and liver microsomal Acyl-CoA:cholesterol acyltransferase (ACAT) activity (Carr et al., 1992) and hepatic cholesterol ester secretion during liver perfusion (Rudel et al., 1997) was found, implicating hepatic ACAT as an important contributor to the atherogenic properties of apolipoprotein B (apoB)-containing lipoproteins. Studies in isolated, perfused livers from monkeys demonstrated that inhibition of ACAT activity was associated with decreased cholesteryl ester and apoB accumulation in recirculating perfusate (Carr et al., 1995). Studies in other animals and cultured cells have also implicated a role for hepatic ACAT in the regulation of apoB-containing lipoprotein particle assembly and secretion (Cianflone et al., 1990; Huff et al., 1994; Wilcox et al., 1999). Pharmacological inhibition of ACAT resulted in reduced diet-induced plasma low-density lipoprotein cholesterol and atherosclerosis in rabbits and minipigs (Bocan et al., 1991, 1993). Such findings highlight the importance of determining the specific roles of ACAT where atherosclerosis prevention is a goal.
Two forms of the enzyme ACAT are known to exist, ACAT1 and ACAT2 (Anderson et al., 1998; Cases et al., 1998; Oelkers et al., 1998). The N-terminal 119 amino acid residues of ACAT1 and 101 amino acid residues of ACAT2 show no sequence similarity; however, the remaining amino acid sequences are highly conserved, consistent with their similar enzymatic activities (Anderson et al., 1998). ACAT-derived cholesterol esters have at least two distinct roles in animal physiology: 1) formation of cytoplasmic lipid droplets for storage and 2) assembly into nascent apoB-containing lipoprotein particles in the endoplasmic reticulum (ER) lumen. An indication that the two functions may be performed by different ACAT enzymes was obtained from analysis of the relative tissue distributions of ACAT1 and ACAT2. By Northern analysis of tissues from monkeys and mice, ACAT1 mRNA was ubiquitous among all tissues examined, whereas ACAT2 mRNA was found primarily in the liver and intestine (Anderson et al., 1998; Cases et al., 1998). Reverse transcription-polymerase chain reaction (PCR) analyses indicated that low levels of ACAT2 mRNA may have also been present in several other tissues, but the physiological significance for this finding remains uncertain because the levels were strikingly lower than the levels seen in liver and intestine by Northern analysis (Cases et al., 1998; Oelkers et al., 1998). Immunostaining in monkey liver revealed that ACAT2 is confined predominately to hepatocytes, whereas ACAT1 is localized to Kupffer cells (Lee et al., 1999). In the intestine, ACAT2 is localized to the apical portion of mucosal cells, the site of chylomicron particle assembly (Lee et al., 1999, 2000). ACAT2 was not identified in tissues other than liver and intestine by immunostaining. Thus, it appears that many cells express ACAT1, consistent with its role in intracellular cholesterol homeostasis, whereas ACAT2 is confined to cells engaged in the assembly and secretion of apoB-containing lipoproteins.
In addition to differences in tissue distribution, initial computer-based prediction methods suggested that ACAT1 and ACAT2 display different topologies in the ER membrane (Anderson et al., 1998). These models placed the serine residue, thought to be required for activity, on opposite sides of the membrane. Consistent with a possible role in lipoprotein assembly, the ACAT2 serine (Ser249) was proposed to reside in the ER lumen, whereas the analogous serine in ACAT1 (Ser269) was cytosolic. Recently, Chang et al. (1998) experimentally analyzed ACAT1 topology (Lin et al., 1999) and suggested that the protein contains seven transmembrane domains, with Ser269 (Cao et al., 1996) positioned on the cytoplasmic side of the membrane. However, an analysis of ACAT2 topology has not been performed nor have studies been performed to specifically address the possibility that the two enzymes display different topologies at the level of the ER membrane.
In the present report, a detailed comparison of the membrane topology of both ACAT1 and ACAT2 was performed by fusing a reporter peptide after each predicted transmembrane domain identified by computer modeling. These studies revealed that the serine residues required for activity in ACAT1 and ACAT2 are indeed positioned on opposite sites of the ER membrane, consistent with the hypothesis that each enzyme may play a distinct role in cholesterol metabolism.
MATERIALS AND METHODS
Generation of ACAT-Reporter Peptide Fusions
A cDNA reporter fragment was constructed that contains amino acids 23–83, of prepro-α-factor followed by the 8-amino acid FLAG epitope tag (DYKDDDDK), and a termination codon. The prepro-α-factor domain contains three sites for N-linked glycosylation. The reporter sequence was produced by PCR with prepro-α−factor cDNA plasmid (Shelness et al., 1993) as template. The sense strand primer (primer A) included an XhoI site at the 5′ end for subcloning. The antisense primer (primer B) included nontemplated sequences that encoded the FLAG epitope tag, a stop codon, and a XbaI restriction site. After XhoI and XbaI digestion, the PCR product was ligated to XhoI and XbaI digested pcDNA3+ (Invitrogen, San Diego, CA). This plasmid was named Ppα-pcDNA3. The ACAT portion of the fusion proteins were produced by PCR with ACAT expression plasmid DNA as template (Anderson et al., 1998) and a vector-encoded T7 5′ sense primer and ACAT-specific 3′ antisense primers (see Figure 1 for 3′ coordinates of ACAT constructs). In most cases, the 3′ primers contained XhoI sites that were used to create in-frame C-terminal fusions with the glycosylation/FLAG reporter sequence present in Ppα-pcDNA3. For this purpose the PCR products were digested at a 5′ vector-encoded KpnI site and the 3′ XhoI site and ligated to KpnI and XhoI-digested Ppα-pcDNA3. ACAT1-185, ACAT1-328, ACAT1-438, and ACAT2-163 truncation fusions were created by overlapping extension PCR (Landt et al., 1990). In this strategy, ACAT regions were amplified with the T7 5′ primer and an ACAT-specific 3′ primer. The 5′ end of the 3′ primer included 35–40 nucleotides derived from the 5′ boundary of the reporter peptide in Ppα-pcDNA3. The reporter region was produced by using prepro-α-factor DNA as template, and a modified form of primer A, which lacked the nontemplated XhoI site, and primer B. The products of the two PCR reactions were mixed together and amplified with the T7 primer and primer B. Final PCR products were digested with KpnI and XbaI and ligated to KpnI and XbaI-digested pcDNA3+. The presence of the correct open reading frame was confirmed by automated DNA sequencing.
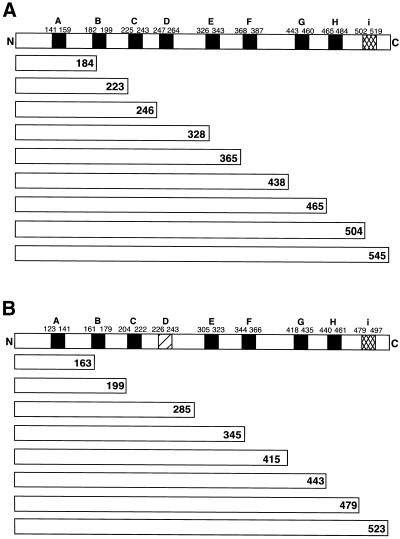
Figure 1.
Truncation-fusion constructs used to assess ACAT1 and ACAT2 transmembrane topology. The transmembrane domains in ACAT1 (A) and ACAT2 (B), as predicted by European Molecular Biology Laboratory web-based protein topology prediction software (Rost and Sander, 1993; Rost et al., 1995, 1996; Rost, 1996) are indicated by solid rectangular boxes. Although ACAT2 was predicted to lack transmembrane domain D, present in ACAT1, the analogous region is indicated with a slashed box. Transmembrane domain i, although not predicted by the software, was identified in ACAT1 experimentally (Lin et al., 1999). This, and the analogous region of ACAT2, are indicated by the cross-hatched boxes. The amino acid coordinates of each domain are indicated. The various truncated constructs used to assess topology are aligned under the appropriate ACAT1 and ACAT2 domain diagrams. The number at the C-terminal end of each refers to the C-terminal amino acid residue. The truncated proteins were fused at their C termini to the glycosylation-FLAG epitope reporter peptide as described in MATERIALS AND METHODS. Each construct is named according to the parent protein and the C-terminal amino acid residue, i.e., “ACAT1-184” refers to truncation of ACAT1 at amino acid residue 184, which is then fused at its C terminus to the glycosylation-FLAG epitope reporter peptide.
Site-directed Mutagenesis and ACAT Activity Assay
The serines putatively required for activity of ACAT1 (Ser269) and ACAT2 (Ser249) were mutated to leucine by a single nucleotide change of the cDNA. The Ser to Leu mutants were generated by using overlapping PCR as described (Landt et al., 1990). The ACAT mutants were subcloned into pcDNA3 and used to transfect ACAT deficient AC29 Chinese hamster ovary (CHO) cells (Cadigan et al., 1988) by using the FuGene 6 transfection reagent (Roche Molecular Biochemicals). Cells were incubated with DNA for ∼48 h at 37°C and then harvested and sonicated. Cell homogenates and/or microsomes were prepared and assayed for ACAT activity as described (Carr et al., 1992).
In Vitro Translations
In vitro transcriptions of the various ACAT constructs with T7 RNA polymerase were carried out by using 10 μg of linear template DNA, 0.5 mM dithiothreitol, 0.05 mM rNTPs, 5× optimized transcription buffer (Promega, Madison, WI), and 30 U T7 RNA polymerase (Promega) at 37°C for 2 h. Newly synthesized RNA was extracted with phenol:CHCl3 (50:50 vol/vol) and then CHCl3, and precipitated with 0.1 volume 3 M NaAc and 2.5 volumes 100% ethanol. The RNA pellet was resuspended in diethylpyrocarbonate-treated H2O to a final concentration of 0.5–1.0 μg/μl. In vitro translations were performed in the presence of nuclease-treated canine pancreatic rough microsomes (Walter and Blobel, 1983). Translations were performed in a 20-μl volume that consisted of 8 μl of nuclease-treated rabbit reticulocyte lysate (Promega), 6 μl of compensation buffer (50 mM tetraethylammonium, 140 mM KOAc, 2.5 mM Mg(OAc)2, 2 μl of Met-deficient amino acid mix (Promega), 10 μCi [35S]methionine (NEN, Boston, MA), and 1 μg of RNA (Scheele, 1983; Nicchitta and Blobel, 1989). Samples were incubated at 30°C for 2 h. Where indicated, reactions were supplemented with 1 mM acetylated tripeptide glycosylation inhibitor (NYT) (Shelness et al., 1993). Trypsin digestion was performed by adjusting samples to 10 mM CaCl2, 20 μg/ml trypsin and incubating at room temperature for 1 h. Where indicated, trypsinization was performed in the presence of 1% Triton X-100. After digestion, soybean trypsin inhibitor was added to a final volume of 2 mg/ml and samples were incubated on ice for 10 min. FLAG-containing fusion proteins and peptides were recovered by immunoprecipitation with anti-FLAG monoclonal antibody M2 as described previously (Shelness et al., 1993). Immunoprecipitates were analyzed by 8–20% SDS-PAGE gels followed by autoradiography. Immunopreciptation with the 5′A1 and 5′A2 antibodies was performed under the same conditions.
Immunocytochemistry
Transient transfection of AC29 cells (ACAT-deficient CHO cells, a gift from T.Y. Chang, Dartmouth College of Medicine, New Hanover, NH) with ACAT constructs was performed by using FuGene 6 transfection reagent. Forty-eight hours after transfection, cells in 35-mm dishes were fixed in 3.7% formaldehyde and washed with BSS. Two dishes of transfected cells from each transfection were treated with 0.25 μg/ml digitonin (60% purity; Fluka) and two dishes were treated with 0.1% saponin for 5 min on ice. The former treatment exposes only cytosolic epitopes, whereas the latter exposes both cytosolic and ER lumenal epitopes. Cells were blocked in phosphate-buffered saline containing 0.1% bovine serum albumin for 30 min at room temperature and washed three times with BSS. Rabbit antibodies to maltose binding protein fusion constructs specific for the amino termini of monkey ACAT1 and ACAT2 were applied at 10 and 25 μg/ml, respectively, to digitonin- and saponin-treated cells. The anti-FLAG M2 monoclonal C-terminal antibody (Sigma, St. Louis, MO) was used at a concentration of 4.4 μg/ml. After a 30-min incubation with primary antibody and three BSS washes, rhodamine-conjugated goat anti-rabbit antibody was applied to cells receiving ACAT antibody (25 μg/ml) and rhodamine-conjugated goat anti-mouse antibody was applied to dishes that received M2 antibody. Cells were incubated with secondary antibody at room temperature for 30 min. Immunostained cells were viewed under an Axial Plane 2 Zeiss microscope by using a rhodamine channel and pictures were taken with the Dage 300 camera and software.
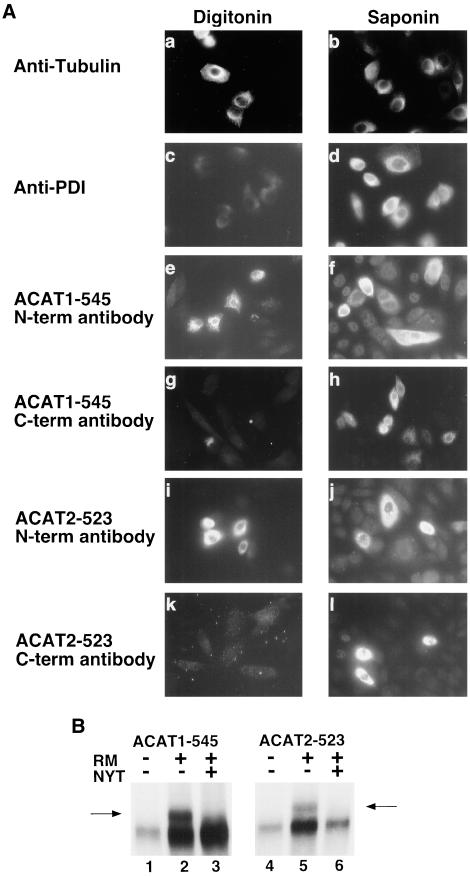
To make sure that these assay conditions were appropriate, control experiments were done by using antibodies to tubulin and protein disulfide isomerase as representative cytosolic and ER lumenal proteins. The correct identification of the cellular location of the two control proteins was observed, validating this experimental approach.
RESULTS
Computer-based Detection of ACAT1 and ACAT2 Transmembrane Domains
Membrane topology software, available on the European Molecular Biology Laboratory (EMBL) protein prediction web site, was used to generate candidate membrane topology models for African green monkey ACAT1 and ACAT2 (Figure 1). The software evaluates the hydrophobicity of the amino acid sequence and compares it with the hydrophobicity of polytopic proteins whose membrane topologies have been solved (Rost and Sander, 1993; Rost et al., 1995, 1996). The ACAT1 model predicted eight transmembrane domains (domains A–H, Figure 1A) with both amino and carboxy termini in the cytoplasm. The ACAT2 model predicted seven transmembrane domains with the amino terminus in the cytoplasm and the carboxy terminus in the ER lumen (Figure 1B). An additional transmembrane domain not predicted by the EMBL model (i) was recently identified by Lin et al. (1999) and added to the model. All of the predicted ACAT1 transmembrane domains were also predicted to occur in ACAT2, with the exception of domain D (Figure 1A). At the time of the topology prediction the observation was made that the presence of transmembrane domain D in ACAT1 placed serine residue 269, previously shown essential for enzyme activity in SRD4 cell ACAT1 (Cao et al., 1996), in the cytosol. The analogous ACAT2 serine (Ser249) was predicted to occur on the opposite side of the membrane in the ER lumen.
Mutational Inactivation of ACAT1 and ACAT2 by Mutagenesis of Conserved Serine Residues
Site-directed mutagenesis was used to explore the role of the serine residues proposed to be essential for ACAT activity. AC29 cells (ACAT-deficient CHO cells) were transiently transfected with cDNAs containing wild-type ACAT1, wild-type ACAT2, ACAT1 Ser269-Leu, ACAT2 Ser249-Leu, or the empty pcDNA3+ vector. Compared with wild type, the Ser to Leu mutations resulted in inactivation of both enzymes (Table 1). In contrast, ACAT1-545 and ACAT2-523, fusion constructs in which a reporter peptide (see below) was fused to the C terminus of each enzyme, exhibited reduced but detectable (≥3-fold higher than background) levels of enzyme activity (Table 1).
Table 1.
Activities among modified forms of ACAT
| ACAT Activity (pmol/min/mg prot) | |
|---|---|
| ACAT1 | 420.0 |
| ACAT1-545 | 53.0 |
| ACAT1 S269 → L | 6.0 |
| ACAT2 | 188.0 |
| ACAT2-523 | 13.0 |
| ACAT2 S249 → L | 4.0 |
| pcDNA3+ | 5.0 |
| Rat microsomes | 641.0 |
ACAT activity data for whole-cell homogenates made from AC29 cells transiently transfected with wild-type African green monkey ACAT1 and ACAT2, ACAT1-545 and ACAT2-523, and ACAT1 Ser269 ⇒ Leu and ACAT2 Ser249 ⇒ Leu. For comparison, homogenates from pcDNA3+ mock-transfected AC29 cells and rat liver microsomes were also assayed.
ACAT1 and ACAT2 Have an Odd Number of Transmembrane Domains and Display Ncyt Cexo Orientations in the ER Membrane
To experimentally explore ACAT1 and ACAT2 membrane topology, a series of truncation-fusions were prepared. Where possible, truncations were made 15–20 amino acids after each of the putative transmembrane domains to ensure that the flanking charge distribution, a property that can potentially affect transmembrane domain function, remained unchanged. In all cases, the truncated ACAT proteins were fused to a reporter peptide containing three N-linked glycosylation sites derived from the pro region of prepro-α-factor and the eight amino acid FLAG epitope tag. N-Linked glycosylation, which occurs exclusively in the ER lumen (corresponding to the interior of ER-derived microsomal vesicles), was used as a reporter for domain-specific translocation into the ER. Accessibility of the FLAG or endogenous epitopes to antibodies or protease served as a reporter for cytosolic localization.
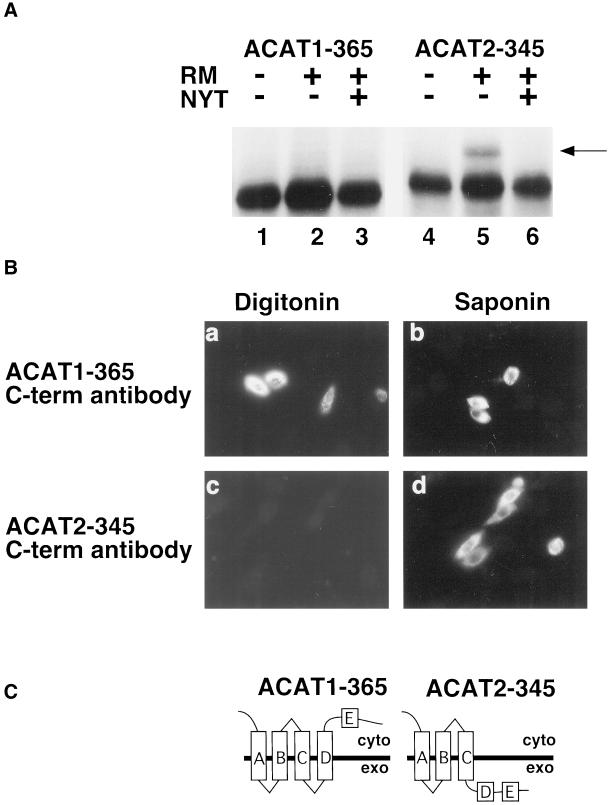
The orientation of the amino and carboxy termini of ACAT1 and ACAT2 across the ER membrane was determined by antibody accessibility in digitonin-permeabilized cells. The plasma membrane of digitonin-treated cells is selectively permeabilized while the ER membrane remains intact. To document this, wild-type AC29 cells were treated with digitonin or saponin and then immunostained with anti-tubulin antibody, a cytosolic marker, or with anti-PDI, an ER lumenal-specific marker. Signal was observed in cells stained with the tubulin antibody after digitonin or saponin treatment (Figure 2A, a and b). An absence of signal was noted in cells treated with digitonin and stained with the ER lumenal-specific PDI antibody, confirming the ER membrane was intact (Figure 2A, c), whereas signal was observed in cells stained with the PDI-specific antibody after saponin treatment (Figure 2A, d). AC29 cells that were transiently transfected with ACAT1-545 were permeabilized with digitonin and stained with either an ACAT1 amino terminal-specific antibody (Figure 2A, e) or with the carboxy-terminal anti-FLAG antibody (Figure 2A, g). Fluorescent signal was observed only in cells stained with the amino terminal antibody, indicating that the amino terminus of ACAT1–545 is located in the cytosol. The anti-FLAG antibody failed to produce a signal in digitonin-treated cells, indicating that the carboxy terminus of the ACAT1 protein is located in the ER lumen. When cells were incubated with saponin, a treatment that disrupts all cellular membranes, signal was observed with both the amino- and carboxy-terminal antibodies (Figure 2A, f and h). Similar results were obtained when ACAT2-523 was analyzed. Digitonin-treated cells were stained with the amino-terminal specific ACAT2 antibody (Figure 2A, i) but not with the carboxy-terminal anti-FLAG antibody (Figure 2A, k). When cells were permeabilized with saponin, both epitopes were accessible (Figure 2A, j and l).
Figure 2.
Orientation of the amino and carboxy termini of ACAT1 and ACAT2 across the ER membrane. (A) Wild-type ACAT-deficient AC29 CHO cells and AC29 cells transiently transfected with either ACAT1-545 or ACAT2-523 (nearly full-length) fusion proteins were treated with digitonin (a, c, e, g, i, and k) to selectively permeabilize the plasma membrane (leaving the ER membrane intact), or with saponin (b, d, f, h, j, and l), to permeabilize all cell membranes. Untransfected cells were treated with digitonin (a and c) or saponin (b and d) and then immunostained with either tubulin (a and b) or PDI (c and d) antibodies. Rhodamine-labeled goat anti-mouse secondary antibody was applied to AC29 cells immunostained with the mouse anti-tubulin antibody and fluorescein-labeled goat anti-rat secondary antibody was applied to AC29 cells immunostained with the rat anti-PDI antibody. Transfected cells were stained with antibodies specific to amino terminal sequences of either ACAT1 (e and f) or ACAT2 (i and j) (N-term antibody) or with the C-terminal-specific (C-term antibody) anti-FLAG M2 antibody (g, h, k, and l). Rhodamine-labeled goat anti-rabbit secondary antibody was applied to cells stained with the ACAT1 or ACAT2 N-term antibodies and rhodamine-labeled goat anti-mouse secondary antibody was applied to cells stained with the anti-FLAG M2 C-term antibody. Cells were viewed microscopically with rhodamine or fluorescein channel fluorescence. Positive signal, where obtained, is indicated by light areas within cells. (B) ACAT1-545 and ACAT2-523 mRNA were translated in rabbit reticulocyte lysate in the presence (+) or absence (−) of canine pancreas rough microsomes (RMs). In lanes 3 and 6, tripeptide glycosylation inhibitor (NYT) was included during the translations. The proteins resulting from each translation were immunoprecipitated by using anti-FLAG M2 antibody and fractionated by 8–20% SDS-polyacrylamide gradient gel electrophoresis. Glycosylated products in lanes 2 and 4 were detected based on their decreased electrophoretic gel mobility (arrows).
To further explore topology by using an independent approach, ACAT1-545 and ACAT2-523 mRNA were translated in vitro in the presence or absence of canine pancreatic rough microsomes. (Figure 2B). Use of the three N-linked glycosylation sites within the carboxy-terminal reporter region is dependent on its colocalization with the active site of oligosaccharyltransferase in the ER (microsomal) lumen. In translations performed in the presence of canine pancreatic rough microsomes (+), forms of the ACAT1-545 and ACAT2-523 fusion proteins were generated with reduced electrophoretic gel mobilities (Figure 2B, arrows). Unglycosylated forms of protein were present in lanes 2 and 4 with a slightly faster mobility than glycosylated forms, and this can be attributed to the inefficiency of the in vitro system to modify all translates present. This was observed in subsequent in vitro translations with the efficiency of glycosylation varying among constructs. When a specific tripeptide inhibitor of glycosylation (NYT) was added to the translation mixtures, the slower migrating forms were eliminated, confirming their identity as glycosylation products. These data, along with those in Figure 2A, indicate that both ACAT1 and ACAT2 possess an odd number of transmembrane domains and are positioned in the ER membrane with their N termini in the cytosol and C termini in the ER lumen.
Detection and Localization of Transmembrane Domains in the N-terminal Half of ACAT1 and ACAT2
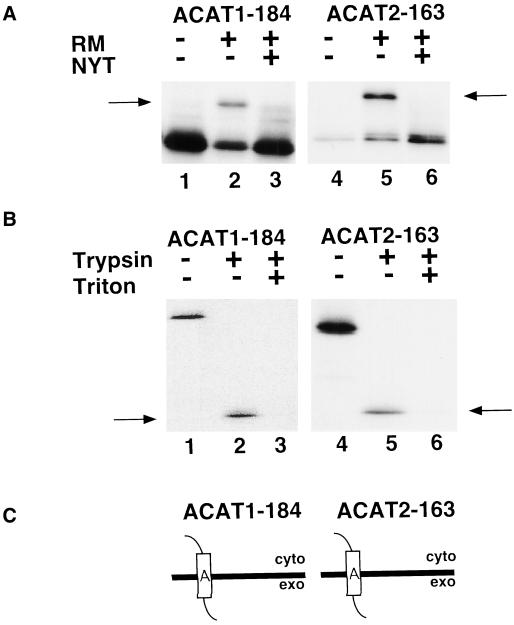
The first predicted transmembrane domains for ACAT1 and ACAT2 include amino acids 141–159 and 123–141, respectively (Figure 1). The data in Figure 3 show that truncated fusion protein constructs ACAT1-184 and ACAT2-163 were both glycosylated during in vitro translation assays in the presence (+) of rough microsomes (Figure 3A, arrows). Furthermore, when the translation reactions were treated with trypsin, both produced protected fragments that were immunoprecipitable with anti-FLAG antibody. These 14-kDa products corresponded in size to peptides extending from the N-terminal boundary of transmembrane domain A to the C terminus of the fusion protein. These trypsinization products were not found when incubations were done in the presence of 1% Triton X-100, confirming that production was dependent on the presence of intact microsomal membranes. These results indicate that ACAT1-184 and ACAT2-163 span the membrane only once with Ncyt Cexo orientations (Figure 3C).
Figure 3.
Orientation of ACAT1-184 and ACAT2-163 in the ER membrane. (A) ACAT1-184 (lanes 1–3) and ACAT2-163 (lanes 4–6) mRNA were translated in vitro in the presence and absence of rough microsomes (RMs) and glycosylation inhibitor (NYT) as described in Figure 2B. Arrows show the position of glycosylated products. (B) Translations of ACAT1-184 and ACAT2-163 were performed in the presence RM and NYT. Reactions were then divided into three equal aliquots and incubated with (+) or without (−) trypsin and Triton X-100. The 14-kDa protected fragments are indicated by arrows. (C) Schematic diagrams of ACAT1-184 and ACAT2-163 exhibiting Ncyt/Cexo membrane topologies.
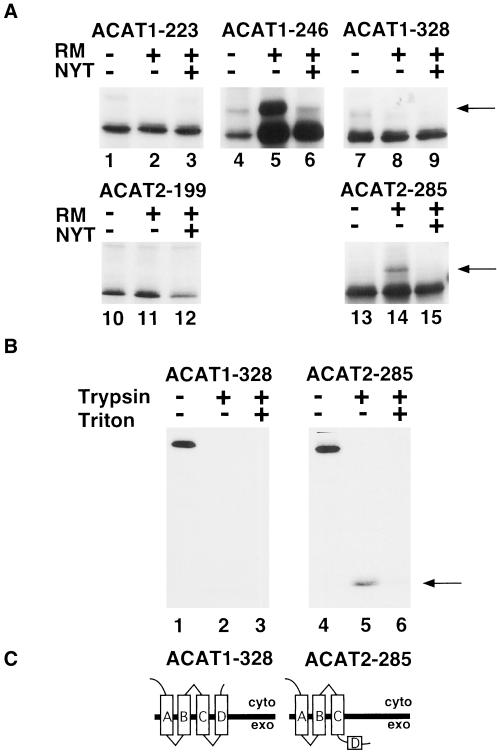
Use of transmembrane domains B, C, and D of ACAT1 and transmembrane domains B and C of ACAT2 was examined by truncating ACAT1 at amino acids 223, 246, and 328, and ACAT2 at amino acids 199 and 285 (Figure 1, A and B). ACAT1-223 and ACAT2-199 were not glycosylated during in vitro translations in the presence of rough microsomes (+), indicating their carboxy termini were on the cytoplasmic side of microsomal membranes (Figure 4A). This result, in conjunction with those in Figure 3, suggests that transmembrane domains A and B function in both ACAT1 and ACAT2. When ACAT1-246 was translated in vitro, glycosylation of the C-terminal epitope was restored, suggesting that domain C spans the membrane in ACAT1 in an Ncyt Cexo orientation. Because computer and visual analysis of the ACAT2 sequence failed to detect a candidate transmembrane domain corresponding to domain D in ACAT1, ACAT2 was truncated well downstream of domain C, at amino acid 285. This fusion underwent C-terminal glycosylation, consistent with the existence of only one transmembrane domain (domain C) between amino acids 199 and 285 in ACAT2 (Figure 4C). However, truncation of ACAT1 at an analogous position (i.e., amino acid 328) abolished C-terminal glycosylation consistent with the presence of two transmembrane domains (i.e., domains C and D; Figure 4C). The validity of this prediction is further supported by protease protection experiments demonstrating protection of the C-terminal epitope in ACAT2-285 but not in ACAT1-328 (Figure 4B). Furthermore, ACAT2-285 but not ACAT1-328 was glycosylated in transiently transfected COS cells (our unpublished results). These data indicate a point of divergence in the membrane topologies of ACAT1 and ACAT2 despite a 61% amino acid identity between transmembrane domain D of ACAT1 and the analogous region of ACAT2. Interestingly, this point of divergence affects the topology of the serines required for activity, placing Ser269 of ACAT1 in the cytosol and Ser249 of ACAT2 in the ER lumen.
Figure 4.
Divergent transmembrane use of domain D in ACAT1 and ACAT2. (A) Indicated ACAT1 and ACAT2 truncation fusion constructs were translated in vitro in the presence and absence of rough microsomes and glycosylation inhibitor as described in Figure 2B. Leftward pointing arrows indicate the position of glycosylated products in lanes 5 and 14. (B) ACAT1-328 and ACAT2-285 were translated in the presence of rough microsomes and glycosylation inhibitor and subjected to trypsin digestion as described in Figure 3B. The 16-kDa protected peptide produced by trypsin digestion of the ACAT2-285 translation is indicated by an arrow. (C) Diagrams of ACAT1-328 and ACAT2-285 membrane topologies.
Detection and Localization of Transmembrane Domains in the C-terminal Half of ACAT1 and ACAT2
The divergence in ACAT1 and ACAT2 membrane topology was maintained through amino acid residue 365 of ACAT1 and 345 of ACAT2. ACAT2-345 was glycosylated during in vitro translation in the presence of microsomes, whereas a glycosylated form of ACAT1-365 was not observed (Figure 5A). The same outcome was observed in transiently transfected COS cells (our unpublished results). Immunofluorescence by using anti-FLAG antibody revealed that the C terminus of ACAT2-345 was inaccessible to anti-FLAG antibody in digitonin-permeabilized cells (Figure 5B, c), whereas reactivity with the A1-365 C-terminal epitope did not require saponin permeabilization (Figure 5B, a). These results indicate that the carboxy terminus of ACAT1-365 remained cytosolic and the carboxy terminus of the ACAT2-345 fusion protein remained in the ER lumen as shown in Figure 5C. Therefore, candidate transmembrane domain E was not used for either ACAT1 or ACAT2.
Figure 5.
The C termini of ACAT1-365 and ACAT2-345 are oriented differently in the ER membrane. (A) ACAT1-365 and ACAT2-345 mRNA were translated in vitro in the presence and absence of rough microsomes and glycosylation inhibitor, as described in Figure 2B. The position of the glycosylated ACAT2-345 product (lane 5) is indicated by the arrow. (B) AC29 cells were transiently transfected with ACAT1-365 (a and b) or ACAT2–345 (c and d). After permeabilization with digitonin (a and c) or saponin (b and d), cells were stained with anti-FLAG M2 C-term antibody as described in Figure 2A. (C) ACAT1-365 and ACAT2-345 membrane topology models.
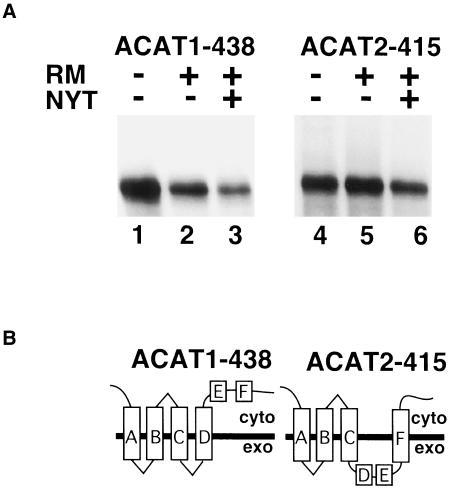
Fusion proteins prepared with a truncation at amino acid residue 438 in ACAT1 and 415 in ACAT2 tested the functionality of predicted transmembrane domain F. Neither of the fusion proteins were glycosylated during in vitro translation in the presence of rough microsomes (Figure 6A). In addition, trypsin digestion of the translation reactions containing rough microsomes failed to produce a protected fragment and neither of the constructs were glycosylated by COS cells (our unpublished results). These data demonstrate that predicted transmembrane domain F is not used in the construct ACAT1-438 (Figure 6B). However, because the C-terminal reporter in ACAT2-345 was glycosylated and rendered inaccessible to antibody (Figure 5B, c), the result in Figure 6 suggests that transmembrane domain F does function to span the membrane in ACAT2-415 (Figure 6B). As a result, the similarity in the topology for the two proteins is reestablished in this region.
Figure 6.
Cytosolic C-terminal localization of ACAT1-438 and ACAT2-415 is achieved by differential transmembrane use of domain F. (A) ACAT1-438 and ACAT2-415 mRNA were translated in vitro in the presence and absence of rough microsomes and glycosylation inhibitor as described in Figure 2B. (B) ACAT1-438 and ACAT2-415 membrane topology models.
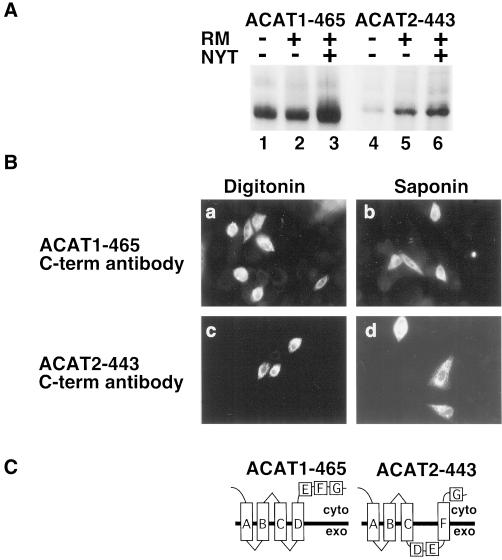
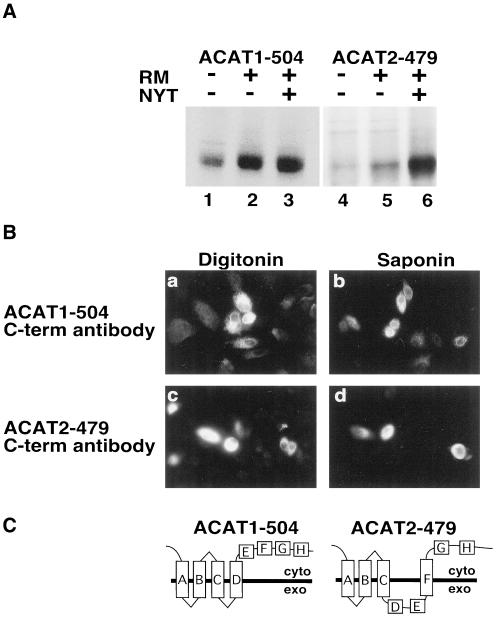
Predicted transmembrane domains G and H of ACAT1 and ACAT2 were analyzed by truncating ACAT1 at amino acids 465 and 504, and ACAT2 at amino acids 443 and 479. ACAT1-465 and ACAT2-443 were not glycosylated during in vitro translation in the presence (+) of rough microsomes (Figure 7A), indicating that domain G of ACAT1 and ACAT2 does not span the membrane (Figure 7C). Likewise, neither ACAT1-504 nor ACAT2-479 were glycosylated during in vitro translation in the presence of rough microsomes (Figure 8A), indicating transmembrane domain H of ACAT1 and ACAT2 also fails to function as a transmembrane domain (Figure 8C). This conclusion was supported by experiments demonstrating that the carboxy terminal FLAG epitopes in ACAT1-465 and ACAT2-443 (Figure 7B) and ACAT1-504 and ACAT2-479 (Figure 8B) all were accessible to anti-FLAG antibodies in digitonin-permeabilized AC29 cells.
Figure 7.
Orientation of ACAT1-465 and ACAT2-443 in the ER membrane. (A) ACAT1-465 and ACAT2-443 mRNA were translated in vitro in the presence and absence of rough microsomes and glycosylation inhibitor as described in Figure 2B. (B) AC29 cells were transiently transfected with ACAT1-465 (a and b) or ACAT2-443 (c and d). After permeabilization with digitonin (a and c) or saponin (b and d), cells were stained with anti-FLAG M2 C-term antibody as described in Figure 2A. (C) ACAT1-465 and ACAT2-443 membrane topology models.
Figure 8.
Orientation of ACAT1-504 and ACAT2-479 in the ER membrane. (A) ACAT1-504 and ACAT2-479 mRNA were translated in vitro in the presence and absence of rough microsomes and glycosylation inhibitor as described in Figure 2B. (B) AC29 cells were transiently transfected with ACAT1-504 (a and b) or ACAT2-479 (c and d). After permeabilization with digitonin (a and c) or saponin (b and d), cells were stained with anti-FLAG M2 C-term antibody as described in Figure 2A. (C) ACAT1-504 and ACAT2-479 membrane topology models.
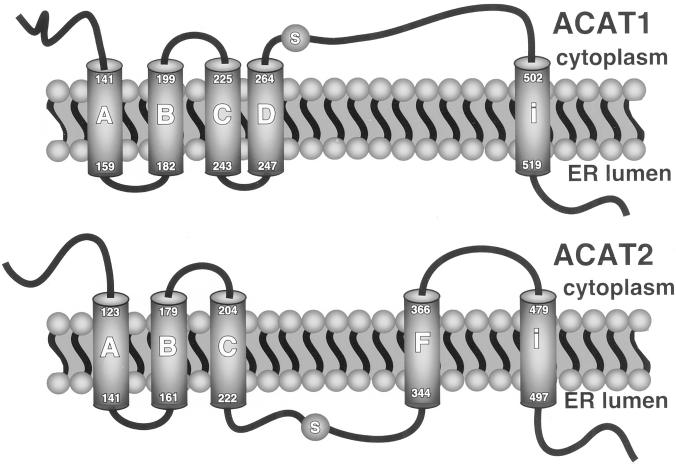
An additional ACAT1 transmembrane domain that was not predicted in the EMBL model was identified by Lin et al. (1999). Because the C terminus of ACAT1-545 resides in the ER lumen (Figure 2), it is clear that ACAT1 transmembrane domain “i” is indeed functional, confirming the previously published results. The analogous domain “i” region in ACAT2 is 65% identical to the ACAT1 amino acid sequence. Because the ACAT2-523 fusion protein was glycosylated (Figure 2B), and ACAT2-479 (Figure 8A) was not, it is clear that domain “i” also is used in ACAT2. Thus, the final model for ACAT1 and ACAT2 is as shown in Figure 9.
Figure 9.
Membrane topology models for ACAT1 and ACAT2. Both enzymes display cytoplasmic amino termini and lumenal carboxy termini. A serine residue (S) critical for activity is shown to occur in the cytoplasm for ACAT1 after its fourth transmembrane domain and in the ER lumen for ACAT2 after its third transmembrane domain.
DISCUSSION
The results presented in this article demonstrate that ACAT1 and ACAT2 are proteins with five transmembrane domains with the serine residues essential for the activity of both enzymes positioned on opposite sides of the ER membrane (Figure 9). The topology model developed for ACAT1 places Ser269 on the cytoplasmic side of the ER membrane, whereas the analogous serine in ACAT2 (Ser249) is positioned in the ER lumen. As indicated by Northern analyses in mice and nonhuman primates, ACAT2 is found predominantly in the liver and intestine, the major sites of apoB-containing lipoprotein assembly and secretion (Anderson et al., 1998; Cases et al., 1998). Hence, as initially hypothesized, it is conceivable that ACAT2 couples cholesterol ester production with the assembly and secretion of apoB-containing lipoprotein particles, whereas ACAT1 functions primarily in maintenance of proper intracellular unesterified cholesterol concentrations through generation of cholesterol esters for storage in cytosolic lipid droplets. The mechanism for coupling of either ACAT to its function is uncertain. Upon synthesis, cholesterol esters, as very hydrophobic molecules, may partition into the phospholipid bilayer of the membrane and were this to happen, sidedness of the active site may not matter. However, cholesterol esters have limited solubility in the phospholipid bilayer and dissolution into the membrane is not necessarily their only fate. In CHO cells devoid of ACAT, we have seen that expression of either ACAT enzyme results in the appearance of cytoplasmic lipid droplets (Lee et al., 2000). In secretion competent cells, cholesterol esters formed in the ER lumen may be readily used by apoB and microsomal triglyceride transfer protein for lipoprotein particle assembly, whereas proteins in the cytosol associated with lipid droplet formation may target cholesterol esters for storage. In other words, it is conceivable that interactions of specific proteins with ACAT2 or ACAT1, respectively, could facilitate lipoprotein particle lipidation versus lipid droplet formation although this remains to be demonstrated.
The topology model of ACAT1 presented here is different from the model recently published by Lin et al. (1999). Their model indicated that ACAT1 possesses seven transmembrane domains, whereas our studies are consistent with the presence of only five transmembrane domains. However, both models agree that ACAT1 displays an Ncyt Cexo orientation and that Ser269 resides on the cytoplasmic face of the ER membrane. The major difference between the two models relates to the use of predicted transmembrane domains E and G, which are used in the Lin et al. (1999) model but did not display functionality in our studies. The underlying basis for this discrepancy may be related to the differences in experimental approaches. Lin et al. (1999) used an approach in which an epitope tag was inserted between proposed transmembrane domains and their orientation relative to the ER membrane established only by immunostaining of digitonin-permeabilized cells. In our studies, ACAT1 and ACAT2 were sequentially truncated downstream of predicted transmembrane domains and the orientation of an engineered C-terminal reporter sequence was determined by glycosylation site use, by antigenic reactivity, as well as by protease accessibility. Both the epitope insertion and truncation fusion techniques have been widely used to explore the topology of polytopic membrane proteins, although both rely on underlying assumptions that may not uniformly apply to the biogenesis of all membrane proteins (Wessels and Spiess, 1988; Lipp et al., 1989). The insertion of antibody epitopes affects the overall charge distribution of a protein, which may influence the use of upstream or downstream transmembrane domains. Likewise, truncating a protein and fusing it to a C-terminal reporter may influence the use of upstream transmembrane domains (Nilsson and von Heijne, 1990; Gafvelin et al., 1997; Turk and Wright, 1997; Zhang et al., 1998). Therefore, the disparity between the ACAT1 models cannot be resolved as of yet. With time, the techniques and methods used to determine membrane topology of proteins will improve and likely will provide clarification on the exact membrane topology of ACAT1. Nevertheless, the comparative approach performed in these studies strongly supports the notion that ACAT1 and ACAT2 display distinct topologies that are consistent with the unique role played by each enzyme in cholesterol metabolism.
Although no single approach may be sufficiently reliable to elucidate membrane topology, the main goal of the present study was to explore the possibility that ACAT1 and ACAT2, although sharing overall sequence similarity, display divergent membrane topologies, consistent with possible differences in their physiological roles in cholesterol metabolism. The truncation fusion approach used in this study clearly indicated a point of divergence between the two enzymes that is best illustrated by comparing the consequences of truncating ACAT1 and ACAT2 after the region of predicted ACAT1 transmembrane domain D. Although ACAT1 truncated in this region places the C-terminal reporter in the cytosol, consistent with use of transmembrane domain D, ACAT2 truncated at the analogous region displays a lumenal C terminus. This point of divergence appears to persist until the region of domain F, which is used in ACAT2 but not in ACAT1. Because ACAT1 and ACAT2 are highly conserved it is unlikely that truncations made in the two proteins would manifest themselves differently unless it reflected an actual divergence in their modes of membrane assembly. Hence, it appears clear that ACAT1 and ACAT2 do indeed display inverted topologies in a region that affects the relative position of the serine residues required for activity (Figure 9).
The current ACAT topology models suggest intracellular compartmentalization of cholesterol esterification. The ACAT1 topology models suggest that ACAT1-catalyzed cholesteryl esterification occurs within the cytosol of the cell. Therefore, ACAT1 may play an important role in intracellular sterol homeostasis, which can be perturbed when macrophages within arterial lesions accumulate excess ACAT1-derived cytoplasmic cholesterol ester droplets, leading to foam cell development and atherosclerosis. However, recent data have also suggested that ACAT1-driven intracellular cholesterol esterification may serve as a mechanism to prolong macrophage viability, in the process actually slowing the development of advanced atherosclerotic lesions. Transplantation of ACAT1 null mouse bone marrow into low-density lipoprotein receptor-deficient mice resulted in the formation of cholesterol-rich lesions (Fazio et al., 1999) and hypercholesterolemic ACAT1 null mice have complicated atherosclerotic lesions (Accad et al., 2000). In vitro studies have demonstrated that the inhibition of ACAT1 in J774 macrophages results in free cholesterol accumulation and crystallization within cells, followed by apoptosis (Kellner-Weibel et al., 1998). Because the absence of ACAT1 could promote the progression of atherosclerosis and may also have deleterious consequences in areas other than the artery wall, selective ACAT2 inhibition, targeted to decreasing plasma apoB-lipoprotein cholesterol concentrations, may be a more desirable means of preventing atherosclerosis and avoiding potentially toxic effects at the level of the artery wall.
The potential localization of the ACAT2 active site in the ER lumen of lipoprotein-producing cells raises several intriguing questions. If cholesterol esterification by ACAT2 occurs in the ER, it would imply that cholesterol obtained from the ER membrane is esterified to a fatty acid derived from an acyl-CoA present in the ER lumen. The presence of a carnitine palmitoyltransferase in the ER membrane, analogous to the carnitine palmitoyltransferase identified in mitochondria, has been postulated to be the mechanism that transfers fatty acids from the acyl-CoA pool in the cytosol to the acyl-CoA pool in the ER lumen (Zammit, 1996). Such a system could supply ACAT2 and other enzymes that function in the lumenal space, such as diacylglycerol acyl transferase (DGAT), with the fatty acyl-CoA substrates required for catalysis. Alternatively, the C-terminal regions of ACAT1, ACAT2, and DGAT possess a conserved Asp-Trp-Trp-Asn (DWWN) domain that, according to our ACAT topology data and to preliminary DGAT computer prediction methods with the Rost model (Rost and Sander, 1993; Rost et al., 1995, 1996; Rost, 1996), are located in the cytosol. Because fatty acyl-CoAs are required substrates for each of these three enzymes, it is possible that this highly conserved region could serve as a fatty acyl-CoA recognition site or binding region. In this scenario, fatty acid binding would occur in the cytosol for each enzyme. ACAT1-catalyzed cholesterol esterification would necessarily occur in a slightly different manner than for ACAT2 if the serines at position 269 and 249 for ACAT1 and ACAT2, respectively, are indeed active site serines. Somehow, for ACAT2, the fatty acid bound in the cytosol would have to be translocated to the lumenal side of the membrane for the reaction to be completed. As the data for ACAT1 suggest (Chang et al., 1998), these enzymes may associate in multimeric subunits to be active. In such a case, the possibility of a channel or pore through the membrane, which could be used for translocating a fatty acid, seems feasible.
In summary the studies described here provide further evidence that ACAT1 and ACAT2 may have distinct roles within cells. ACAT1 is ubiquitously expressed in all tissues and seems to have a role in intracellular cholesterol homeostasis via formation of cytosolic cholesterol ester droplets. ACAT2 expression, which is limited to tissues that express apoB, may produce cholesterol ester that is incorporated into newly forming lipoprotein particles. As such, ACAT2 may play a prominent role in determining the lipid composition and any associated atherogenicity of apoB-containing lipoproteins.
ACKNOWLEDGMENTS
The AC29 cell line, a modified CHO cell without ACAT, was a generous gift from T.-Y. Chang, Dartmouth College of Medicine, New Hanover, NH. This work was supported in part by the National Institutes of Health, National Heart, Lung, and Blood Institute, Grant HL-49373. C.W.J. was supported by predoctoral trainee grant HL-07115 from the National Heart, Lung, and Blood Institute, National Institutes of Health. G.S.S. was supported in part as an Established Investigator of the American Heart Association.
Abbreviations used:
- ACAT
Acyl-CoA:cholesterol acyltransferase
- ApoB
apolipoprotein B
- exo
exoplasmic
- ER
endoplasmic reticulum
- DGAT
diacylglycerol acyltransferase
- M2 FLAG
epitope for anti-flag M2 antibody
- cyt
cytosolic
- NYT
asparagine-tyrosine-threonine tripeptide
- ppα
yeast pre-pro α factor
- PCR
polymerase chain reaction
REFERENCES
- Accad M, Smith SJ, Newland DL, Sanan DA, King LE, Jr, Linton MF, Fazio S, Farese RL., Jr Massive xanthomatosis and altered composition of atherosclerotic lesions in hypercholesterolemic mice lacking acyl CoA:cholesterol acyltransferase 1. J Clin Invest. 2000;105:711–719. doi: 10.1172/JCI9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RA, Joyce C, Davis M, Reagan JW, Clark M, Shelness G, Rudel LL. Identification of a form of acyl-CoA:cholesterol acyltransferase specific to liver and intestine in nonhuman primates. J Biol Chem. 1998;273:26747–26754. doi: 10.1074/jbc.273.41.26747. [DOI] [PubMed] [Google Scholar]
- Bocan TMA, Mueller SB, Uhlendorf PD, Brown EQ, Mazur MJ, Black AE. Inhibition of acyl-CoA cholesterol O-acyltransferase reduces the cholesteryl ester enrichment of atherosclerotic lesions in the Yucatan micropig. Atherosclerosis. 1993;99:175–186. doi: 10.1016/0021-9150(93)90020-u. [DOI] [PubMed] [Google Scholar]
- Bocan TMA, Mueller SB, Uhlendorf PD, Newton RS, Krause BR. Comparison of CI-976, an ACAT inhibitor, and selected lipid-lowering agents for antiatheroscleotic activity in iliac-femoral and thoracic aortic lesions. A biochemical, morphological and morphometric evaluation. Arterioscler Thromb. 1991;11:1830–1843. doi: 10.1161/01.atv.11.6.1830. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Heider JG, Chang T-Y. Isolation and characterization of Chinese hamster ovary cell mutants deficient in acyl-coenzyme A:cholesterol acyltransferase activity. J Biol Chem. 1988;263:274–282. [PubMed] [Google Scholar]
- Cao GQ, Goldstein JL, Brown MS. Complementation of mutation in acyl-CoA:cholesterol acyltransferase (ACAT) fails to restore sterol regulation in ACAT-defective sterol-resistant hamster cells. J Biol Chem. 1996;271:14642–14648. doi: 10.1074/jbc.271.24.14642. [DOI] [PubMed] [Google Scholar]
- Carr TP, Hamilton RL, Jr, Rudel LL. ACAT inhibitors decrease secretion of cholesteryl esters and apolipoprotein B by perfused livers of African green monkeys. J Lipid Res. 1995;36:25–36. [PubMed] [Google Scholar]
- Carr TP, Parks JS, Rudel LL. Hepatic ACAT activity in African green monkeys is highly correlated to plasma LDL cholesteryl ester enrichment and coronary artery atherosclerosis. Arterioscler Thromb. 1992;12:1274–1283. doi: 10.1161/01.atv.12.11.1274. [DOI] [PubMed] [Google Scholar]
- Cases S, Novak S, Zheng Y-W, Myers HM, Lear SR, Sande E, Welch CB, Lusis AJ, Spencer TA, Krause BR, Erickson SK, Farese RV., Jr ACAT-2, A second mammalian acyl-CoA:cholesterol acyltransferase. Its cloning, expression, and characterization. J Biol Chem. 1998;273:26755–26764. doi: 10.1074/jbc.273.41.26755. [DOI] [PubMed] [Google Scholar]
- Chang CCY, Lee C-YC, Chang ET, Cruz JC, Levesque MC, Chang T-Y. Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner. J Biol Chem. 1998;273:35132–35141. doi: 10.1074/jbc.273.52.35132. [DOI] [PubMed] [Google Scholar]
- Cianflone KM, Yasruel Z, Rodriquez MA, Vas D, Sniderman AD. Regulation of apoB secretion from HepG2 cells: evidence for a critical role for cholesteryl ester synthesis in the response to a fatty acid challenge. J Lipid Res. 1990;31:2045–2055. [PubMed] [Google Scholar]
- Fazio S, Liu L, Major AS, Gleaves LA, Accad M, Linton MF, Farese RV., Jr Accelerated atherosclerosis in LDL receptor null mice reconstituted with ACAT negative macrophage. Circulation. 1999;100:1–613. [Google Scholar]
- Gafvelin G, Sakaguchi M, Anderson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- Huff MW, Telford DE, Barrett PHR, Billheimer JT, Gillies PJ. Inhibition of hepatic ACAT decreases apoB secretion in miniature pigs fed a cholesterol-free diet. Arterioscler Thromb. 1994;14:1498–1508. doi: 10.1161/01.atv.14.9.1498. [DOI] [PubMed] [Google Scholar]
- Kellner-Weibel G, Jerome WG, Small DM, Warner GJ, Stoltenborg JK, Kearney MA, Corjay MH, Phillips MC, Rothblat GH. Effects of intracellular free cholesterol accumulation on macrophage viability - A model for foam cell death. Arterioscler Thromb Vasc Biol. 1998;18:423–431. doi: 10.1161/01.atv.18.3.423. [DOI] [PubMed] [Google Scholar]
- Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- Lee RG, Willingham MC, Davis MA, Reagan JW, Jr, Skinner KA, Anderson RA, Rudel LL. ACAT1 and ACAT2 have unique cellular distributions in nonhuman primates. Circulation. 1999;100(suppl I):1–612. [Google Scholar]
- Lee, R.G., Willingham, M.C., Davis, M.A., Skinner, K.A., and Rudel, L.L. (2000) Differential expression of ACAT1 and ACAT2 among cells within liver, intestine, kidney and adrenal of nonhuman primates. J. Lipid Res. (in press). [PubMed]
- Lin S, Cheng D, Liu MS, Chen J, Chang TY. Human Acyl-CoA:cholesterol acyltransferase-1 in the endoplasmic reticulum contains seven transmembrane domains. J Biol Chem. 1999;274:23276–23285. doi: 10.1074/jbc.274.33.23276. [DOI] [PubMed] [Google Scholar]
- Lipp J, Flint N, Haeuptle M-T, Dobberstein B. Structural requirements for membrane assembly of proteins spanning the membrane several times. J Cell Biol. 1989;109:2013–2022. doi: 10.1083/jcb.109.5.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Nascent secretory chain binding and translocation are distinct processes: differentiation by chemical alkylation. J Cell Biol. 1989;108:789–795. doi: 10.1083/jcb.108.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Behari A, Cromley D, Billheimer JT, Sturley SL. Characterization of two human genes encoding acyl coenzyme A:cholesterol acyltransferase-related enzymes. J Biol Chem. 1998;273:26765–26771. doi: 10.1074/jbc.273.41.26765. [DOI] [PubMed] [Google Scholar]
- Rost B. Predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- Rost B, Casadio R, Fariselli P, Sander C. Prediction of helical transmembrane segments at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;7:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Rudel LL, Haines J, Sawyer JK, Shah R, Wilson MS, Carr TP. Hepatic origin of cholesteryl oleate in coronary artery atherosclerosis in African green monkeys. Enrichment by dietary monounsaturated fat. J Clin Invest. 1997;100:74–83. doi: 10.1172/JCI119524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele G. Methods for the study of protein translocation across the RER membrane using the reticulocyte lysate translation system and canine pancreatic microsomal membranes. New York, NY: Academic Press; 1983. pp. 94–110. [DOI] [PubMed] [Google Scholar]
- Shelness GS, Lin L, Nicchitta CV. Membrane topology and biogenesis of eukaryotic signal peptidase. J Biol Chem. 1993;268:5201–5208. [PubMed] [Google Scholar]
- Turk E, Wright EM. Membrane topology motifs in the SGLT contrasporter family. J Membr Biol. 1997;159:1–20. doi: 10.1007/s002329900264. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Methods in Enzymology. New York: Academic Press; 1983. Preparation of microsomal membranes for cotranslational protein translocation; pp. 84–93. [DOI] [PubMed] [Google Scholar]
- Wessels HP, Spiess M. Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell. 1988;55:61–70. doi: 10.1016/0092-8674(88)90009-8. [DOI] [PubMed] [Google Scholar]
- Wilcox LJ, Barrett PHR, Newton RS, Huff MW. ApoB100 secretion from HepG2 cells is decreased by the ACAT inhibitor CI-1011 - An effect associated with enhanced intracellular degradation of ApoB. Arterioscler Thromb Vasc Biol. 1999;19:939–949. doi: 10.1161/01.atv.19.4.939. [DOI] [PubMed] [Google Scholar]
- Zammit VA. Role of insulin in hepatic fatty acid partitioning: emerging concepts. Biochem J. 1996;314:1–14. doi: 10.1042/bj3140001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-T, Chen M, Han E, Wang C. Dissection of de novo membrane insertion activities of internal transmembrane segments of ATP-binding-cassette transporters: toward understanding topological rules of membrane assembly of polytopic membrane proteins. Mol Biol Cell. 1998;9:853–863. doi: 10.1091/mbc.9.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]