Abstract
Acinetobacter ursingii has not been reported in infectious processes apart from its recent description as a new species. A bacteremia caused by A. ursingii in a patient with a pulmonary adenocarcinoma confirms that this microorganism is an opportunistic human pathogen. The isolate was susceptible to imipenem, aminoglycosides, rifampin, and fluoroquinolones.
CASE REPORT
A 63-year-old man presenting with a pulmonary adenocarcinoma diagnosed in July 2001 was admitted to the University Hospital Hôtel Dieu, Paris, France. A fifth course of intravenous chemotherapy consisting of cisplatin (50 mg/m2 of body area) and vinorelbine (30 mg/m2) was initiated through a catheter chamber, which had been implanted 12 weeks earlier. The patient had received corticosteroids for 2 months because of thoracic pain due to tumoral compression. On the day following admission (day 1), his condition deteriorated with fever (39.5°C) and chills associated with an increase of white blood cell count (15 × 103 cells per μl with 90% neutrophils), C-reactive protein (9.5 mg/dl), and fibrinogen (0.51 mg/dl). Thus, he received cefotaxime (3 g per day) and gentamicin (180 mg per day) for 2 days. Urine analysis was normal. The chest X ray and the abdominal and cardiac echography were not modified. A strain of an Acinetobacter sp. was isolated from five blood samples, including two obtained from the catheter chamber. The catheter chamber was removed, but its culture was sterile. On day 3, the antimicrobial therapy was changed to imipenem (2 g per day) and amikacin (900 mg per day) for 2 weeks according to the susceptibility of the strain. On day 5, rifampin (1.2 g per day) was added as the patient remained febrile. On day 7, the patient was apyretic, with normalization of white blood cell count and C-reactive protein.
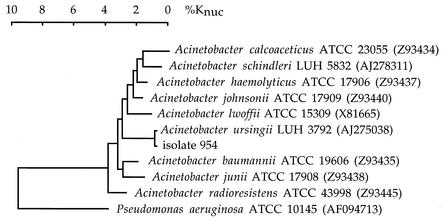
Blood samples were inoculated in aerobic and anaerobic blood culture vials (BACTEC PLUS; BD Diagnostic Systems, Sparks, Md.). Aerobic vials were positive and were subcultured on nutrient agar at 37°C. After 24 h of incubation, colonies were 1 to 1.5 mm in diameter, circular, convex, smooth, and slightly opaque with entire margins. Staining of the bacteria showed gram-negative coccobacilli. Growth in brain heart infusion (BHI) broth was observed at 37°C but not at 41 and 44°C. The microorganism (isolate 954) was nonmotile, strictly aerobic, and oxidase negative. It grew on MacConkey agar (colorless colonies), was nonhemolytic on sheep blood agar, did not oxidize d-glucose, did not reduce nitrate, and was urease and gelatinase negative. In an attempt to identify this isolate, the strips API 20 NE and API ID 32 GN (bioMérieux, Marcy l'Etoile, France) were used as recommended by the manufacturer. The repeated bacterial identifications obtained with API 20 NE and API ID 32 GN strips were Acinetobacter junii or Acinetobacter johnsonii (code no. 0000071; percentage of identification [p] = 63.5%; index of typicity [T] = 0.77) and A. johnsonii (code no. 00270063062; p = 90.5%; T = 0.87), respectively. These results prompted us to determine the 16S rRNA gene (16S ribosomal DNA [rDNA]) sequence of the isolate as previously described (3, 6). Briefly, the 16S rDNA was amplified by PCR with the primers Ad (5′-AGAGTTTGATC[A,C]TGGCTCAG-3′) and rJ (5′-GGTTACCTTGTTACGACTT-3′). A total of 1,484 continuous nucleotides of 16S rDNA were determined. The complete 16S rDNA sequence of the isolate was compared to all bacterial sequences available from the GenBank database by using the Blast program (National Center for Biotechnology Information) and showed 99% similarity to that of the type strain of Acinetobacter ursingii (GenBank accession no. AJ275038). 16S rDNA sequences from phylogenetically related strains were obtained from the GenBank database. All 16S rDNA sequences were aligned with CLUSTAL X, and a phylogenetic tree was constructed by using DENDROGRAF, a program of the Taxotron package (Taxolab Institut Pasteur, Paris, France) (Fig. 1). Antimicrobial susceptibility of the isolate was determined by the agar diffusion method using the Epsilometer test (E test; AB BIODISK, Solna, Sweden) on Mueller-Hinton agar as recommended by the manufacturer. MIC results were as follows: amoxicillin, 16 μg/ml; piperacillin, 12 μg/ml; cefotaxime, 32 μg/ml; cefepime, 24 μg/ml; ceftazidime, 128 μg/ml; imipenem, 0.125 μg/ml; gentamicin, 0.25 μg/ml; amikacin, 1 μg/ml; tobramycin, 0.5 μg/ml; rifampin, 3 μg/ml; ciprofloxacin, 0.19 μg/ml. An assay for the detection of beta-lactamase by using a nitrocefin disk (BD Cefinase; BD Diagnostic Systems) was positive.
FIG. 1.
Phylogenetic tree of isolate 954 of A. ursingii and the type strains of related species based on comparative analysis of 16S rRNA gene sequences. Sequence alignment was by the CLUSTAL method. The dendrogram was generated by using the neighbor-joining algorithm and choosing Pseudomonas aeruginosa as the outgroup. %Knuc, percentage of nucleotide substitution.
Members of the genus Acinetobacter belong to the gamma subdivision of the class Proteobacteria. In 1986, Bouvet and Grimont described four new species of Acinetobacter, namely, Acinetobacter baumannii, Acinetobacter haemolyticus, A. johnsonii, and A. junii (2). In addition, these authors emended the descriptions of two other species, Acinetobacter calcoaceticus and Acinetobacter lwoffii. In 1988, Acinetobacter radioresistens, from the environment, was described (9). More recently, A. ursingii and Acinetobacter schindleri, isolated from human clinical specimens, have been delineated (7, 8). Thus, at present, the genus Acinetobacter consists of the above nine species. However, this remains insufficient to name all the members of this genus, as shown by the 21 different DNA groups (genomospecies) previously reported (5). In clinical laboratories, the identification of nonfermentative gram-negative rods is usually carried out by using identification systems such as the API 20 NE and the API ID 32 GN strips (bioMérieux). A. baumannii, the most frequent species of Acinetobacter involved in nosocomial infections, is easily identified with these systems. However, the discriminative power of the tests using API strips has been shown to be insufficient for accurate identification of the other species of Acinetobacter (1). In the present case, the preliminary misidentification was due to the absence of A. ursingii in the databases of the API 20 NE and API ID 32 GN strips. Additional phenotypic tests, such as growth in BHI broth at 37 and 41°C and oxidization of glutarate and l-aspartate, may help to differentiate A. ursingii from A. junii and A. johnsonii (Table 1).
TABLE 1.
Phenotypic tests for differentiating between A. ursingii, A. junii, and A. johnsoniia
| Test | Resultb for:
|
||
|---|---|---|---|
| A. ursingii | A. junii | A. johnsonii | |
| Growth at 37°C in BHI broth | + | + | − |
| Growth at 41°C in BHI broth | − | + | − |
| Oxidization of glutarate | + | − | − |
| Oxidization of l-aspartate | + | − | V |
Data are from Nemec et al. (8).
+, positive for 90 to 100% of strains; −, positive for 0 to 10% of strains; V, positive for 11 to 89% of strains.
Acinetobacter species are commonly isolated from the environment and may also be isolated from humans (skin and mucous membranes) (10). During the last 2 decades, they have emerged as nosocomial pathogens. In debilitated patients, they may be responsible for severe and fatal infections involving the respiratory tract, the urinary tract, and wounds (including catheter sites). Risk factors for infection include serious underlying disease such as cancer, intravascular or intravesical catheterization, treatment with broad-spectrum antibiotics or corticosteroids, prolonged hospital stay, and stay in intensive care units. Due to their prolonged survival in the environment, Acinetobacter species may spread among patients and cause hospital-associated outbreaks (4, 11). In the present case, the pulmonary adenocarcinoma and the corticosteroid administration were two major risk factors for the dissemination of A. ursingii in blood. Although A. ursingii has been isolated solely from humans, its natural habitat is not known. We assume that this isolate colonized the patient's skin and that the intravascular catheterization triggered its spread to the bloodstream. The accurate identification of the species of Acinetobacter is important for epidemiological and therapeutic reasons. Improvement of the taxonomy and of the methods of identification must be taken into account when analyzing previous studies, which often used names or methods no longer valid. The Acinetobacter species involved in human infections and their antimicrobial susceptibilities remain partly undetermined. A. ursingii has not been reported in infectious processes apart from its recent description as a new species (8). However, this species may have the same medical importance as our isolate. Indeed, out of the 29 strains reported in the literature, 13 were isolated from the blood of patients suffering from severe underlying disease. Furthermore, A. ursingii strains may have the potential to spread to other patients, as demonstrated by molecular typing (7). For these reasons, clinical microbiologists must be aware of the opportunistic pathogenicity of this newly described species, which deserves further studies to determine its prevalence in humans.
REFERENCES
- 1.Bernards, A. T., J. van der Toorn, C. P. van Boven, and L. Dijkshoorn. 1996. Evaluation of the ability of a commercial system to identify Acinetobacter genomic species. Eur. J. Clin. Microbiol. Infect. Dis. 15:303-308. [DOI] [PubMed] [Google Scholar]
- 2.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., and Acinetobacter junii sp. nov. and emended description of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 3.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouby, A., M.-J. Carles-Nurit, N. Bouziges, G. Bourg, R. Mesnard, and P. J. M. Bouvet. 1992. Use of pulsed-field gel electrophoresis for investigation of hospital outbreaks of Acinetobacter baumannii. J. Clin. Microbiol. 30:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibrahim, A., P. Gerner-Smidt, and W. Liesack. 1997. Phylogenetic relationship of the twenty-one DNA groups of the genus Acinetobacter as revealed by 16S ribosomal DNA sequence analysis. Int. J. Syst. Bacteriol. 47:837-841. [DOI] [PubMed] [Google Scholar]
- 6.Janvier, M., and P. A. D. Grimont. 1995. The genus Methylophaga, a new line of descent within phylogenetic branch γ of Proteobacteria. Res. Microbiol. 146:543-550. [DOI] [PubMed] [Google Scholar]
- 7.Nemec, A., L. Dijkshoorn, and P. Jezek. 2000. Recognition of two novel phenons of the genus Acinetobacter among non-glucose-acidifying isolates from human specimens. J. Clin. Microbiol. 38:3937-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemec, A., T. De Baere, I. Tjernberg, M. Vaneechoutte, T. J. K. van der Reijden, and L. Dijkshoorn. 2001. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. E vol. Microbiol. 51:1891-1899. [DOI] [PubMed] [Google Scholar]
- 9.Nishimura, Y., T. Ino, and H. Hzuka. 1988. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int. J. Syst. Bacteriol. 38:209-211. [Google Scholar]
- 10.Seifert, H., L. Dijkshoorn, P. Gerner-Smidt, N. Pelzer, I. Tjernberg, and M. Vaneechoutte. 1997. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J. Clin. Microbiol. 35:2819-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendt, C., B. Dietze, E. Dietz, and H. Rüden. 1997. Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 35:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]