Abstract
Given the diversity of human immunodeficiency virus type 1 (HIV-1) subtypes and the emergence of subtypes other than HIV-1 subtype B in the United States, genotypic assays must be capable of delivering sequence data on diverse HIV-1 subtypes. We evaluated the performance of Visible Genetics TRUGENE HIV-1 genotyping kit and Applied Biosystems ViroSeq HIV-1 genotyping system on a panel of 34 well-characterized HIV-1 viral stocks (subtypes A through H). Both assays perform well on diverse HIV-1 subtypes despite being designed for HIV-1 subtype B. The TRUGENE assay produced sequence data for 31 isolates but not for one C and two G isolates. The TRUGENE assay using prototype 1.5 RT-PCR primers and the ViroSeq assay were both successful for all variants tested, although five isolates lacked double-strand sequence coverage in the ViroSeq assay. The availability of standardized HIV-1 genotyping kits that perform reliably with all HIV subtypes will facilitate broad implementation of HIV-1 resistance testing.
The human immunodeficiency virus (HIV) population is continuously evolving, generating genetically distinct viral variants (3, 9, 14). The propagation of novel mutants is a function of environmental selective pressures created by immunological responses of the host and drug therapy. Thus, mutants that confer a survival advantage will propagate and emerge as a major quasispecies (3). The incomplete inhibition of HIV replication can lead to the emergence of virus that has reduced susceptibility to antiretroviral drugs, limiting the efficacy of antiretroviral treatment regimens. Drug-resistant HIV type 1 (HIV-1) can also be transmitted from one individual to another (4, 7, 8, 13, 20, 22). Almost all of the information on the characterization of HIV-1 drug resistance mutation sites has come from studies of HIV-1 group M, subtype B infection, the most common subtype found in the United States and Europe. However, HIV-1 infections caused by other HIV-1 subtypes predominate in other regions of the world. With an increase in prevalence of non-B HIV-1 subtypes in the United States and the availability of antiretroviral drugs in regions of the world where HIV-1 infections other than those of subtype B are prevalent, research on the drug resistance patterns in non-B HIV-1 subtypes and the clinical monitoring of patients having non-B HIV-1 infections is becoming increasingly important (1, 9, 10, 17, 19, 21). The global deployment of U.S. military personnel has increased the incidence of infections with non-B HIV-1 subtypes in military personnel, further underscoring the need for HIV-1 resistance genotyping tests that work reliably with diverse HIV-1 subtypes (1, 2, 6, 8, 12, 16, 17-19). This study assesses the ability of Visible Genetics TRUGENE HIV-1 genotyping test and Applied Biosystems ViroSeq HIV-1 genotyping system to successfully amplify HIV-1 viral RNA and produce DNA sequences for non-B HIV-1 subtypes.
MATERIALS AND METHODS
HIV-1 subtype panel.
A panel of 34 HIV-1 isolates (Table 1) of group M subtypes A through H (two of subtype A, two of subtype A/G, seven of subtype B, six of subtype C, two of subtype D, eight of subtype E, four of subtype F, two of subtype G, and one of subtype H) was used to assess the performance of the TRUGENE and ViroSeq HIV-1 genotyping tests (11, 15, 18). Twenty-nine of the viral stocks were from the Walter Reed Army Institute of Research. Five of these viral stocks were obtained from BBI-Biotech Research Laboratories, Inc. (Gaithersburg, Md.). All viral stocks were characterized by p24 antigen testing, electron microscopy particle counts, viral RNA testing, and sequence (15).
TABLE 1.
Performance of genotypic drug resistance tests on HIV-1 subtypes A through H
| HIV-1 subtype | Country of origin | Isolate identification no. | Consensus derived with TRUGENE primersa
|
TRUGENE primer failured | Consensus derived with ViroSeq version 2.0 | ViroSeq primer failuree | |
|---|---|---|---|---|---|---|---|
| 1.0 RT-PCR | 1.5 RT-PCR | ||||||
| A | Uganda | UG273 | Y | Y | Protease | Y | D |
| A/G | Djibouti | DJ258 | Y | Y | Protease | Y | D |
| A/G | Djibouti | DJ263 | Y | Y | Protease | Y | D |
| A | Uganda | UG275c | Y | Y | None | Y | D |
| B | United States | US1 | Y | Y | None | Y | None |
| B | United States | US2 | Y | Y | None | Y | None |
| B | United States | US3 | Y | Y | None | Y | None |
| B | United States | US4 | Y | Y | None | Yb | F |
| B | Thailand | CM237 | Y | Y | None | Y | D |
| B | Thailand | BK132 | Y | Y | None | Y | D |
| B | Brazil | BZ167 | Y | Y | None | Y | None |
| C | Zambia | ZAM18 | Y | Y | Protease | Yb | A & D |
| C | Uganda | UG268 | Y | Y | Protease | Y | D |
| C | Ethiopia | ETH2220 | Y | Y | Protease | Yb | D & H |
| C | Senegal | SE364 | Y | Y | Protease | Y | D |
| C | Somalia | SM145 | N | Y | Protease + 1.0 RT-PCR | Y | D |
| C | Djibouti | DJ259c | Y | Y | Protease | Y | D |
| D | Senegal | SE365 | Y | Y | None | Y | None |
| D | Uganda | UG270 | Y | Y | None | Y | None |
| E | Thailand | CM235 | Y | Y | Protease | Y | D |
| E | Thailand | CM238 | Y | Y | Protease | Y | D |
| E | Thailand | CM240 | Y | Y | Protease | Yb | A & D |
| E | Thailand | CM243 | Y | Y | Protease | Y | D |
| E | Thailand | POC30506 | Y | Y | Protease | Y | None |
| E | Indonesia | ID12 | Y | Y | Protease | Y | D |
| E | Indonesia | ID17 | Y | Y | Protease | Y | D |
| E | Thailand | NP1465 | Y | Y | Protease | Y | D |
| F | Brazil | BZ126 | Y | Y | Protease | Y | None |
| F | Brazil | BZ162 | Y | Y | None | Y | None |
| F | Brazil | BZ163 | Y | Y | Protease | Y | None |
| F | Romania | BCI-R07c | Y | Y | None | Y | D |
| G | Zaire | HH8793 | N | Y | Protease + 1.0 RT-PCR | Y | None |
| G | Kenya | BCF-DIOUMc | N | Y | 1.0 RT-PCR | Y | None |
| H | Zaire | BCP-KITAc | Y | Y | Protease | Yb | A & D |
Abbreviations: Y, consensus sequence derived from both strands; N, No TRUGENE sequence data were generated for these HIV-1 isolates using version 1.0 RT-PCR primers supplied in the FDA-cleared kit.
Consensus sequence derived from partial single-strand data.
Isolate obtained from BBI-Biotech Research Laboratories, Inc. All of the other isolates were from stocks on site.
The protease (PR) sequencing primers did not deliver sequence data for isolates noted. The 1.0 RT-PCR primers did not generate sufficient amplicon for CLIP sequencing reactions.
ViroSeq sequencing primers A, D, F, and H did not generate readable sequence data for isolates noted.
Initial studies were performed using viral stocks having greater than 20,000 HIV-1 RNA copies/ml in order to ensure that sufficient template was available to allow for a successful amplification. Panel members that were not successfully sequenced by either method were repeat tested using viral stocks at concentrations greater than 100,000 RNA copies/ml. Due to the inability of the TRUGENE assay to deliver sequence data for three of the HIV-1 panel members, we obtained the prototype 1.5 RT-PCR primers (research use only) from Visible Genetics. These reverse transcription (RT)-PCR primers were designed to amplify the protease and RT gene regions for all known HIV subtypes. Since limited quantities of the original viral stocks remained, new dilutions targeted at 5,000 RNA copies/ml were prepared for use in assessing the performance of the prototype 1.5 RT-PCR primers and the quality of ViroSeq sequence data generated by capillary electrophoresis. HIV viral RNA concentrations (1,800 to 20,000 RNA copies/ml) were determined using the Amplicor HIV-1 MONITOR test (versions 1.0 and 1.5; Roche Diagnostics, Branchburg, N.J.).
HIV-1 genotype testing.
Viral RNA was extracted from each panel member, amplified, and sequenced following procedures recommended by the manufacturers of the tests. Consensus sequences were generated for each panel member using software provided by the manufacturer of each test (Visible Genetics OpenGene DNA sequencing system and Applied Biosystems ViroSeq HIV genotyping system software, version 2.2). Software programs for each assay compared the consensus sequence generated to HIV-1 reference sequences (TRUGENE, LAV-1; ViroSeq, pNL4-3). An interpretative report was generated for the TRUGENE assay while a research report was issued for the ViroSeq assay.
RESULTS
The Visible Genetics TRUGENE HIV-1 genotyping kit and Applied Biosystems ViroSeq HIV-1 genotyping system were assessed for their ability to (i) generate RT-PCR amplicons for non-B HIV-1 subtypes, (ii) provide double-stranded nucleotide sequence data on each subtype panel member, and (iii) generate equivalent sequence data.
Performance of the TRUGENE HIV-1 genotyping kit.
The Food and Drug Administration (FDA)-approved version of the TRUGENE HIV-1 genotyping kit successfully genotyped 31 out of 34 (91%) of the HIV-1 subtype panel (Table 1). This assay did not produce sequence data for one C (SM145) and two G HIV-1 subtypes (HH8793 and BCF-DIOUM). The sequencing reaction profiles for SM145 and HH8793 were negative independent of viral RNA stock input. A dirty sequence profile was observed for BCF-DIOUM when the original TRUGENE RT-PCR (1.0 RT-PCR) primers were used in the amplification reactions. When Visible Genetics prototype 1.5 RT-PCR primers were substituted for the 1.0 RT-PCR primers supplied in the kit, sequence data were generated for all of the HIV-1 subtypes using the viral stocks with 5,000 RNA copies/ml.
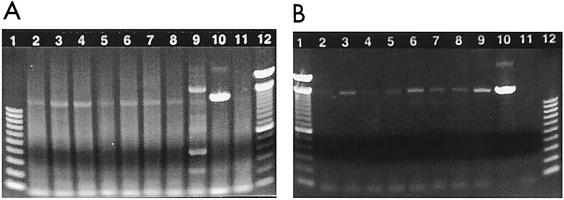
The improvement in the performance of the TRUGENE assay when the 1.5 RT-PCR primers were used to produce amplicons suggested that the prototype 1.5 RT-PCR primers were more efficient than the 1.0 RT-PCR primers. In order to determine if the improved performance was due to better primer efficiency, one panel member was selected from each HIV subtype and tested using both of the RT-PCR primer sets. Amplification products were fractionated electrophoretically through 1.2% agarose E-Gels (Invitrogen, Carlsbad, Calif.) at 70 V for 30 min. The electrophoretic gel profiles of the RT-PCR products generated using the FDA-cleared TRUGENE HIV-1 genotyping kit (with 1.0 RT-PCR primers) are shown in Fig. 1A. A smear of nonspecific background amplification is visible for all RT-PCRs using the RT-PCR primers supplied in the TRUGENE kit. The gel profile for the RT-PCR products generated using the prototype 1.5 RT-PCR primers shows little to no nonspecific amplification and an amplicon of ∼1,300 bp (Fig. 1B). The amount of amplified product generated varies for each sample and this difference is clearly visible by electrophoretic separation through agarose gels. The TRUGENE HIV-1 assay does not require large quantities of RT-PCR product for sequencing since additional amplification (nested) of the target occurs in the CLIP sequencing reactions. Amplified product may not be clearly visible in the agarose gel (for example, A subtype, Fig. 1B, lane 2), but a sufficient amount of amplified DNA is available for successful amplification in CLIP sequencing reactions. The CLIP bidirectional sequencing reaction is a unique feature of the TRUGENE HIV-1 assay system.
FIG. 1.
Electrophoretic gel profile of RT-PCR products generated using the Visible Genetics TRUGENE HIV-1 genotyping test. One HIV-1 isolate was selected from each HIV-1 subtype for analysis using the TRUGENE kit (version 1.0) and prototype 1.5 RT-PCR primers. The 1.5 RT-PCR primers were substituted for the kit RT-PCR primers and cycling parameters were modified. Lanes 1 and 12 contain 100-bp DNA ladders. Lanes 2 through 9 contain subtypes A, B, C, D, E, G, F, and H. Lane 10 contains the positive control from the kit. Lane 11 contains a negative specimen control. (A) Gel profile of amplified product generated by the TRUGENE kit (version 1.0). (B) Gel profile of amplified product generated using the prototype 1.5 RT-PCR primers with the TRUGENE kit.
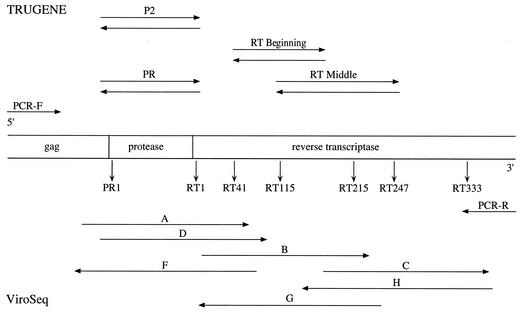
The TRUGENE assay provides two primer sets in the kit for sequencing of the HIV-1 protease gene (PR and P2) (Fig. 2). The protease (PR) primers were found to fail on some HIV subtype B specimens, which resulted in the manufacture of a second set of protease primers for use in sequencing reactions (P2). Both of these primer sets are supplied in the FDA-cleared kit and are used routinely. The PR sequencing primers did not generate sequence data for 21 (61%) of the specimens (Table 1). The PR primers performed well on subtype B and D isolates, while the P2 sequencing primers performed better on all of the other HIV subtypes. The P2 sequencing primer pair successfully generated sequence data for all of the HIV subtypes tested in this study. Sequence data generated only by the P2 primer pair is found to have short regions (less than 35 bases) of single-strand sequence coverage at the beginning of the protease gene. The E subtype isolates had larger regions of single-strand sequence (up to 50 bases) at the beginning of the protease gene and at the 3′ end of the RT gene compared to the other subtypes. The resolution of the bases in the reverse transcriptase gene region was not as clean for HIV subtypes E and G. Fewer mixed-base calls were observed when prototype 1.5 RT-PCR primers were used to generate amplicons, suggesting that the use of an amplicon with less background of nonspecific product in sequencing reactions resulted in better resolution. Overall, double-strand sequence data were generated for all of the HIV-1 subtypes across regions of interest used to determine HIV drug resistance.
FIG. 2.
Schematic of primers used in DNA sequencing reactions for the protease and reverse transcriptase gene regions. The top region shows the names of the TRUGENE primers and their orientation with respect to the HIV-1 gene region sequence. The bottom half of the diagram shows the names and orientation of the primers used in the ViroSeq test.
The TRUGENE test generates an interpretative report for use by clinicians and a research report that lists all of the drug resistance mutation sites, synonymous (silent) mutations, polymorphisms, and unexpected mutations. Nucleotide mutations that do not result in a change in the translated amino acid are denoted as synonymous mutations. A mean average of the synonymous mutations was calculated for each HIV-1 subtype. Five to fourteen additional synonymous mutations are observed in the protease gene for all subtypes other than HIV-1 subtype B. The reverse transcriptase gene has 14 to 30 additional synonymous mutations for subtypes other than HIV-1 subtype B. All of the subtypes other than HIV-1 subtype B have significantly larger numbers of synonymous mutations. Subtype D has the lowest number of synonymous mutations, while subtypes C and G have the largest number of mutations. A long list of synonymous mutations may suggest that the sequence obtained for a clinical specimen is derived from a non-B subtype infection.
Performance of the ViroSeq HIV-1 genotyping system.
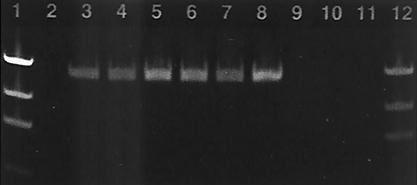
The ViroSeq HIV-1 genotyping system generated sufficient amplicon for all subtypes tested and delivered sequence data on all of the panel members. Problems were encountered in the clarity of DNA sequence data produced on the model 377 DNA sequencer, where gels had to be repeated due to poor gel resolution. Approximately 50% of the panel members exhibited high background fluorescence in profiles generated on the model 377 DNA sequencer. Four of the subtypes other than HIV-1 subtype B (ZAM18, ETH2220, CM240, and BCP-KITA) and one B subtype (US4) had significant lengths of single-strand sequence due to the poor performance of one or two sequencing primers. Due to the clarity problems observed in the sequence data obtained from the model 377 DNA sequencer, 15 panel members were selected for retesting using the HIV-1 viral stocks with 5,000 RNA copies/ml. DNA sequencing reaction products for these 15 specimens were fractionated by capillary electrophoresis. Sequencing profiles generated using the model 3100 genetic analyzer showed well-defined peaks with little to no background fluorescence. All RT-PCR products generated using the ViroSeq RT-PCR module showed a single band amplicon of ∼1,800 bp. Background contributed by nonspecific amplification as observed for the 1.0 RT-PCR primers used in the TRUGENE kit was not observed when purified amplicons from the ViroSeq test were fractionated through 1.2% agarose gels (Fig. 3). Amplification yields were excellent and in most cases exceeded the amount of amplified product generated in the TRUGENE assay. All HIV-1 subtypes produced at least 18 ng of amplicon in 5 μl of reaction mixture. However, the ViroSeq assay requires significantly higher quantities of amplicon (at least 12 ng/5 μl) for execution of the sequencing reactions than the TRUGENE assay since a nested amplification does not occur in the ViroSeq sequencing reactions.
FIG. 3.
Electrophoretic gel profile of RT-PCR products generated using the Applied Biosystems ViroSeq HIV-1 genotyping system. Fifteen members of the HIV-1 subtype panel at 5,000 RNA copies/ml were subjected to sequence analysis by the ViroSeq assay using the model 3100 genetic analyzer. The gel profile for purified amplified product is shown and is representative of all of the HIV-1 subtypes tested. Lanes 1 and 12 contain two amounts of the DNA mass ladder. Lanes 3 and 4 contain the two subtype D isolates (46 and 44 ng). Lanes 5, 6, and 7 contain subtypes F, H, and A (74, 70, and 53 ng). Lane 8 contains the positive control provided in the kit (84 ng). Lane 10 contains the negative control.
A summary of sequencing primer performance is shown in Table 1. Primers A and D sequence the same region of the protease gene and are sense directional primers (Fig. 2). Both primers are used routinely in our laboratory since one or the other primer may not generate readable sequence data. Primer D did not successfully generate sequence data for 22 (65%) of the panel members. Primer A did not produce sequence data for three panel members (C subtype from Zambia [ZAM18], E subtype from Thailand [CM240], and H subtype from Zaire [BCP-KITA]). Primer D also did not generate sequence data for the protease gene for each of these three subtypes. Since primer D is a backup for primer A, single-strand sequence coverage was obtained for these three panel members in the protease gene region. Single-strand sequence data were also obtained for the protease gene region in one B subtype (US4) due to the failure of primer F in producing sequence data across this region. The failure of primer F to deliver sequence data on a B subtype was reproducible and observed both on the 377 DNA sequencer and the 3100 genetic analyzer. Primer H did not deliver sequence data for an Ethiopian C subtype (ETH2220), resulting in single-strand sequence coverage at the three prime end of the RT gene. The lack of successful amplification by sequencing primers A, D, F, and H results in the generation of single-strand sequence regions where mixed-base calls cannot be discerned. Rules for calling bases ambiguous (mixed) require that both strands of the sequence show a mixed base in that particular position. Thus, sequences generated using data from single-strand coverage do not allow for the determination of mixed bases and may not fully reflect the pool of virus in the specimen tested.
Poor resolution or high background fluorescence were observed for sequencing primers A, B, D, G, and H for three, five, two, seven, and six panel members, respectively, when sequencing reactions were fractionated on the model 377 DNA sequencer. Such poor resolution may reflect problems with gel polymerization or the quality of the sequencing reactions generated by the primers. The resolution improved when the sequencing reactions were repeated and analyzed by capillary electrophoresis (model 3100 genetic analyzer), suggesting that the resolution problems encountered previously were due in part to the polymerization of the gel used on the model 377 DNA sequencer. We observed a higher variability in the quality of the sequence data delivered on the model 377 DNA sequencer compared to the data produced by the model 3100 genetic analyzer.
Comparative analysis.
The DNA sequences generated for each panel member were aligned using the MegAlign program of Lasergene Navigator (version 1.6; DNASTAR Inc., Madison, Wis.). The electropherograms were examined for all highlighted discrepancies in order to confirm sequence calls. Approximately 97% of the differences noted were due to differences in ambiguity calls where sites are called as a mixture of two or more bases versus single-base calls. The FDA-cleared TRUGENE assay where the 1.0 RT-PCR primers are used had twice as many ambiguities as the ViroSeq assay. Fewer ambiguities were observed in the TRUGENE assay when the prototype 1.5 RT-PCR primers were used to generate the amplicon. This suggests that the use of PCR products having lower levels of background contaminants results in sequencing reactions which have better resolution. Of 57 site differences, 27 differences were due to ambiguities in the TRUGENE reading (prototype 1.5 RT-PCR primers) where one of the calls associated with the mixed-base call was equivalent to the single-base call given in the ViroSeq assay. The ViroSeq test accounted for 28 ambiguities that were in agreement with the defined base call given by the TRUGENE test. Two of the scored differences were due to different base calls between the two assays. These calls were not located at drug resistance sites and therefore did not alter the interpretation of the sequence data.
All sequences were checked for uniqueness and subtype grouping by phylogenetic tree analysis using neighbor-joining, maximum-likelihood, and distance analyses (Wisconsin Package, version 10; Genetics Computer Group). This analytic tool allows for the identification of HIV subtype and can be used to monitor for contamination or mix-up of specimens during genotyping test procedures. Sequence results obtained for each panel member branched with its corresponding result obtained using the other test procedure. Each panel member grouped within the region of the tree that corresponded with its cognate subtype.
DISCUSSION
The Visible Genetics TRUGENE HIV-1 genotyping kit and the Applied Biosystems ViroSeq genotyping system are both capable of delivering HIV-1 resistance data for clinical specimens infected with non-B virus. The FDA-cleared TRUGENE HIV-1 genotyping assay does not make any claims as to performance on non-B HIV-1 subtypes. The TRUGENE assay was unsuccessful in generating sequence data for one C and both G subtypes using the 1.0 RT-PCR primers supplied in the test kit. The prototype 1.5 RT-PCR primers (research use only) provided by Visible Genetics and used with the TRUGENE HIV-1 genotyping kit improved the performance of the test where DNA sequence was successfully generated on all of the panel members. These RT-PCR primers are available from Visible Genetics (research use only) and can be substituted for the primers included in the kit.
The ViroSeq HIV-1 genotyping system was successful in generating sequence data for all members of the HIV-1 subtype panel with no modifications. Amplification was excellent for this assay based upon the amount of RT-PCR product generated for each viral isolate. Performance issues exist for the sequencing primers where regions of single-strand sequences are generated due to poor primer performance or in regions were sequence data had high background fluorescence. All regions in which single-strand sequences were generated had excellent resolution, but without the sequence of the complementary strand base call ambiguities cannot be fully discerned. Improved resolution in the sequencing profiles was observed when sequencing reactions were fractionated using the model 3100 genetic analyzer compared to electropherograms generated on the model 377 DNA sequencer. The quality of the data delivered on the model 3100 genetic analyzer was superior to that of the data obtained on the model 377 DNA sequencer.
Essentially equivalent nucleotide sequence data are generated by the TRUGENE and ViroSeq assays. Differences observed are due to ambiguities resulting in mixed-base sequence calls in one assay compared to defined single-base calls in the other assay system. These ambiguities can result in differences in the interpretation of resistance data if the ambiguity occurs at drug-associated resistance sites. The ambiguities observed in this study are similar to those reported by other laboratories (5, 6, 16).
The expanded access to antiretroviral drugs in populations where non-B HIV-1 subtypes predominate and the increasing diversity of the viral population require continuous monitoring of HIV-1 genotyping tests so as to allow for improvements in assay performance. The information obtained in this study can be used to improve the design of the primers used in both kits, leading to better optimization and broader utility of the assays. The extensive sequence diversity in the RT gene region and the potential for coinfection with two or more subtypes in regions where multiple HIV subtypes prevail, leading to recombinant viral strains, provide a challenge in designing assays that can successfully amplify and sequence all HIV subtypes. As more HIV-1 isolates are sequenced from geographical regions where subtypes other than HIV-1 subtype B prevail, isolates will be found which are not sequenced by either the TRUGENE or ViroSeq assays. Modified versions of HIV-1 genotyping tests will be required to address evolving HIV-1 sequence diversity.
Acknowledgments
We thank Visible Genetics, Inc., for providing the prototype 1.5 RT-PCR primers and two test kits.
The views and opinions expressed herein are those of the authors and do not purport to reflect the official policy or position of the U.S. Army or the Department of Defense.
This work was supported in part by Cooperative Agreement DAMD17-93-V-3004, between the U.S. Army Medical Research and Materiel Command and the Henry M. Jackson Foundation for the Advancement of Military Medicine.
REFERENCES
- 1.Brodine, S. K., J. R. Mascola, P. J. Weiss, S. I. Ito, K. R. Porter, A. W. Artenstein, F. C. Garland, F. E. McCutchan, and D. S. Burke. 1995. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet 346:1198-1199. [DOI] [PubMed] [Google Scholar]
- 2.Brodine, S. K., R. A. Shaffer, M. J. Starkey, S. A. Tasker, J. L. Gilcrest, M. K. Louder, A. Barile, T. C. VanCott, M. T. Vahey, F. E. McCutchan, D. L. Birx, D. D. Richman, and J. R. Mascola. 1999. Drug resistance patterns, genetic subtypes, clinical features, and risk factors in military personnel with HIV-1 seroconversion. Ann. Intern. Med. 131:502-506. [DOI] [PubMed] [Google Scholar]
- 3.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 4.Colgrove, R. C., J. Pitt, P. H. Chung, S. L. Welles, and A. J. Japour. 1998. Selective vertical transmission of HIV-1 antiretroviral resistance mutations. AIDS 12:2281-2288. [DOI] [PubMed] [Google Scholar]
- 5.Erali, M., S. Page, L. G. Reimer, and D. R. Hillyard. 2001. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J. Clin. Microbiol. 39:2157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontaine, E., C. Riva, M. Peeters, J. C. Schmit, E. Delaporte, K. Van Laethem, K. Van Vaerenbergh, J. Snoeck, E. Van Wijngaerden, E. De Clercq, M. Van Ranst, and A. M. Vandamme. 2001. Evaluation of two commercial kits for the detection of genotypic drug resistance on a panel of HIV-1 subtypes A through J. J. Acquir. Immune Defic. Syndr. 28:254-258. [DOI] [PubMed] [Google Scholar]
- 7.Hecht, F. M., R. M. Grant, C. J. Petropoulos, B. Dillon, M. A. Chesney, H. Tian, N. S. Hellman, N. I. Bandrapalli, L. Difilio, B. Brandson, and J. O. Kahn. 1998. Sexual transmission of an HIV-1 variant resistant to multiple reverse-transcriptase and protease inhibitors. N. Engl. J. Med. 339:307-311. [DOI] [PubMed] [Google Scholar]
- 8.Hirsch, M. S., F. Brun-Vezinet, R. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an international AIDS society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 9.Hu, D. J., T. J. Dondero, M. A. Rayfield, J. R. George, G. Schochetman, H. W. Jaffe, C.-C. Luo, M. L. Kalish, B. G. Weniger, C.-P. Pau, C. A. Schable, and J. W. Curran. 1996. The emerging genetic diversity of HIV: the importance of global surveillance for diagnostics, research, and prevention. JAMA 275:210-216. [PubMed] [Google Scholar]
- 10.Irwin, K. L., C. P. Pau, D. Lupo, D. Pientazek, C. C. Luo, N. Olivo, M. Rayfield, D. J. Hu, J. T. Weber, R. A. Respess, R. Janssen, P. Minor, J. Ernst, and the Centers for Disease Control and Prevention-Bronx Lebanon HIV Serosurvey Team. 1997. Presence of human immunodeficiency virus (HIV) subtype A infection in a New York community with high HIV prevalence: a sentinel site for monitoring HIV genetic diversity in North America. J. Infect. Dis. 176:1629-1633. [DOI] [PubMed] [Google Scholar]
- 11.Jagodzinski, L. L., D. L. Wiggins, J. L. McManis, S. Emery. J. Overbaugh, M. Robb, S. Bodrug, and N. L. Michael. 2000. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J. Clin. Microbiol. 38:1247-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasky, M., J. L. Perret, M. Peeters, F. Bibollet-Ruche, F. Liegeois, D. Patrel, S. Molinier, C. Gras, and E. Delaporte. 1997. Presence of multiple non-B subtypes and divergent subtype B strains of HIV-1 in individuals infected after overseas deployment. AIDS 11:43-51. [DOI] [PubMed] [Google Scholar]
- 13.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 14.McCutchan, F. E., M. O. Salminen, J. K. Carr, and D. S. Burke. 1996. HIV-1 genetic diversity. AIDS 10(Suppl. 3):S13-S20. [PubMed]
- 15.Michael, N. L., S. A. Herman, S. Kwok, K. Dreyer, J. Wang, C. Chistopherson, J. P. Spadoro, K. K. Y. Young, V. Polinis, F. E. McCutchan, J. Carr, J. R. Mascola, L. L. Jagodzinski, and M. L. Robb. 1999. Development of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 and performance of an improved AMPLICOR HIV-1 MONITOR test with isolates of diverse subtypes. J. Clin. Microbiol. 37:2557-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mracna, M., G. Becker-Pergola, J. Dileanis, L. A. Guay, S. Cunningham, J. B. Jackson, and S. H. Eshleman. 2001. Performance of Applied Biosystems ViroSeq HIV-1 genotyping system for sequence-based analysis of non-subtype B human immunodeficiency virus type 1 from Uganda. J. Clin. Microbiol. 39:4323-4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pieniazek, D., M. Rayfield, D. J. Hu, J. Nkengasong, S. Z. Wiktor, R. Downing, B. Biryahwaho, T. Mastro, A. Tanuri, V. Soriano, R. Lal, T. Dondero, and the HIV variant working group. 2000. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naïve individuals worldwide. AIDS 14:1489-1495. [DOI] [PubMed] [Google Scholar]
- 18.Vahey, M., M. Nau, S. Barrick, J. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. A. Wegner. 1999. Performance of the Affymetrix GeneChip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vergne, L., M. Peeters, E. Mpoudi-Ngole, A. Bourgeois, F. Liegeois, C. Toure-Kane, S. Mboup, C. Mulanga-Kabeya, E. Saman, J. Jourdan, J. Reynes, and E. Delaporte. 2000. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naïve patients. J. Clin. Microbiol. 38:3919-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naïve HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 21.Weidle, P. J., C. E. Ganea, K. L. Irwin, D. Pieniazek, J. P. McGowan, N. Olivo, A. Ramos, C. Schable, R. B. Lal, S. D. Holmberg, and J. A. Ernst. 2000. Presence of human immunodeficiency virus (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J. Infect. Dis. 181:470-475. [DOI] [PubMed] [Google Scholar]
- 22.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]