Abstract
A total of 56 C. difficile strains were selected from 310 isolates obtained from different hospitals in Japan and Korea and from healthy infants from Indonesia. Strains that had been previously typed by pulsed-field gel electrophoresis and PCR ribotyping, were characterized by toxinotyping and binary toxin gene detection. When toxinotyped, 35 strains were determined to be toxinotype 0, whereas 21 strains showed variations in toxin genes and could be grouped into 11 variant toxinotypes. Six of the toxinotypes had been described before (I, III, IV, VIII, IX, and XII). In addition, five new toxinotypes were defined (XVI to XX). Three of the new toxinotypes (XVIII, XIX, and XX) vary only in repetitive regions of tcdA and produce both toxins. In two strains from toxinotypes XVI and XVII, the production of TcdA could not be detected with commercial immunological kits. Strain J9965 (toxinotype XVII) was in PaLoc similar but not identical to another known A−B+ strain, C. difficile 8864. Strain SUC 36 (toxinotype XVI), on the other hand, was similar to well-defined group consisting of toxinotypes V, VI, and VII, which thus far includes only A+B+ strains. Toxinotypes XVI and XVII represent two new groups of A−B+ strains. Strains of the well-known A−B+ group from toxinotype VIII have a nonsense mutation at the beginning of tcdA gene, and the introduction of a stop codon at amino acid position 47 results in nonproduction of TcdA. The 5′-end sequence of tcdA in two newly described A−B+ strains does not contain an identical mutation. The prevalence of variant C. difficile strains varied greatly among nine hospitals. Only five strains from four different hospitals were positive in PCR for amplification of the binary toxin gene.
Clostridium difficile causes intestinal infections that range from mild self-curing diarrhea to pseudomembraneus colitis. The main virulence factors for C. difficile are two toxins: toxin A (TcdA, enterotoxin) and toxin B (TcdB, cytotoxin). Toxin production allows one to differentiate nontoxigenic C. difficile strains (TcdA and TcdB nonproducing [A−B−]); toxinogenic strains (producing both toxins [A+B+]); and A−B+ strains, which produce only toxin TcdB (3, 5, 18).
Toxins TcdA and TcdB are encoded by two genes, tcdA and tcdB, which together with three additional genes form a pathogenicity locus, PaLoc (4, 10). Certain variant C. difficile strains differ from the reference strain VPI 10463 in the length and restriction sites in toxin genes, as well as in other regions of PaLoc. Currently, 15 groups of such variant strains are recognized and defined as toxinotypes I to XV (22, 23).
The most studied and well-known representatives of variant C. difficile are the A−B+ strains. Most A−B+ strains belong to toxinotype VIII and are characterized by a 1.8-kb deletion within repetitive regions of tcdA gene (1, 13, 14, 19, 23, 25). Toxinotype X, on the other hand, contains only one strain, C. difficile 8864. This strain has a large 5.9-kb deletion at the 3′ end of the tcdA gene and a 1.1-kb insertion between tcdE and tcdA (23, 26). Strain 8864 was virulent in a hamster model (3), whereas strains from toxinotype VIII were first reported to be frequently isolated from asymptomatic children and type strain C. difficile 1470 did not cause the disease in axenic mice (5). However, many reports were published recently on the isolation of A−B+ strains of toxinotype VIII from symptomatic patients (2, 14, 25), from outbreaks (1, 16), and from patients with pseudomembraneus colitis (2, 11, 17, 25).
Production of the third toxin, binary toxin CDT, was found in some strains (20, 27), but its role in pathogenesis is not yet known. CDT is composed of two nonlinked components: CDTa (enzymatic component) and CDTb (binding component). Genes coding for both components, cdtA and cdtB, have been sequenced (20). They are located at the C. difficile chromosome, but their relative position with respect to PaLoc is not known (9).
Toxinotyping of C. difficile strains has already been applied to two large European strain collections from the Université Catholique de Louvain in Brussels, Belgium (23), and the Anaerobe Reference Unit, PHLS, in Cardiff, Wales (22). In the present study, we screened a collection of strains isolated mainly in Japan, along with some isolates from Korea and Indonesia, for the presence of variant toxinotypes. In addition, we also tested for the presence of genes for binary toxin. We found several types of variant C. difficile strains, some of which are new. In two strains with new types of variant toxin genes, only TcdB could be detected.
MATERIALS AND METHODS
Strains.
Fifty-six C. difficile strains included in the study were selected from 310 isolates from two large groups studied previously (Table 1).
TABLE 1.
Distribution of strains from two groups of Asian C. difficile isolates (group I and II) selected for toxinotyping and testing for the presence of the binary-toxin gene cdtBa
| Strain group | No. of strains fromb:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group I
|
Group II
|
Total (groups I and II) | |||||||||||
| A | B | C | D | E | F | Indon | Total | Kor | H | I | Total | ||
| Total | 34 | 28 | 25 | 22 | 13 | 13 | 8 | 143 | 61 | 57 | 49 | 167 | 310 |
| Strains distributed by toxin productionc | |||||||||||||
| A+B+ | 33 | 25 | 24 | 18 | 11 | 12 | 2 | 125 | 61 | 29 | 30 | 120 | 245 |
| A−B+ | 1 | 3 | 1 | 4 | 2 | 1 | 6 | 18 | 0 | 28 | 19 | 47 | 65 |
| Strains selected for toxinotyping and testing for binary-toxin gene | 5 | 5 | 8 | 11 | 11 | 6 | 5 | 51 | 1 | 2 | 2 | 5 | 56 |
| Variant strains found | |||||||||||||
| A+B+ | 0 | 1BT | 1 | 1 + 1BT | 0 | 0 | 0 | 4 | 1 | 1BT | 2 | 4 | 8 |
| A−B+ | 1 | 2 | 1 | 1 + 1BT | 2 | 1 | 2 + 1BT | 12 | 0 | 1 | 0 | 1 | 13 |
| All variant strains (%)d | 2.9 | 14.2 | 8 | 27.2 | 15.3 | 7.7 | 75 | 15.4 | 1.6 | 50.8 | 42.8 | 30.5 | 23.5 |
Group I was previously typed by PCR ribotyping and PFGE; group II was previously tested based on changes in repetitive regions of the tcdA gene.
A to I, different hospitals from Japan; Indon, healthy infants from Indonesia; Kor, Korean hospital. A superscript “BT” indicates a binary-toxin-gene-positive strain.
A+B+, strains producing both toxins; A−B+, strains producing only TcdB.
The percentage for each location was calculated as the percentage of variant A+B+ strains found plus all A−B+ strains from a given location. All A−B+ strains were regarded as variant strains in spite of the fact that not all of them were actually toxinotyped. However, strains that, in a previous study (characterization of groups I and II), were determined to have amplified repetitive regions with lengths identical to that of the toxinotype VIII type strain and that were in the toxin production test positive only for TcdB were regarded as toxinotype VIII.
The first group included 143 isolates recovered from patients admitted to six hospitals (hospitals A to F) located in diverse areas in Japan between 1996 and 2000 and from healthy infants in Indonesia in 1993. All isolates were typed by two typing systems (PCR ribotyping and pulsed-field gel electrophoresis [PFGE]); results for the strains from hospitals A, B, and C were published by Kato et al. (12). From this first group, 51 isolates were selected; these isolates represented 39 different types according to a combination of two typing systems (Tables 1 and 2).
TABLE 2.
Distribution of toxinotypes and binary-toxin-gene-positive C. difficile strains within PCR ribotypes and PFGE types
| Toxin production | PCR ribotype | PFGE type | Strain sourcea | No. of strains per type/no. of strains studiedf | Toxinotype | Presence (+) or absence (−) of binary toxin gene cdtB |
|---|---|---|---|---|---|---|
| A+B+ | cr | SUC78 | Ind | 1/1 | 0 | − |
| cr | SUC89 | Ind | 1/1 | 0 | − | |
| g-r | NTb | B, D, E, F | 11/4 | 0 | − | |
| hr | Hr-ac | C, D, E | 8/3 | 0 | − | |
| hr | Hr-b | C | 2/1 | 0 | − | |
| hr | SG22-a | F | 1/1 | 0 | − | |
| hr | SG22-b | F | 1/1 | 0 | − | |
| hr | Y11-a | C, E | 2/2 | 0 | − | |
| hr | Y11-b | D | 1/1 | 0 | − | |
| j41 | J9941 | D | 1/1 | 0 | − | |
| j49 | NT | D | 1/1 | 0 | − | |
| j52 | J9952 | D | 1/1 | IV | + | |
| kgy | HR10 | E | 1/1 | 0 | − | |
| ktn | HR02 | E | 2/1 | 0 | − | |
| ktn | HR11-a | E | 1/1 | 0 | − | |
| ktn | HR11-b | E | 1/1 | 0 | − | |
| og39 | J9945 | D | 1/1 | 0 | − | |
| og39 | OG39 | B | 1/1 | 0 | − | |
| og45 | OG45 | B | 1/1 | III | + | |
| okm | HR07 | E | 1/1 | 0 | − | |
| okz | NK09 | A | 1/1 | 0 | − | |
| smz | NT | A, D, E, F, I | 68/5 | 0; XIX (1) | − | |
| ud | NK103 | A | 3/1 | 0 | − | |
| y02 | Y2 | C | 1/1 | XII | − | |
| y05 | Y5 | C | 1/1 | 0 | − | |
| y32 | Y32 | C | 1/1 | 0 | − | |
| yok | OG02-a | A, D, F | 10/3 | 0 (2); I (1) | − | |
| yok | OG02-b | C | 1/1 | 0 | − | |
| g9376 | NDd | H | ∗/1 | IX | + | |
| hr | ND | Kor | ∗/1 | XVIII | − | |
| tr14 | TR14 | I | ∗/1 | XX | − | |
| A−B+ | j65 | J9965 | D | 1/1 | XVII | + |
| suc36 | SUC36 | Ind | 1/1 | XVI | + | |
| fr | ND | H | ∗/1 | VIII | − | |
| fr | Fr-c | A, D | 5/2 | VIII | − | |
| fr | Fr-d | Ind | 1/1 | VIII | − | |
| fr | Fr-e | Ind | 4/1 | VIII | − | |
| fr | Fr-f | B | 1/1 | VIII | − | |
| fr | OG57 | B | 1/1 | VIII | − | |
| fr | Y30 | C | 1/1 | VIII | − | |
| fr | HR16-a | E | 1/1 | VIII | − | |
| fr | HR16-b | E | 1/1 | VIII | − | |
| sgf | SG31 | F | 1/1 | VIII | − |
See Table 1, footnote b.
NT, nontypeable.
The isolates were assigned to major types when they had more than three fragment differences and to subtypes (a to f) when they had three or fewer than three fragment differences.
ND, not done.
An asterisk indicates that the majority of strains from a group of 167 isolates (see Materials and Methods) were not typed by PCR ribotyping and PFGE; therefore, they could not be assigned to a type.
The second group, from which five more C. difficile strains were selected for the present study, consisted of 167 isolates recovered from patients with C. difficile-associated diarrhea who were admitted to two Japanese hospitals (hospitals H and I) and one hospital in Korea between 1980 and 2000 (Table 1). The five strains were found to show atypical reactions in PCRs detecting the nonrepeating and repeating sequences of the tcdA gene during a screening of 167 clinical isolates (14, 15).
Determination of toxigenicity.
All 310 C. difficile isolates were screened for toxigenicity by PCR detection of the nonrepeating and repeating sequences of the tcdA gene (14, 15). The strains which were found to be positive by PCR for the nonrepeating sequences with the primer set NK3-NK2 and generated a PCR product of ca. 1,200 bp by PCR for the repeating sequences with primer set NK11-NK9 were identified as toxin A positive and toxin B positive (A+B+) (14). The strains that were positive by PCR for the nonrepeating sequences but yielded either a segment shorter than 1,200 bp or no amplification products of the repeating sequences were further tested for TcdA and TcdB production as described bellow.
All 56 C. difficile strains included in toxinotyping analyses were retested for TcdA and TcdB production. Strains were cultured anaerobically in brain heart infusion broth for 5 to 7 days. An enzyme-linked immunosorbent assay (ELISA) kit, Tox-A TEST (TechLab, Blacksburg, Va.), was used to detect TcdA. TcdB was detected by cell culture assay with Vero cells. C. difficile goat anti-toxin B serum (TechLab) was used for the cytotoxin neutralization assay.
Toxinotyping.
Strains were grown on brain heart infusion medium for 24 h, and crude DNA was prepared with Chelex-100 as described previously (23). In strains where all 10 PCRs had to be done, pure DNA was isolated (23).
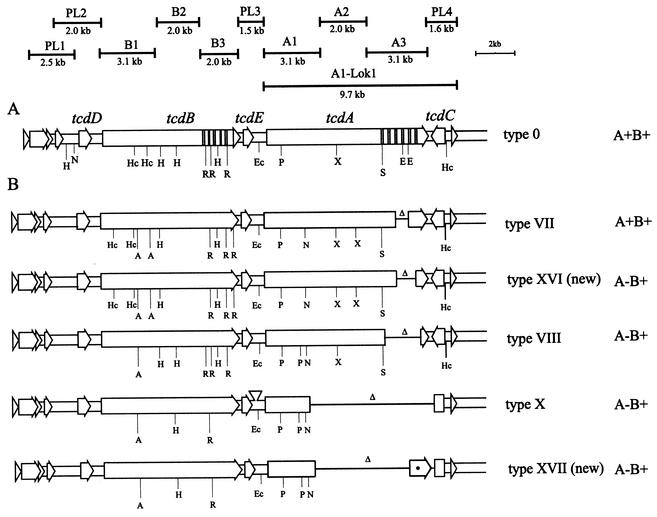
For toxinotyping, two PCR fragments were used, B1 and A3; these were subsequently cut with HincII and AccI (for B1) or with EcoRI (for A3) (24). The toxinotype was then determined according to the combination of restriction patterns in PCR fragments B1 and A3 (22, 23; http://www.uni-lj.si/∼bfbcdiff). For five strains representing new toxinotypes we checked the entire tcdB and tcdA gene with PCRs and restriction analyses as described previously (Fig. 3) (23). The PL3 fragment, which should contain the typical insertion (23, 26), was amplified by PCR for confirmation of toxinotype X.
FIG. 3.
Schematic presentation of the PaLoc in two new A−B+ toxinotypes. (A) PaLoc in the representative strain VPI 10463 from toxinotype 0 is ca. 19-kb region with two toxin genes (tcdA and tcdB) and three additional genes (tcdC, tcdD, and tcdE). Hatched areas represent the repetitive sequences of both toxin genes. Ten PCR fragments used for amplification of entire PaLoc, as well as A1-Lok1 PCR used for amplification of the region including tcdA and tcdC, are shown. (B) Comparison of new toxinotypes XVI and XVII with known A−B+ types (VIII and X). The most prevalent and studied A−B+ toxinotype (VIII) differs from new toxinotypes XVI and XVII in the restriction sites in both toxin genes and in the length of the deletion (Δ) in gene tcdA. Toxinotype XVI is similar to the TcdA-positive group of toxinotypes V, VI, and VII (shown here as representative). Toxinotype XVII is similar to toxinotype X in tcdB but lacks the insertion between tcdE and tcdA and has less of the tcdA gene deleted. The exact position of the deletion was not determined with the PCRs. The presence of the 3′ end of tcdA was not confirmed and is therefore indicated with an asterisk. Restriction enzymes: A, AccI; Ec, EcoRV; H, HindIII; Hc, HincII; N, NsiI; P, PstI; R, RsaI; S, SpeI; X, XbaI.
In four strains—VPI 10463 (toxinotype 0), 8864 (toxinotype X), SUC 36 (new toxinotype XVI), and J9965 (new toxinotype XVII)—the entire region within the PaLoc that includes tcdA and tcdC genes was amplified (Fig. 3A, A1-Lok1 PCR). We used primers A1C (5′-GGAGGTTTTTATGTCTTTAATATCTAAAGA-3′ [24]) and LOK1 (5′-AAAATATACTGCACATCTGTATAC-3′ [4]). PCR was performed in a 50-μl volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.001% gelatin, 2 mM MgCl2, 0.2 mM concentrations of deoxynucleoside triphosphates, 5 pmol of each primer, and 0.5 U of Taq polymerase (Perkin-Elmer). The PCR program started with a 3-min denaturation at 93°C, followed by 35 cycles of 8 min at 56°C and 4 s at 93°C, and then finished with a 10-min incubation at 56°C.
Detection of binary toxin gene.
PCR as described by Stubbs et al. (27) was used for detection of the gene cdtB coding for the binding component of the binary toxin CDT.
Sequencing of the 5′ end of the tcdA gene.
Sequencing analysis of the 5′ end of the gene tcdA was performed as described elsewhere (13). Primers A1C and NK34 were used for PCR, and the PCR product was sequenced directly with ABI Prism dye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, Calif.). Primer A1C (24) originates 10 nucleotides upstream from the start codon of the tcdA gene, and primer NK34 (5′-TAGCTGTAATGCTTCAGTGG-3′, positions 446 to 465) was derived from the published sequences of the tcdA gene in VPI 10463 (6).
RESULTS
Of 56 studied toxinogenic strains, we found 35 strains to be similar to the C. difficile VPI 10463, and there were therefore included in toxinotype 0. The remaining 21 strains showed differences in their toxin genes and could be distributed into 11 toxinotypes (Table 2). Six of the toxinotypes were described before (I, III, IV, VIII, IX, and XII) and five toxinotypes were new (XVI to XX [see below]). From more prevalent ribotypes and PFGE groups, up to five strains were chosen from different sources and toxinotyped (Table 2). Variant strains were mostly found in ribotypes and PFGE types with only one known isolate. The exception was toxinotype VIII, which included 11 strains (Table 2). Isolates within two ribotype-PFGE groups belonged to more than one toxinotype. In group yok/OG02-a (for ribotype/PFGE type), three isolates were tested. One of them belonged to toxinotype I, and other two belonged to toxinotype 0. Group smz/NT was reported to be the most prevalent group among Japanese strains (12). Of five tested strains, one was of new toxinotype XIX and others were VPI 10463-like (toxinotype 0).
When the toxinogenicity of strains was checked by the ELISA kit for TcdA detection and cell culture assay for TcdB detection, 43 strains were determined to be A+B+ and 13 strains were determined to be A−B+. Of 13 A−B+ strains, 11 were from toxinotype VIII and 2 were from previously unknown toxinotypes. Twelve A−B+ strains, including eleven toxinotype VIII strains and toxinotype XVII strain J9965, caused variant cytotoxic effects that differed from the cytopathic effect obtained with VPI 10463 TcdB. Such a variant cytopathic effect had already been described not only for A−B+ toxinotypes VIII and X (3, 8, 21) but also for A+B+ toxinotype IX (21). One A−B+ strain, SUC 36, had a cytotoxic effect similar to that of reference strain VPI 10463.
All eight A+B+ variant strains described here were isolated from symptomatic patients (antibiotic-associated diarrhea or colitis). A strain of the new toxinotype XVI (A−B+) was isolated from a healthy child, and a strain of the new toxinotype XVII (A−B+) was isolated from a severely ill adult. Of 11 strains from toxinotype VIII (A−B+), 7 were isolated from hospitalized symptomatic adult patients, 1 was from a hospitalized asymptomatic adult patient, and 3 were from asymptomatic children.
The prevalence of variant strains differed among nine hospitals (Table 1; Indonesian strains were isolated from healthy infants and do not represent a hospital). In all hospitals (except the hospital in Korea), A−B+ strains were found, whereas A+B+ variants were found in only six of nine hospitals. The binary toxin genes were found in 5 of 56 strains (Table 2). Three of these belonged to toxinotypes III, IV, and IX, which had previously been described as binary-toxin producers. Two binary-toxin-positive strains belonged to the two new A−B+ toxinotypes XVI and XVII. These five binary-toxin-gene-positive strains represented 8.9% of strains selected for study but only 1.6% of all 310 strains.
Description of new A+B+ toxinotypes.
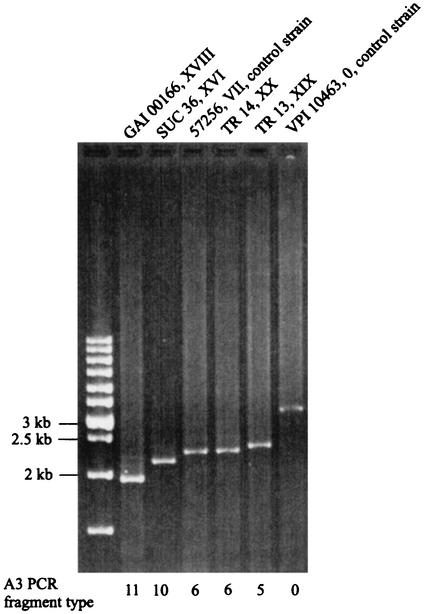
Three new A+B+ toxinotypes were found (XVIII, XIX, and XX; Table 3). These were all characterized by changes in repetitive regions of tcdA gene (A3 PCR fragment), whereas the A1 and A2 PCR fragments, as well as the tcdB gene, are the same as in reference strain VPI 10463. The A3 PCR fragment was shorter in all three strains than in the reference strain. None of the shorter A3 PCR fragments could be cut with EcoRI. The A3 PCR fragment of the reference strain VPI 10463 results in two restriction fragments after EcoRI digestion. In Fig. 1 this PCR fragment is presented uncut to allow comparison with the shorter fragments.
TABLE 3.
Characteristics used for the determination of new toxinotypes
| Toxinotype | B1 PCR fragment type (HincII/AccI restriction pattern) | A3 PCR fragment type (EcoRI restriction pattern) | Toxin production | Representative strain |
|---|---|---|---|---|
| XVI | 3 | 10 | A−B+ | SUC 36 |
| XVII | 5 | Nega | A−B+ | J9965 |
| XVIII | 1 | 11 | A+B+ | GAI00166 |
| XIX | 1 | 5 | A+B+ | TR 13 |
| XX | 1 | 6 | A+B+ | TR 14 |
Neg, negative.
FIG. 1.
PCR amplification of tcdA fragment A3 in the new toxinotypes XVI and XVIII to XX. (In new toxinotype XVII the A3 fragment could not be amplified.) The strain designation and toxinotype are shown above the gel lanes. A3 PCR fragment types as determined by size and restriction patterns (data not shown) are indicated below the gel lanes. All shown A3 PCR fragments were shorter than the fragment obtained with reference strain VPI 10463. Strains GAI 00166 and SUC 36 represent two new deleted A3 fragment types, 11 and 10, respectively. Strains TR 13 and TR 14 are identical in length to the known A3 PCR fragment types (5 and 6). Strain 57267 was used here as a control for differentiation between A3 PCR fragments pattern 6 and 5, which are very similar in deletion size.
Toxinotype XVIII represents a new type of A3 deletion (Fig. 1, A3 fragment type 11). Deletions found in toxinotypes XIX and XX were similar in length to those found in toxinotypes VI and VII. Therefore, we also designated them as types 5 and 6 of PCR fragment A3 (Fig. 1 and Table 3).
Description of new A−B+ toxinotypes.
Two new types of PaLoc were found in two strains in which TcdA could not be detected with commercial kit.
Strain SUC 36 (new toxinotype XVI) was isolated from a healthy 8-month-old child. In gene tcdB and in the 5′ part of gene tcdA this strain was similar to toxinotypes V, VI, and VII (Fig. 3). However, the A3 fragment of the tcdA gene was shorter than in these toxinotypes (Fig. 1, A3 fragment types 10 and 6). A still larger new deletion was found in toxinotype XVIII (Fig. 1, A3 fragment type 11). Unlike toxinotypes V, VI, and VII, which produce TcdA and TcdB (23), strain SUC 36 produced no TcdA as determined by ELISA.
Strain J9965 (new toxinotype XVII) was isolated from a severely ill adult patient with chronic renal failure, diabetes mellitus, and neurogenic bladder. The patient had diarrhea but no prior antibiotic therapy. The restriction fragment length polymorphisms of amplified B1 fragment from strain J9965 were identical to those of the B1 fragment from the previously characterized A−B+ strain 8864 (toxinotype X). The A3 fragment could not be amplified from either strain (Table 3 and Fig. 3). Restriction sites checked in B2 and B3 PCR fragments of gene tcdB were also identical in both strains. Differences were found in tcdA gene and in PL3 PCR fragment. One of the characteristics of strain 8864 is a large insertion in the region between tcdE and tcdA gene (covered with PL3-PCR fragment [23]). This insertion was not found in strain J9965.
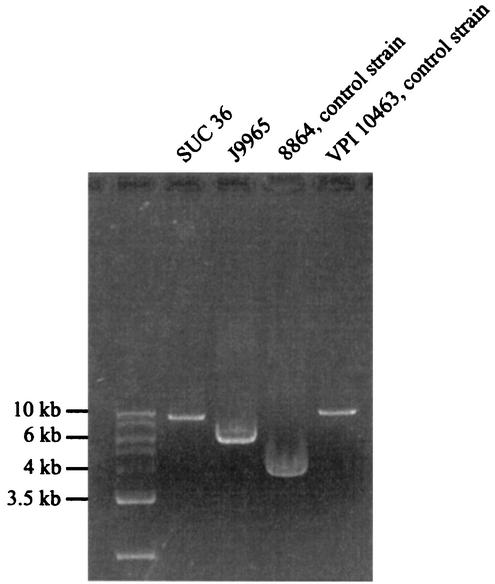
An entire region from the beginning of tcdA gene to the end of PaLoc was amplified in strains representing two new A−B+ toxinotypes and in the two control strains VPI 10463 and C. difficile 8864 (Fig. 2 and Fig. 3, A1-Lok1 PCR). In strain VPI 10463 this fragment has 9,763 bp, as deduced from the sequence. Strain SUC 36 differed from VPI 10463 by <1 kb. This finding was also in agreement with length differences in A3 PCR fragments (Fig. 1). In strain 8864, an A1-Lok1 fragment of ∼4 kb was obtained that corresponds to a published deletion of 5.9 kb within the amplified region (24). Strain J9965 had fragment that was ∼1.9 kb larger than in C. difficile 8864. Therefore, the deletion in strain J9965 was somewhat smaller than in C. difficile 8864. Since none of the fragments covering tcdA gene (A1 to A3) could be obtained in strain J9965, we speculate that the deleted region starts within the A1 fragment (which is probably the same as in strain 8864) and spans within A3 fragment, but at least a part of A3 fragment is probably present (Fig. 3). However, the exact position of the deletion has to be confirmed by sequencing.
FIG. 2.
PCR fragments obtained with primer pair A1C-Lok1 that covers the region from the gene tcdA to the 3′ end of PaLoc. Fragments were amplified in three A−B+ strains (8864, SUC 36, and J9965) and compared to the reference strain VPI 10463. Strains VPI 10463 and 8864 were controls representing a full-length PaLoc and 3′-deleted form of PaLoc, respectively. Strain SUC 36, a new representative of A−B+ strains, has only a small deletion in this region. In another new type of A−B+ strain (J9965), the deletion is larger but ∼2 kb more are present than in C. difficile 8864.
In strains J9965 and SUC 36 we also sequenced the 5′ end of tcdA gene to determine whether the same mutation is present that is responsible for the absence of TcdA in strains of toxinotype VIII. A comparison of the first 130 amino acids showed that strains J9965 and SUC 36 were, at position 47, identical to strain VPI 10463 but not to strain GAI 95601 of toxinotype VIII (Fig. 4). However, all tested A−B+ strains had an identical point mutation at position 64, whereas strains J9965 and 8864 had additional but not identical point mutations (Fig. 4).
FIG. 4.
Comparison of the first 130 N-terminal amino acids of TcdA in C. difficile reference strain VPI 10463 and four A−B+ strains from toxinotypes VIII, X (C. difficile 8864; GenBank accession number AF035716), XVI, and XVII. The nonsense mutation at position 47 (indicated with an asterisk), which was shown to cause the lack of production of the TcdA in toxinotype VIII, was not found in the other three A−B+ strains.
DISCUSSION
We have studied the presence of C. difficile variant strains among isolates from patients of eight Japanese hospitals, one Korean hospital, and healthy infants living in Surabaya, Indonesia, and compared the results to those obtained on strains from the UCL Brussels collection and the ARU Cardiff collection (22, 23).
In all three collections, we found common types of variant strains. However, in each collection new toxinotypes were also found. Whereas the UCL Brussels and ARU Cardiff collections share almost all of the 15 known toxinotypes, only 6 of these 15 toxinotypes were found among Japanese strains. Five of them were the most prevalent types of variant strains (toxinotypes III, IV, VIII, IX, and XII) and were numerous in both the Brussels and the Cardiff collections (22, 23). A sixth toxinotype found in all three collections was toxinotype I. Interestingly, no strains from toxinotypes V, VI, VII, and XI were found in the Japanese collection. These toxinotypes were previously shown by molecular typing and partial 16S rRNA sequencing to be related one to another and to represent an independent branch of variant C. difficile strains (22, 28). This group of variant strains might have developed and spread in Europe but not in Asia. However, one of the new toxinotypes (XVI) was in the PaLoc identical to groups V, VI, VII, and XI and differed from them only in the length of the A3 PCR fragment (Fig. 1).
Among A+B+ variant strains we found three new toxinotypes. However, these were all minor toxinotypes because only one part of the tcdA gene was changed and the rest of PaLoc was the same as in reference strain VPI 10463. Interestingly, two strains showed deletions in the repetitive regions that were similar in length to the one observed in toxinotypes VI and VII. By restriction fragment length polymorphism analysis it could be shown that a different region of approximately the same length was deleted (data not shown).
We also describe here two new types of A−B+ C. difficile strains. Up to now, A−B+ strains seemed to be a widespread and coherent group. Many studies have confirmed that these strains differ slightly in serogroup (F and X), PFGE results, and ribotype markers (three ribotypes) but that they all have the same PaLoc and belong to toxinotype VIII. Moncrief et al. (19) tested 48 A−B+ strains from various locations, and these strains all had a typical 1.8-kb deletion in the tcdA gene. The same deletion was found in 50 strains examined by Kato et al. (13, 14) and was confirmed by sequencing in 6 strains. Another A−B+ strain (CF2) was characterized by Sambol et al. (25). The tcdA gene again had a typical 1,821-bp deletion, and the tcdB gene differed in 16 nucleotides from the tcdB gene of strain 1470 (type strain of toxinotype VIII). Strain HSC98, which is responsible for a nosocomial outbreak, was also characterized as toxinotype VIII (1). Only one A−B+ strain with different PaLoc is known: strain 8864 (3, 18, 21, 26).
Of the 310 isolates from the two Asian collections, 65 had been previously identified as A−B+ by PCR amplification of the repetitive regions of the tcdA gene. Of these 65 strains, 13 representing different ribotype-PFGE types were included in the present study for toxinotyping (Table 2). Sixty-three A−B+ strains belonged to toxinotype VIII, whereas two strains had a different PaLoc. Interestingly, one of them, defined as toxinotype XVII, was in PaLoc similar to the strain 8864 and to A+B+ strains from toxinotype IX and could represent an intermediate type. Toxinotype XVI, representing another new type of A−B+ strain, is in its PaLoc similar to toxinotypes V, VI, and VII. Two differences noted were (i) the length of the repetitive sequences of the tcdA gene and (ii) the production of TcdA, which was present in toxinotypes V, VI, VII and absent in toxinotype XVI.
Strains SUC 36 and J9965 are described here as A−B+ because they did not react with TcdA-specific commercial ELISA test. For two already-described types of A−B+ strains, toxinotypes VIII and X, the molecular basis of TcdA nonproduction is also known. Strain 8864, the only representative of toxinotype X, has a huge deletion within PaLoc that affects TcdA production at the transcriptional level (26). Strains from toxinotype VIII have a mutation within the tcdA gene that introduces a stop codon at amino acid position 47; therefore, TcdA is not produced (7). The same mutation was not found in the new types of A−B+ strains described here. The molecular mechanism causing the absence of TcdA in new A−B+ strains, which would confirm their A−B+ status, has yet to be determined.
The prevalence of all variant strains (A+B+ and A−B+) among the 310 strains studied here was calculated to be 23.5% (Table 1). Only 56 representative strains were toxinotyped, but it was previously shown that toxinotypes correlate well with PCR ribotypes (22). A given toxinotype could be found in many different ribotypes (e.g., toxinotype 0), but all strains within a given ribotype belong to the same or closely related toxinotypes. Therefore, it is enough to check just one isolate per ribotype, as we did in group I. Variant strains were found mainly in ribotype-PFGE types with a single representative. In two cases (smz/NT and yok/OG02-a; Table 2), variant strains were found in the same group as the VPI 10463-like strains (toxinotype 0). In both cases variant strains had changes only in repetitive regions of the tcdA gene, and such strains were often found in the same groups as ordinary strains (22). Group II was screened with a PCR specific for repetitive regions of the tcdA gene whereby one can detect 13 of 15 known toxinotypes (21). Only five isolates that showed variations were further toxinotyped.
The percentage of variant strains varied greatly between the two groups (I and II) and between different hospitals (Table 1). The presence of strains with binary toxin was very low, and such strains were found only in four hospitals. Hospital D was somewhat unique, because it had a high proportion of variant strains, despite the fact that the strains were well distributed into several PCR ribotypes and PFGE types and the number of A−B+ isolates was low.
Generally, the high percentage of variant strains is a consequence of the high number of A−B+ strains. This is especially true for hospitals H and I, which both probably might have an outbreak of A−B+ strain (H. Kato, unpublished data). Therefore, we suggest that the prevalence of variant strains estimated for group I (15%) is closer to the real situation in Japan. It is comparable to the prevalence of variant strains estimated for the ARU Cardiff collection (8.8%) (22). The study of strains from the Brussels collection revealed that the 21% of strains were variant; however, a large number of A−B+ strains was included and, because toxinotypes correlated poorly with serogroups, no real estimation for the entire collection could be calculated (23).
In the present study we describe the prevalence and types of variants among C. difficile strains isolated in Japan, Korea, and Indonesia. The percentage of variant strains can vary between hospitals and is mainly influenced by the number of A−B+ strains. We have also described two new types of A−B+ strains that differed from the most widespread A−B+ group of toxinotype VIII in the organization of PaLoc and in the presence of binary toxin genes. Further studies would show whether those new types are characteristic for this geographic region or whether they can be found in other countries.
Acknowledgments
We thank T. Yamamoto and K. Suzuki of Nagoyashi Koseiin Geriatric Hospital, S. Ishigo of Ogaki Municipal Hospital, S. Kunihiro and I. Nakamura of Yamaguchi Prefectural Hospital, T. Oguri and S. Misawa of Juntendo University Hospital, K. Yoshimoto of Shirasagi Hospital, M. Komatsu and M. Aihara of Tenri Hospital, E. B. Wasito of Airlangga University, and Y. Chong of Yonsei University College of Medicine for sample and strain collection.
REFERENCES
- 1.Alfa, M. J., A. Kabani, D. Lyerly, S. Moncrief, L. M. Neville, A. Al-Barak, G. K. H. Harding, B. Dyck, K. Olekson, and J. M. Embil. 2000. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile responsible for a nosocomial outbreak of Clostridium difficile-associated diarrhea. J. Clin. Microbiol. 38:2706-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbut, F., V. Lalande, B. Burghoffer, H. V. Thien, E. Grimprel, and J.-C. Petit. 2002. Prevalence and genetic characterization of toxin A variant strains of Clostridium difficile among adults and children with diarrhea in France. J. Clin. Microbiol. 40:2079-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borriello, S. P., B. W. Wren, S. Hyde, S. V. Seddon, P. Sibbons, M. M. Krishna, S. Tabaqchali, S. Manek, and A. B. Price. 1992. Molecular, immunological, and biological characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4192-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., T. Hundsberger, P. Leukel, M. Sauerborn, and C. V. Eichel-Streiber. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29-38. [DOI] [PubMed] [Google Scholar]
- 5.Depitre, C., M. Delmée, V. Avesani, R. L'Haridon, A. Roels, M. Popoff, and G. Corthier. 1993. Serogroup F strains of Clostridium difficile produce toxin B but not toxin A. J. Med. Microbiol. 38:434-441. [DOI] [PubMed] [Google Scholar]
- 6.Dove, C. H., S. Z. Wang, S. B. Price, C. J. Phelps, D. M. Lyerly, T. D. Wilkins, and J. L. Johnson. 1990. Molecular characterization of the Clostridium difficile toxin A gene. Infect. Immun. 58:480-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eichel-Streiber, C. V., I. Zec-Pirnat, M. Grabnar, and M. Rupnik. 1999. A nonsense mutation abrogates production of functional enterotoxin A in Clostridium difficile toxinotype VIII strains of serogroups F and X. FEMS Microbiol. Lett. 178:163-168. [DOI] [PubMed] [Google Scholar]
- 8.Eichel-Streiber, C. V., P. Boquet, M. Sauerborn, and M. Thelestam. 1996. Large clostridial cytotoxins: a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 4:375-382. [DOI] [PubMed] [Google Scholar]
- 9.Gibert, M., S. Perelle, G. Daube, and M. R. Popoff. 1997. Clostridium spiroforme toxin genes are related to C. perfringens iota toxin genes but have a different genomic localization. Syst. Appl. Microbiol. 20:337-347. [Google Scholar]
- 10.Hammond, G. A., and J. L. Johnson. 1995. The toxinogenic element of Clostridium difficile strain VPI 10463. Microb. Pathog. 19:203-213. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, S., S. A. Kent, K. J. O'Leary, M. M. Merrigan, S. P. Sambol, L. R. Peterson, and D. N. Gerding. 2001. Fatal pseudomembranous colitis associated with a variant Clostridium difficile strain not detected by toxin A immunoassay. Ann. Intern. Med. 135:434-438. [DOI] [PubMed] [Google Scholar]
- 12.Kato, H., N. Kato, K. Watanabe, T. Yamamoto, K. Suzuki, S. Ishigo, S. Kunihiro, I. Nakamura, G. E. Killgore, and S. Nakamura. 2001. Analysis of Clostridium difficile isolates from nosocomial outbreaks at the three hospitals in diverse areas of Japan. J. Clin. Microbiol. 39:1391-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kato, H., N. Kato, S. Katow, T. Maegawa, S. Nakamura, and D. M. Lyerly. 1999. Deletions in the repeating sequences of the toxin A gene of toxin A-negative, toxin B-positive Clostridium difficile strains. FEMS Microbiol. lett. 175:197-203. [DOI] [PubMed] [Google Scholar]
- 14.Kato, N. H., Kato, K. Watanabe, N. Iwai, H. Nakamura, T. Yamamoto, K. Suzuki, S. M. Kim, Y. Chong, and E. B. Wasito. 1998. Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J. Clin. Microbiol. 36:2178-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato, N., C. Y. Ou, H. Kato, S. L. Bartley, C. C. Luo, G. E. Killgore, and K. Ueno. 1993. Detection of toxigenic Clostridium difficile in stool specimens by the polymerase chain reaction. J. Infect. Dis. 167:455-458. [DOI] [PubMed] [Google Scholar]
- 16.Kuijper, E. J., J. de Weerdt, H. Kato, N. Kato, A. P. van Dam, E. R. van der Vorm, J. Weel, D. van Rheenen, and J. Dankert. 2001. Nosocomial outbreak of Clostridium difficile-associated diarrhoea due to a clindamycin-resistant enterotoxin A-negative strain. Eur. J. Clin. Microbiol. Infect. Dis. 20:528-534. [DOI] [PubMed] [Google Scholar]
- 17.Limaye, A. P., D. K. Turgeon, B. T. Cookson, and T. R. Fritsche. 2000. Pseudomembranous colitis caused by a toxin A−B+ strain of Clostridium difficile. J. Clin. Microbiol. 38:1696-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyerly, D. M., L. A. Barroso, T. D. Wilkins, C. Depitre, and G. Corthier. 1992. Characterization of a toxin A-negative, toxin B-positive strain of Clostridium difficile. Infect. Immun. 60:4633-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moncrief, J. S., L. Zheng, L. M. Neville, and D. M. Lyerly. 2000. Genetic characterization of a toxin A-negative, toxin B-positive Clostridium difficile isolates by PCR. J. Clin. Microbiol. 38:3072-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perelle, S., M. Gilbert, P. Bourlioux, G. Corthier, and M. R. Popoff. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect. Immun. 65:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupnik, M. 2001. How to detect Clostridium difficile variant strains in a routine laboratory. Clin. Microbiol. Infect. 7:417-420. [DOI] [PubMed] [Google Scholar]
- 22.Rupnik, M., J. S. Brazier, B. I. Duerden, M. Grabnar, and S. L. J. Stubbs. 2001. Comparison of toxinotyping and PCR ribotyping of Clostridium difficile strains and description of novel toxinotypes. Microbiology 147:439-447. [DOI] [PubMed] [Google Scholar]
- 23.Rupnik, M., V. Avesani, M. Janc, C. V. Eichel-Streiber, and M. Delmée. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rupnik, M., V. Braun, F. Soehn, M. Janc, M. Hofstetter, R. Laufenberg-Feldman, and C. V. Eichel-Streiber. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197-202. [DOI] [PubMed] [Google Scholar]
- 25.Sambol, S. P., M. M. Merrigan, D. Lyerly, D. N. Gerding, and S. Johnson. 2000. Toxin gene analysis of a variant strain of Clostridium difficile that causes human clinical disease. Infect. Immun. 68:5480-5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soehn, F., A. Wagenknecht-Wiesner, P. Leukel, M. Kohl, M. Weidman, C. V. Eichel-Streiber, and V. Braun. 1998. Genetic rearrangements in the pathogenicity locus of Clostridium difficile strain 8864: implications for transcription, expression and enzymatic activity of toxins A and B. Mol. Gen. Genet. 258:222-232. [DOI] [PubMed] [Google Scholar]
- 27.Stubbs, S., M. Rupnik, M. Gibert, J. Brazier, B. Duerden, and M. Popoff. 2000. Production of actin-specific ADP-ribosyltransferase (binary toxin) by strains of Clostridium difficile. FEMS Microbiol. Lett. 186:307-312. [DOI] [PubMed] [Google Scholar]
- 28.Wozniak, G., P. Trontelj, and M. Rupnik. 2000. Genomic relatedness of Clostridium difficile strains from different toxinotypes and serogroups. Anaerobe 6:261-267. [Google Scholar]