Abstract
Resistance to antiretroviral agents often results from mutations within the human immunodeficiency virus (HIV) pol gene. Moreover, insertions within the p6 gag-pol region have recently been found to be involved with resistance to nucleoside analogs. Overall, we found that 21% of 156 specimens collected from HIV-infected individuals (17.6% from 74 drug-naive patients and 24.4% from 82 pretreated patients) harbored these insertions. Insertions around the KQE (Lys-Gln-Glu) motif were found in 12.2% of the pretreated patients but in none of the drug-naive subjects (P = 0.002). In contrast, insertions around the PTAP (Prol-Thre-Ala-Prol) motif were seen at similar rates (∼15%) among drug-naive and pretreated patients, which supports the idea that they may be natural polymorphisms.
Combination drug regimens consisting of reverse transcriptase (RT) and protease inhibitors (PIs) have proven to be highly effective in suppressing human immunodeficiency virus (HIV) replication for a sustained period of time (2; www.hivatis.org). However, the effectiveness of these therapies is often blunted after the emergence of drug-resistant viruses, which frequently show extensive cross-resistance within each drug class (4, 5, 10, 11).
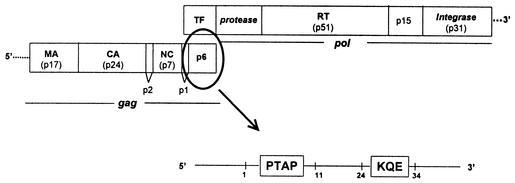
Reduced sensitivity to antiretroviral drugs may be associated not only with mutations within protease and RT genes but also with genotypic changes at the p6gag region localized immediately upstream of the pol gene (Fig. 1) (13, 16). Changes in the p6 region lead to a complex viral phenotype that includes increased infectivity and resistance to nucleoside analog RT inhibitors (NRTIs) and PIs. Insertions in the p6 region have been associated with loss of susceptibility to NRTIs, particularly with loss of susceptibility to stavudine (d4T), zalcitabine, and didanosine (ddI). This is of particular interest given our limited understanding of the mechanisms of HIV type 1 resistance to d4T and ddI (3, 14).
FIG. 1.
The HIV p7-p1-p6 genomic region. TF, trans frame; MA, matrix; CA, capsid; NC, nucleocapsid.
In this study, we evaluated the rate and conditions of selection of insertions within the p6 region. All plasma samples referred to our reference laboratory for drug resistance testing during the last trimester of 2001 were examined. They belonged to HIV-infected individuals attending several HIV clinics across Spain. At the time of analysis, all samples had viral loads of >1,000 HIV RNA copies/ml. Genotypic analyses were performed by using an RT-PCR with C35 (5′-AAGAAATGATGACAGCATGTCAGGGAGTAGGAGGA-3′) and NE135 (5′-CTTACT AACTTCTGTATGTCATTGACAGTCCAGCT-3′) as outer primers. A nested PCR was then performed with DP15 (5′-AAGGGCTGTTGGAAATGTGG-3′) and DP17 (5′-AAAATTTAAAGTGCAGCCAAT-3′) as inner primers. Genetic sequencing was performed with an ABI Prism 3100 automated sequencer (Applied Biosystems, Foster City, Calif.). Consensus sequences for the virus population were aligned with those belonging to HIV type 1 HXB2.
Genotypic results were obtained from 156 specimens, 74 of which were collected from drug-naive individuals and the remaining 82 of which were collected from patients on antiretroviral therapy. Both insertions and deletions were identified in the p7-p1-p6gag region. A total of 33 subjects (21%) carried viruses with insertions in the p6gag region. They were found in 13 (17.6%) of 74 samples from drug-naive subjects and in 20 (24.4%) of 82 treatment-experienced patients (no statistically significant difference). Insertions were recognized preferentially in the first 11 amino acids, involving the polyproline motif PTAP (Pro-Thre-Ala-Prol), and between amino acids 24 and 34, including the KQE (Lys-Gln-Glu) motif, of the p6 protein (Fig. 1).
The duplication of the PTAP motif appeared at similar rates among drug-naive and pretreated individuals (13 [17.6%] versus 10 [12.2%], respectively), which supports the idea that it may be a natural polymorphism (W. Dong, Z. Brumme, K. Chan, R. S. Hogg, V. O'Shaughnessy, J. Montaner, and P. R. Harrigan, 9th Conf. Retrovir. Opportunistic Infect., abstr. 572-T, p. 265; D. Smith, S. J. Little, E. S. Daar, R. Koup, N. S. Hellmann, K. Dawson, J. W. Wong, D. Richman, and A. J. Leigh Brown, 9th Conf. Retrovir. Opportunistic Infect., abstr. 357-M, p. 189). In contrast, duplications of the KQE motif of the p6gag protein were found in 10 samples (12.2%) belonging to antiretroviral therapy-experienced subjects but in none from drug-naive individuals (P = 0.002).
Nearly 80% of samples from treatment-experienced patients showed resistant genotypes at the pol gene, a rate similar to that reported in previous studies conducted with subjects whose antiretroviral treatment failed (6, 7, 15). All samples harboring insertions within the p6gag region showed classical mutations associated with resistance to PIs (median of 7 [range, 4 to 9] mutations in the PRO gene) and to NRTI (median of 7 [range, 4 to 12] mutations in the RT gene). All of these subjects had been extensively exposed to antiretroviral drugs for a median of 77 (range, 53 to 108) months.
Extensive polymorphisms were observed in the gag region throughout the p7, p1, and p6 proteins. However only four point mutations localized immediately upstream of the pol gene, in the cleavage sites, were more frequently found among treatment-experienced subjects (Table 1). The A54I/V mutation in the p7 region appeared in 14 pretreated patients but in only 1 drug-naive patient (17 versus 1.3%; P = 0.001). This change appeared to be associated with specific PI resistance mutations, such as L90M in nine (64.3%), V82A/F/T in eight (57%), and M46I/L in seven (50%) of the cases, respectively. On the other hand, the I5F/L/V change in the p1 region was found in 14 treatment-experienced patients and in 4 drug-naive patients (17 versus 5.4%; P = 0.023). This amino acid substitution appeared in 71.4% of samples associated with L90M, a key PI resistance mutation. These findings are in agreement with those of Bally et al. (1). Finally, two other substitutions in the p6 region were associated with exposure to antiretroviral agents; P5I/L/T and E20A/G/K were found in 34 and 8.5% of the pretreated subjects but in 11% and none of the drug-naive subjects, respectively (P = 0.001 and 0.01, respectively). Both mutations in p6 were associated with L90M at the PRO gene (46.4 and 85.7% of the cases, respectively). Overall, the recognition of a significant association between mutations at gag cleavage sites and primary PI resistance mutations supports the concept that they serve as compensatory changes increasing the kinetics of virus-mutated proteases and therefore contributing to enhance PI resistance (16, 17).
TABLE 1.
Insertions in the p6gag region and mutations at cleavage sites in antiretroviral-naive and pretreated HIV-infected patients
| p7-p1-p6gag genotype | No. of patients
|
P value | |
|---|---|---|---|
| Drug naive (n = 74) | Pretreated (n = 82) | ||
| Insertion at PTAP motif | 13 | 10 | 0.345 |
| Insertion at KQE motif | 0 | 10 | 0.002 |
| Total insertions in p6gag | 13 | 20 | 0.300 |
| Point mutations | |||
| A541/V in p7gag | 1 | 14 | 0.001 |
| I5F/LV in p1gag | 4 | 14 | 0.023 |
| P5I/L/T in p6gag | 8 | 28 | 0.001 |
| E20A/G/K in p6gag | 0 | 7 | 0.01 |
Peters et al. (13) reported recently that the presence of duplications in the polyproline (PTAP) motif was associated with exposure to antiretroviral drugs, mainly with exposure to d4T and ddI. Hypothetically, viruses with insertions in the PTAP motif could package an excess of RT enzymes and, by an inoculum effect, cause loss of phenotypic susceptibility to all NRTIs, although to different extents (d4T > zalcitabine > ddI > zidovudine > abacavir). This circumstance could be explained by the different effective intracellular concentrations achieved by these compounds, which, for some drugs, are only marginally above the level necessary to inhibit virus replication (13).
In our study, we did not find an association between PTAP motif duplications and more frequent current or past exposure to d4T and/or ddI. In contrast, insertions in the PTAP motif appeared at similar rates among drug-naive and pretreated patients, independently of the treatment regimen. Similar results have been reported recently by others (4; Smith et al., 9th Conf. Retrovir. Opportunistic Infect.). Therefore, PTAP duplications may more appropriately be considered natural polymorphisms rather than drug-resistant genotypes.
In contrast, we found insertions between amino acids 24 and 34 in the p6gag region in 10 treatment-experienced patients but in none of the drug-naive patients (P = 0.002). We further analyzed the influence of d4T and/or ddI on the development of these insertions within the KQE motif of the p6 protein and found no relationship. The presence of insertions in the C-terminal end of the p6gag protein could have therapeutic implications, increasing drug resistance to a wide range of antiretrovirals drugs. Accordingly, preliminary data from Kaufmann et al. (9) have recently underlined the fact that alterations in the C-terminal end of the p6gag protein contributed to the failure of a quadruple regimen including two NRTIs plus saquinavir and ritonavir.
The analysis of specific associations between gag cleavage site polymorphisms and particular PRO substitutions provides additional evidence of the demands of adapting a mutated active protease site to a modified cleavage site. It is worth noting that mutations at cleavage sites are mainly selected in the presence of protease mutations at residue 90 and less frequently in the presence of mutations at codons 46 and 82.
In conclusion, insertions in the p6gag region appear in a significant proportion (21%) of HIV-infected individuals. Duplications in the PTAP region appear to be much more common (17.6%) in drug-naive individuals than previously reported. Only insertions in the KQE motif in the p6 protein, as well as point mutations at codon 54 in the p7 protein, codon 5 in the p1 protein, and codons 5 and 20 in the p6 protein, are significantly associated with exposure to antiretroviral drugs. The implications of these gag mutations for HIV drug resistance and antiviral therapy failure need to be further investigated.
Acknowledgments
We thank Juan González-Lahoz for continuous support.
This work was funded in part by grants from the Asociación Investigación y Educación en SIDA (AIES), the Comunidad Autónoma de Madrid (CAM), and the Instituto de Salud Carlos III.
REFERENCES
- 1.Bally, F., R. Martinez, S. Peters, P. Sudre, and A. Telenti. 2000. Polymorphism of HIV type 1 Gag p7/p1 and p1/p6 cleavage sites: clinical significance and implications for resistance to protease inhibitors. AIDS Res. Hum. Retroviruses 16:1209-1213. [DOI] [PubMed] [Google Scholar]
- 2.Carpenter, C. J., D. A. Cooper, M. A. Fischl, J. M. Gatell, B. Gazzard, S. M. Hammer, M. S. Hirsch, D. Jacobsen, D. A. Katzenstein, J. Montaner, D. D. Richman, M. S. Saag, M. Schechter, R. Schooley, M. A. Thompson, S. Vella, P. Yeni, and P. Volberding. 2000. Antiretroviral therapy in adults: update recommendations of the International AIDS Society-USA Panel. JAMA 283:381-390. [DOI] [PubMed] [Google Scholar]
- 3.Coakley, E. P., J. M. Gillis, and S. Hammer. 2000. Phenotypic and genotypic resistance patterns of HIV-1 isolates derived from individuals treated with didanosine and stavudine. AIDS 14:F9-F15. [DOI] [PubMed] [Google Scholar]
- 4.Deeks, S. 2001. International perspectives on antiretroviral resistance: nonnucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):25-33. [DOI] [PubMed] [Google Scholar]
- 5.De Mendoza, C., V. Soriano, C. Briones, O. Gallego, P. Barreiro, A. Alvarez, and J. González-Lahoz. 2000. Emergence of zidovudine resistance in HIV-infected patients receiving stavudine. J. Acquir. Immune Defic. Syndr. 23:279-281. [DOI] [PubMed] [Google Scholar]
- 6.Gallego, O., L. Ruiz, A. Vallejo, E. Ferrer, A. Rubio, B. Clotet, M. Leal, V. Soriano. 2001. Changes in the rate of genotypic resistance to antiretroviral drug in Spain. AIDS 15:1894-1896. [DOI] [PubMed] [Google Scholar]
- 7.Gómez-Cano, M., A. Rubio, T. Puig, M. Pérez-Olmeda, L. Ruiz, V. Soriano, J. A. Pineda, L. Zamora, N. Xaus, B. Clotet, and M. Leal. 1998. Prevalence of genotypic resistance to nucleoside analogues in antiretroviral-naive and antiretroviral-experienced HIV-infected patients in Spain. AIDS 12:1015-1020. [PubMed] [Google Scholar]
- 8.Hirsch, M., F. Brun-Vézinet, R. D'Aquila, S. M. Hammer, V. A. Johnson, D. Kuritzkes, C. Loveday, J. Mellors, B. Clotet, B. Conway, L. Demeter, S. Vella, D. Jacobsen, and D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV infection. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann, G. R., K. Suzuki, P. Cunningham, M. Mukaide, M. Kondo, M. Imai, J. Zaunders, and D. A. Cooper. 2001. Impact of HIV type 1 protease, reverse transcriptase, cleavage site, and p6 mutations on the virological response to quadruple therapy with saquinavir, ritonavir, and two nucleoside analogs. AIDS Res. Hum. Retrovir. 17:487-497. [DOI] [PubMed] [Google Scholar]
- 10.Loveday, C. 2001. International perspectives on antiretroviral resistance: nucleoside reverse transcriptase inhibitor resistance. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):10-24. [DOI] [PubMed] [Google Scholar]
- 11.Miller, V. 2001. International perspectives on antiretroviral resistance: resistance to protease inhibitors. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):34-50. [DOI] [PubMed] [Google Scholar]
- 12.Perrin, L., and A. Telenti. 1998. HIV treatment failure: testing for HIV resistance in clinical practice. Science 280:1871-1873. [DOI] [PubMed] [Google Scholar]
- 13.Peters, S., M. Muñoz, S. Yerly, V. Sanchez-Merino, C. López-Galindez, L. Perrin, B. Larder, D. C. Marko, S. Fakan, P. Meylan, and A. Telenti. 2001. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by HIV type 1 p6 protein. J. Virol. 75:9644-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petropoulos, C., N. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for a HIV type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puig, T., M. Pérez-Olmeda, A. Rubio, L. Ruiz, C. Briones, J. M. Franco, M. Gómez-Cano, L. Stuyver, L. Zamora, C. Alvarez, M. Leal, B. Clotet, and V. Soriano. 2000. Prevalence of genotypic resistance to nucleoside analogues and protease inhibitors in Spain. AIDS 14:727-732. [DOI] [PubMed] [Google Scholar]
- 16.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in HIV type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang, Y.-M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]