Abstract
CDC eugonic oxidizer group 2 (EO-2) is a group of unclassified gram-negative bacterial strains isolated from various human sources. As determined by biochemical tests and analyses of fatty acid compositions, these organisms form a homogeneous group that appears to be distinct from but related to other Paracoccus species. Molecular studies were performed on a set of 13 EO-2 strains from various clinical sources and geographic locations in the United States and Canada to determine their relationship to the Paracoccus genus. Control strains were Paracoccus denitrificans ATCC 17741T, P. versutus ATCC 25364T, P. aminophilus ATCC 49673T, P. solventivorans ATCC 700252T, and Psychrobacter immobilis ATCC 43116T, which are phenotypically similar to EO-2. Nearly complete (1,500-base) 16S rRNA gene sequencing of eight EO-2 strains showed a high level of sequence similarity (>99.3%) within the group, and a BLAST search of GenBank placed the EO-2 cluster in close proximity to Paracoccus species (95 to 97% similarity). DNA-DNA hybridization studies of 13 of the EO-2 strains showed all to be related at the species level, with >70% relatedness under stringent conditions and a divergence within the group of less than 2%. None of the Paracoccus control strains hybridized at >54% with any of the EO-2 strains. These results indicate that EO-2 represents a new Paracoccus species, the first isolated from human clinical specimens. A new species, Paracoccus yeeii, is proposed for the EO-2 strains. The type strain of P. yeeii is CDCG1212 (ATCC BAA-599 and CCUG 46822), isolated in Pennsylvania from dialysate of a 77-year-old male with peritonitis.
Since the early 1960s, the Centers for Disease Control and Prevention (CDC) Special Bacteriology Reference Laboratory (SBRL) has given the designation EO-2 to an unidentified group of gram-negative bacteria that are eugonic oxidizers. This group includes aerobic bacteria, with forms ranging from coccoid to short, thick rods, often appearing vacuolated or peripherally stained (O shape) and found in pairs and short chains or packets, which are strongly oxidase positive, nonmotile, and indole negative and utilize glucose, xylose, and lactose. In 1987 Hudson et al. determined that a large number of the strains in this group are related to Psychrobacter immobilis (4), and the remaining strains were later placed in two distinct groups on the basis of their cellular fatty acid (CFA) compositions (8). Fifteen were retained in a homogenous grouping designated EO-2, and five formed another group, which was designated EO-3. Molecular studies were performed on 13 EO-2 strains from various clinical sources and geographical locations in the United States and Canada. The results indicate that these EO-2 strains are genetically related to the genus Paracoccus, and a new species of this genus is described here, for which the name Paracoccus yeeii sp. nov. is proposed.
Bacterial strains.
The EO-2 strains studied, along with their sources and geographical origins, are presented in Table 1. The type strains of Psychrobacter immobilis (ATCC 43116), Paracoccus alcaliphilus (ATCC 51199), P. aminophilus (ATCC 49673), P. denitrificans (ATCC 17741), P. solventivorans (ATCC 700252), and P. versutus (ATCC 25364) were obtained from the American Type Culture Collection. The strains studied were cultured by using the methods of the SBRL of the CDC (15).
TABLE 1.
Sources of P. yeeii isolates and demographics of and clinical information on patients from whom they were isolated
| Strain number(s)a | Date received | Sent from: | Patient's sex, ageb | Source, clinical diagnosis |
|---|---|---|---|---|
| G1212, ATCC BAA-599T, CCUG 46822T | 1988 | Pennsylvania | M, 77 yrs | Abdominal dialysate, peritonitis |
| G1968 | 1988 | Colorado | F, NG | Ankle, ankle wound |
| G3060 | 1989 | Pennsylvania | M, 67 yrs | Ankle, ankle wound |
| G4446 | 1990 | Hawaii | NG | Bile, biliary tract obstruction |
| G4878 | 1990 | Hawaii | F, 60 yrs | Toe, NG |
| G6155 | 1991 | California | M, 6 wks | CSF,c NG |
| G9205 | 1994 | Rhode Island | M, 11 mos | Leg lesion, wound |
| H13 | 1996 | California | M, NG | Blood, NG |
| H1426, NML88-938A | 1988 | Ontario, Canada | F, 64 yrs | Skin, NG |
| H1427, NML89-0416 | 1989 | Ontario, Canada | F, NG | Ear, otitis media |
| H1428, NML90-0347 | 1990 | Prince Edward Island, Canada | F, 64 yrs | Eye, NG |
| H1429, NML90-0484 | 1990 | Ontario, Canada | F, 56 yrs | Neck incision drainage, NG |
| H1430, NML91-0691 | 1991 | New Brunswick, Canada | F, 8 yrs | Ear, NG |
NML indicates strain numbers assigned at the National Microbiology Laboratory, Winnipeg, Canada.
M, male; F, female; NG, not given.
CSF, cerebrospinal fluid.
Phenotypic tests.
Biochemical testing was done by using the methods of the SBRL of the CDC (15). With the exception of the optimum growth temperature tests, all biochemical tests were performed at 35°C in an aerobic incubator. The oxidase, catalase, and growth temperature tests were read after 1 day of incubation. All other tests were read at 1, 2, and 7 days of incubation. A final reading of the gelatin tests was done after 14 days of incubation.
CFA analysis.
Cells were saponified, and the liberated fatty acids were methylated and analyzed by capillary gas-liquid chromatography (GLC) (15). CFA profiles were identified by using a commercially available system (MIDI, Newark, Del.). The identification of fatty acids and the determination of double-bond positions in monounsaturated acids were accomplished by GLC and GLC-mass spectrometry.
DNA relatedness and determination of percent G+C.
Cells were harvested and lysed, and the DNA was isolated and purified according to the method of Brenner et al. (2). DNA from strain G1212 was labeled with [32P]dCTP by using a commercial nick translation kit (Invitrogen Life Technologies, Carlsbad, Calif.) and tested for reassociation to unlabeled DNA from the same strain (homologous reaction) as well as to DNAs from other EO-2 strains and the type strains (heterologous reactions). The G+C content of G1212 was determined to be 62%; therefore, 65°C was chosen as the optimal reassociation temperature. Relative binding ratios (percent heterologous DNAs bound to hydroxyapatite to percent homologous DNA bound to hydroxyapatite, multiplied by 100) and percent divergence (percentage of unpaired bases in related DNA sequences) were calculated as described by Brenner et al. (2). Divergence was calculated to the nearest 0.5%, with each decrease of 1°C in the thermal stability of a heterologous DNA duplex due to an increase of approximately 1% in the percentage of unpaired bases within related DNAs (1). All reactions were done in duplicate at the optimal temperature of 65°C. The percent G+C was determined for strains G1212, G9205, and H1430 by the thermal denaturation method of Mandel et al. (7).
16S rRNA gene sequencing.
Purified genomic DNA was diluted to 1 μg ml−1 in sterile water. Diluted DNA (10 μl) was used in a 100-μl PCR mixture containing 200 μM deoxynucleoside triphosphates, 1 mM MgCl2, 1X PCR buffer II (Perkin-Elmer, Foster City, Calif.), 0.1 μM FD1 primer, 0.1 μM RD1 primer, and 2.5 U of Expand High-Fidelity PCR System Taq DNA polymerase mixture (Perkin-Elmer). The primers FD1 and RD1 were originally described by Weisburg et al. as suitable for amplifying the 16S rRNA genes of many eubacteria (14). The parameters for amplification were 94°C for 5 min, 35 cycles of 15 s at 94°C, 15 s at 50°C, 1.5 min at 72°C, and an extension for 5 min at 72°C before cooling to 4°C. The results of the PCR were checked by running 10 μl of each reaction mixture on a 1.2% (wt/vol) agarose gel. The amplicon was then purified and concentrated by using a Qia Quick PCR purification kit from Qiagen (Valencia, Calif.). Approximately 60 ng (∼4 μl) of PCR product was used for each sequencing reaction. The sequencing reaction mixture consisted of DNA, 8 μl of ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin-Elmer), and 3.2 pmol of primer; sterile water was added to a final volume of 20 μl. The primer set used for sequencing was derived from primers designed by Stackebrandt and Charfreitag (10). The thermocycler conditions were according to the manufacturer's instructions for the cycle sequencing kit. Amplifications were performed on a Perkin-Elmer 9700 thermocycler. The extension products from each reaction were purified through a Centrisep column (Princeton Separations, Adelphia, N.J.) and dried in a vacuum centrifuge for 20 min. The sequencing reaction products were resolved on a 4.2% (wt/vol) acrylamide-8 M urea gel electrophoresed on an ABI 377 automated sequencer (Perkin-Elmer). The sequence data were edited and compiled with the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.). The 16S rRNA gene sequences were aligned by using the PILEUP program. The multiple sequence alignments were edited manually. The edited alignments were used with TREECON (Version 1.3b; Yves Van de Peer) to derive a phylogenetic dendrogram by using the nucleotide substitution model of Jukes and Cantor (5) and the neighbor-joining method of Saitou and Nei (9).
The CDC group EO-2 includes 13 similar human clinical isolates received from seven different states in the United States and from three different provinces in Canada. Two of the isolates were from ears, two were from ankles, and one each was from blood, cerebrospinal fluid, eye, neck incision drainage, bile, abdominal dialysate, leg lesion, toe, and skin (Table 1). The source of these organisms and their route of entry into the body are not known. Since all of the EO-2 isolates known to date were submitted from clinical sources, the natural habitat (or habitats) of this species has not yet been identified. The geographic information available indicates that these strains may be isolated from patients in a wide variety of locations, ranging from Hawaii to Canada. Limited clinical information was available on patients, and no common underlying syndrome was indicated; however, wounds were the most frequent condition. The ages of the patients ranged from 6 weeks to 77 years; 8 of 13 patients were female and the remaining patients were male.
Cells grown on heart infusion agar at 35°C had forms ranging from coccoid or diplococcoid to coccobacillary, often appearing vacuolated or peripherally stained (O shape), and stained gram negative, sometimes appearing gram variable (15). All grew well on blood agar that was incubated aerobically or in a candle jar under CO2 atmosphere for 18 to 24 h at 35°C, and the growth was frequently mucoid to extremely mucoid. No growth was obtained under anaerobic conditions. Isolated colonies were usually punctate. No distinct hemolytic reaction was observed after overnight incubation. The phenotypic characteristics of the CDC group EO-2 are given in Table 2. All strains were positive for oxidase, urease, citrate utilization (sometimes delayed 3 to 7 days), acid production from xylose and lactose (often delayed), growth on MacConkey agar, and growth at 35°C. All except one strain produced acid from glucose and reduced nitrate. All strains were negative for motility, indole, lysine decarboxylase, H2S (triple sugar iron [TSI] butt), insoluble pigment, production of gas from glucose, hydrolysis of esculin and gelatin, and acid production from sucrose and TSI.
TABLE 2.
Phenotypic characteristics of P. yeeiia
| Characteristic(s) | % Positive | No. positive/total no. of strains |
|---|---|---|
| Motility | 0 | 0/13 |
| Gas from glucose | 0 | 0/13 |
| Action in blood | 0 | 0/13 |
| Acid from (OF base): | ||
| d-Glucose | 92 | 12/13 |
| d-Xylose | 100 | 13/13 |
| Lactose | 54 (46) | 7 (6)/13 |
| Sucrose | 0 | 0/13 |
| Maltose | (13 w) | (1 w)/8 |
| d-Mannitol | 8 | 11/13 |
| Catalase | 84 | 11/13 |
| Oxidase | 100 | 13/13 |
| Growth on: | ||
| MacConkey agar | 75 (25) | 6 (2)/8 |
| SS | 0 | 0/8 |
| Cetrimide | 0 | 0/8 |
| Simmons citrate | 60 (38) | 5 (3)/8 |
| Urea, Christensen's | 76 (25) | 6 (2)/8 |
| Nitrate reduction | 92 | 12/13 |
| Gas from nitrate | 8 | 1/13 |
| Nitrite | 13 | 1/8 |
| Indole | 0 | 0/13 |
| TSI slant, acid | 0 | 0/13 |
| TSI butt, acid | 0 | 0/13 |
| H2S (TSI butt) | 0 | 0/13 |
| H2S (PbAc paper) | 25 (63) | 2 (3, 2 w)/8 |
| Gelatin hydrolysisb | 0 | 0/13 |
| Litmus milk | 13 k (37 k) 25 w k (25 IR) | 1 k (3 k) 2 w k (2 IR)/8 |
| Growth at: | ||
| 25°C | 78 | 9, 1 w/13 |
| 35°C | 100 | 13/13 |
| 42°C | 38 | 1, 2w/8 |
| Esculin hydrolysis | 0 | 0/13 |
| Growth in nutrient broth, 0% NaCl | 25 (13) | 2 (1)/8 |
| Growth in nutrient broth, 6% NaCl | 25 (13) | 2 (1 w)/8 |
| Lysine decarboxylase | 0 | 0/8 |
| Arginine dihydrolase | 13 | 1/8 |
| Ornithine decarboxylase | 13 | 1/8 |
| Pigment | ||
| Insoluble | 0 | 0/8 |
| Soluble | 38 yel, 25 brown, 25 amber, 13 olive | 3 yel, 2 brown, 2 amber, 1 olive/8 |
Number of strains tested, 13. Growth is frequently mucoid. Symbols and abbreviations: (), delayed 3 to 7 days; OF, oxidation-fermentation; w, weak reaction; TSI, triple sugar iron; SS, salmonella-shigella agar; yel, yellow; k, alkaline; IR, indicator reduction; PbAc, lead acetate.
Incubation of 7 to 14 days.
The results of DNA relatedness studies are given in Table 3. According to the established molecular criteria for species-level relatedness (strains whose DNAs are 70% or more related at optimal conditions and whose related sequences show 2% or less divergence) (13), all the EO-2 strains were a single species separate from the previously described Paracoccus species. The relatedness within this group was greater than 89%, and the divergence was less than 5%. None of the phenotypically similar taxa, i.e., Psychrobacter immobilis, P. alcaliphilus, P. aminophilus, P. denitrificans, P. solventivorans, or P. versutus, were related to EO-2 strain G1212 at greater than 54%. The percent G+C was determined for a representative strain of CDC group EO-2. The percent G+C for G1212 was 62%.
TABLE 3.
DNA relatedness and percent 16S similarity between P. yeeii, Psychrobacter immobilis, P. alcaliphilus, P. aminophilus, P. denitrificans, P. solventivorans, and P. versutus
| Source of unlabeled DNAa | % Relatedness to labeled DNA from P. yeeii strain G1212
|
% Similarity of 16S to 16S of P. yeeii strain G1212 | |
|---|---|---|---|
| RBR at 65°Cb | Dc | ||
| P. yeeii G1212 (AY014173) | 100 | 0 | 100 |
| P. yeeii G1968 (AY014171) | 97 | 2 | 99.7 |
| P. yeeii G3060 (AY014179) | 90 | 2 | 99.9 |
| P. yeeii G4446 (AY014169) | 97 | 2 | 99.9 |
| P. yeeii G4878 (AY014168) | 91 | 1.5 | 100 |
| P. yeeii G6155 (AY014170) | 95 | 1 | 100 |
| P. yeeii G9205 (AY014172) | 97 | 1 | 99.6 |
| P. yeeii H13 (AY014178) | 89 | 0.5 | 99.6 |
| P. yeeii H1426 | 96 | 0.5 | NDd |
| P. yeeii H1427 | 94 | 0 | ND |
| P. yeeii H1428 | 94 | 0.5 | ND |
| P. yeeii H1429 | 99 | 0 | ND |
| P. yeeii H1430 | 91 | 0.5 | ND |
| Psychrobacter immobilis ATCC 43116 (U39399) | 0 | NDd | 75.3 |
| P. alcaliphilus ATCC 51199 (AY014177) | 38 | 15 | 96.5 |
| P. aminophilus ATCC 49673 (AY014176) | 27 | 14.5 | 96.9 |
| P. denitrificans ATCC 17741 (Y16927) | 27 | 13.5 | 97.2 |
| P. solventivorans ATCC 700252 (AY014175) | 20 | 14.5 | 95.4 |
| P. versutus ATCC 25364 (AY014174) | 54 | 12.5 | 95.4 |
Numbers in parentheses are GenBank accession numbers.
RBR, relative binding ratio: the amount of double-stranded DNA formed between labeled and unlabeled DNAs from different strains divided by the amount of double-stranded DNA formed between labeled and unlabeled DNA from the same strain. RBR is expressed as a percentage.
D, the amount of divergence, determined by the number of unpaired bases, in DNA sequences held in common between two bacteria. D is calculated to the nearest 0.5%.
ND, not calculated.
The CFA composition of saponified whole cells of CDC group EO-2 is given in Table 4. This CFA profile is characterized by very large amounts of 18:1ω7c (71%) and small amounts of 3-OH-10:0 and 3-OH-14:0. It also contains moderate amounts of 16:0 (13%) and small amounts of 12:1ω7c, 17:0, 18:2, 18:1ω9c, and 18:0. The overall CFA profile of EO-2 is similar to that of Paracoccus species (Table 4). Furthermore, the overall CFA profile of the EO-2 group is most like that of the CFA group including Methylobacterium species, Roseomonas fauriae, and Roseomonas genomospecies 6 except for the presence of 3-OH-10:0 (3% versus 0%) (8, 12, 15). EO-2 has about 3% of 12:1ω7, which is absent in all Methylobacterium species and all Roseomonas species and three genomospecies (8, 12, 15). In addition, EO-2 contains ubiquinone with eight isoprene units (Q-8) as the major isoprenolog (8). All the Paracoccus species examined to date contain ubiquinone-10 (Q-10) as the respiratory quinone (6). This difference indicates that CDC group EO-2 can be distinguished from related organisms on the basis of its isoprenoid quinone content.
TABLE 4.
CFA compositions of P. yeeii, P. alcaliphilus, P. aminophilius, P. denitrificans, P. solventivorans, and P. versutusa
| Fatty acidb | Organism
|
|||||
|---|---|---|---|---|---|---|
| P. yeeii | P. alcaliphilus | P. aminophilus | P. denitrificans | P. solventivorans | P. versutus | |
| 3-OH-10:0 | 3 | 6 | 4 | 2 | 4 | 4 |
| 12:1ω7c | 3 | 6 | 4 | 2 | 4 | 4 |
| 3-OH-14:0 | 1 | 2 | 2 | 1 | T | ND |
| 16:0 | 13 | 1 | 21 | 12 | ND | 9 |
| 17:0 | 1 | ND | ND | ND | T | ND |
| 18:2 | 1 | ND | ND | ND | ND | ND |
| 18:1ω9c | 1 | 2 | 2 | 1 | 1 | 1 |
| 18:1ω7c | 71 | 71 | 38 | 71 | 81 | 78 |
| 18:0 | 5 | 7 | 4 | 5 | 6 | 2 |
| 19:0cyc11-12 | ND | 4 | 24 | 4 | ND | 1 |
| 20:1ω9t | ND | ND | ND | ND | 2 | ND |
Values are percentages of total fatty acids and are arithmetic means. T, trace; ND, not detected.
The number before the colon indicates the number of carbons; the number after the colon is the number of double bonds. ω, position of the double bond counting from the hydrocarbon end of the carbon chain; OH, hydroxy group at the 2(α) or 3(β) position from the carboxyl end; c, cis isomer; t, trans isomer; cyc, cyclopropane ring structure.
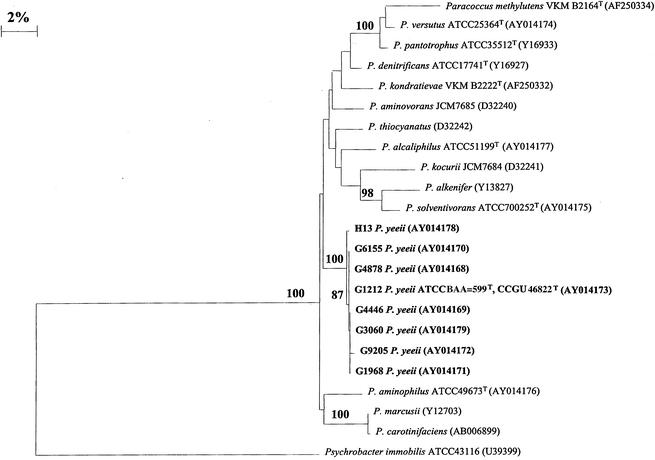
The 16S rRNA gene sequences of eight members of the CDC EO-2 group (AY014168, AY014169, AY014170, AY014171, AY014172, AY014173, AY014178, and AY014179) and five Paracoccus spp. were determined and aligned with a sequence of Psychrobacter immobilis (strain ATCC 43116T; GenBank accession number, U39399) from GenBank. The resulting phylogenetic tree (Fig. 1) demonstrates the close relationships between the members of the EO-2 group and the relationship between the EO-2 strains and other members of the Paracoccus genus. The eight gene sequences from EO-2 strains were 98.9 to 100% identical across 1,450 bp of their 16S rRNA gene sequences. The 16S data correlate well with the DNA-DNA hybridization results. The sequences of the EO-2 strains were 94.6 to 97.2% similar to the sequences of P. aminophilus (ATCC 49673), P. alcaliphilus (ATCC 51199), P. solventivorans (ATCC 700252), P. denitrificans (ATCC 17741), and P. versutus (ATCC 25364). The bootstrap value of 100 indicates the EO-2 strains grouped separately from the Paracoccus spp. when the tree was generated 100 separate times. All eight of the 16S rRNA gene sequences from the EO-2 isolates were very similar to one another, ranging from 99.0 to 100% identical. The 16S rRNA gene sequences from three EO-2 isolates, G6155, G4878, and G1212, were 100% identical, and sequences from a second group of strains, G1968 and G4446, were 100% identical by pairwise comparison.
FIG. 1.
Phylogenetic tree based on 1,450-bp 16S rRNA gene sequences and showing the position of P. yeeii strains. The tree was rooted by using Psychrobacter immobilis as the outgroup. Bootstrap analysis was done with 100 resamplings; bootstrap values are indicated at some branch points. The scale bar represents 2% difference in DNA sequences. The accession numbers for the sequences used in the study are shown.
The genus Paracoccus comprises a group of aerobic, gram-negative bacteria that are spherical or in the form of short rods, nonmotile, and catalase and oxidase positive and reduce nitrate. These occur in soil and presumably in natural and artificial brines. Fourteen species of this genus have been identified, including five phenotypically similar species (3, 6). These species include P. alcaliphilus, P. aminophilus, P. denitrificans, P. solventivorans, and P. versutus (6). The findings presented in this paper indicate that CDC group EO-2 represents a previously undescribed Paracoccus species associated with human infection. The variable growth in 6% NaCl suggests that these organisms are unlikely to be found in the brine environments that have yielded some Paracoccus species.
The assignment of the CDC group EO-2 to the genus Paracoccus as a new species, P. yeeii, is supported by phenotypic and phylogenetic considerations. The cellular morphology and basic biochemical profile of these organisms are consistent with the reference description of this genus. The CFA profile of the EO-2 group is also similar to CFA profiles of other Paracoccus type strains studied. Comparative analysis of 16S rRNA gene sequences of eight members of EO-2 strains places all strains analyzed in a highly related cluster within the Paracoccus region of the phylogenetic tree. Whole chromosomal DNA-DNA hybridization analysis with Paracoccus type strains indicates all EO-2 strains studied to be related at the species level (>70% relatedness under stringent conditions). No other type strains of Paracoccus species studied produced DNA relatedness values of greater than 54% when compared with the proposed type strain of the new species (G1212, ATCC BAA-599, CCUG 46822). The description of this group as a new species of the genus Paracoccus is as follows:
P. yeeii sp. nov. (yeeii′. N.L. gen. m. yeeii, in recognition of the contributions of Robert B. Yee, University of Pittsburgh Graduate School of Public Health, to the characterization of pathogenic bacteria [11, 16, 17]). Gram negative (sometimes appearing gram variable), non-spore forming, nonmotile, with coccoid or diplococcoid to coccobacillary forms, often appearing vacuolated or peripherally stained (O shape). Frequently mucoid to extremely mucoid, nonpigmented colonies on heart infusion agar with 5% rabbit blood at 35°C. All strains were positive for oxidase, urease, citrate utilization, lactose, growth on MacConkey agar, and acid production from xylose. All except one strain produced acid from glucose and reduced nitrate. All strains were negative for motility, indole, lysine decarboxylase, acid production from sucrose, and hydrolysis of esculin and gelatin. All strains shared similar CFA profiles characterized by major amounts of 18:1ω7c, moderate amounts of 16:0, and minor amounts of 3-OH-10, 12:1ω7c, 3-OH-14, 17:0, 18:2, 18:1ω9c, and 18:0. Strain G1212, isolated in Pennsylvania from abdominal dialysate, is the type strain of the species and has been designated in the American Type Culture Collection as ATCC BAA-599 and in the Culture Collection University of Goteborg as CCUG 46822. The percent G+C content of the DNA for G1212 was 62%.
Nucleotide sequence accession number.
The newly determined 16S rRNA gene sequence of G1212 has been submitted to GenBank and assigned the accession number AY014173.
Acknowledgments
We give special thanks to Leta Helsel for submitting the type strain to ATCC and CCUG.
REFERENCES
- 1.Bonner, T. I., D. J. Brenner, B. R. Neufeld, and R. J. Britten. 1973. Reduction in the rate of DNA reassociation by sequence divergence. J. Mol. Biol. 31:123-135. [DOI] [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. C. McWhorter, J. K. Leete-Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doronina, N. V., Y. A. Trotsenko, B. B. Kuznetzov, and T. P. Tourova. 2002. Emended description of Paracoccus kondratievae. Int. J. Syst. Evol. Microbiol. 52:679-682. [DOI] [PubMed] [Google Scholar]
- 4.Hudson, M. J., D. G. Hollis, R. E. Weaver, and C. G. Galvis. 1987. Relationship of CDC group EO-2 and Psychrobacter immobilis. J. Clin. Microbiol. 25:1907-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. 3. Academic Press, New York, N.Y.
- 6.Kelly, D. P., F. A. Rainey, and A. P. Wood. 2000. The genus Paracoccus. In M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. [Online.] Springer, New York, N.Y. http://www.prokaryotes.com.
- 7.Mandel, M., L. Igambi, J. Bergendahl, M. L. Dodson, Jr., and E. Scheltgen. 1970. Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J. Bacteriol. 101:333-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moss, C. W., P. L. Wallace, D. G. Hollis, and R. E. Weaver. 1988. Cultural and chemical characterization of CDC groups EO-2, M-5, and M-6, Moraxella (Moraxella) species, Oligella urethralis, Acinetobacter species, and Psychrobacter immobilis. J. Clin. Microbiol. 26:484-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 10.Stackebrandt, E., and O. Charfreitag. 1990. Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J. Gen. Microbiol. 136:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Wadowsky, R. M., and R. B. Yee. 1985. Effect of non-Legionellaceae bacteria on the multiplication of Legionella pneumophila in potable water. Appl. Environ. Microbiol. 40:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace, P. L., D. G. Hollis, R. E. Weaver, and C. W. Moss. 1990. Biochemical and chemical characterization of pink-pigmented oxidative bacteria. J. Clin. Microbiol. 28:689-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad hoc committee on reconciliation of the approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 14.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. J. Jordan, E. C. Cook, and M. I. Daneshvar. 1996. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria, 2nd ed. The Williams & Wilkins Co., Baltimore, Md.
- 16.Yee, R. B. 1958. Studies on the metabolism of Shigella. II. The oxidation of glutamate by Shigella flexneri. J. Bacteriol. 75:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee, R. B., and C. L. Buffenmeyer. 1970. Infection of cultured mouse macrophage with Shigella flexneri. Infect. Immun. 1:459-463. [DOI] [PMC free article] [PubMed] [Google Scholar]