Abstract
Two extrachromosomal double-stranded RNA (dsRNA) elements occur in Cryptosporidium parvum. A heteroduplex mobility assay (HMA) was developed for the rapid characterization of sequence diversity in a 173-bp fragment of the small dsRNA element of Cryptosporidium with either a natural sequence from Cryptosporidium meleagridis or a synthetic sequence as reference DNA. The 173-bp fragment was generated from 265 samples of whole feces (242 from humans and 18 from livestock with C. parvum genotype 1 or 2, 4 from humans with Cryptosporidium felis, and 1 from a human with C. meleagridis). The HMA method identified 21 patterns in C. parvum (8 in genotype 1, 12 in genotype 2, and a type common to both genotypes), 4 patterns in C. felis, and 1 pattern in C. meleagridis. All patterns were confirmed as distinct by DNA sequencing. For genotype 1, a single HMA type was found in 89% of samples: 64 of 65 cases from three waterborne outbreaks, all 16 cases from eight intrafamilial outbreaks, and 17 of 28 sporadic cases. Among the remaining 11 sporadic cases due to genotype 1, seven other HMA types were detected. For genotype 2, a single HMA type was found in 72% of samples: 36 of 43 cases from three waterborne outbreaks, 11 of 15 cases from seven intrafamilial outbreaks, 44 of 75 sporadic cases, and all 18 samples from livestock. Within the intrafamilial outbreaks, two other HMA types were identified: the same HMA type was detected in samples from cases within the same outbreak. Among the sporadic cases due to genotype 2, 10 additional HMA types were detected.
Protozoan organisms of the genus Cryptosporidium are enteric parasites responsible for gastrointestinal disease in humans and animals (28). In immunocompetent individuals, cryptosporidiosis generally results in an acute, self-limited diarrheal disease, while in immunocompromised individuals, infection can be more severe, prolonged, and invasive (11). Infection spreads from an infected host via the oocyst by fecal-oral transmission, and multiple routes of infection occur and include person-to-person contact; animal-to-person contact; ingestion of contaminated water, food, or beverages; recreational bathing; and contact with environmentally contaminated sources (12, 25). However, the exact modes of transmission for humans usually remain unclear for the majority of cases.
The application of molecular methods for studying the epidemiology of cryptosporidiosis has identified Cryptosporidium parvum as the predominant species infecting humans (22, 24, 44, 49). Two different genotypes of C. parvum are most often associated with human disease and can be identified by the analysis of polymorphic gene loci, including those for the Cryptosporidium oocyst wall protein (COWP) (40), thrombospondin adhesive proteins TRAP-C1 (39) and TRAP-C2 (36, 44), and β-tubulin (8), as well as 18S rRNA gene sequences (27). Genotype 1 (or human type) is isolated predominantly from humans, while genotype 2 (or calf type) has a broader host range, including humans, livestock, and rodents (28). Experimental infections of calves and mice were successful with C. parvum genotype 2 isolates but not with C. parvum genotype 1 isolates (36). A multilocus analysis of different C. parvum genes in isolates from different hosts showed no recombination between genotypes (38). A study conducted in the United Kingdom with 136 isolates genotyped with the COWP and TRAP-C1 genes demonstrated complete segregation of these two independent loci (24). Differences in host range and the absence of recombination between different markers of the two C. parvum genotypes support the existence of different populations with distinct and exclusive transmission cycles (36). Other Cryptosporidium species have been reported to be associated with human infections, extending knowledge about the range of species that can cause cryptosporidiosis in humans. Cryptosporidium felis (16, 31, 37, 45), Cryptosporidium meleagridis (16, 26, 34, 45), Cryptosporidium canis (37), Cryptosporidium muris (14, 45), Cryptosporidium andersoni (16), and Cryptosporidium pig genotype (48) have been identified in samples of feces from immunocompromised individuals. C. meleagridis (34, 49), C. felis (31, 49), C. canis (31), and a novel C. parvum cervine genotype (30) have been detected in immunocompetent individuals.
Molecular epidemiological analyses of C. parvum genotypes 1 and 2 not only have identified different transmission cycles (36) but also have shown seasonal and geographical differences in their distributions (22), together with a suggestion of biological differences in the behavior of these parasites (46). However, more precise epidemiological tracking of these parasites with discriminatory markers is required to distinguish subgenotypes within each genotype. Genetic polymorphisms within genotypes 1 and 2 have been described for microsatellite loci (7, 9, 13), a glycoprotein (15, 35, 41, 42), and two extrachromosomal linear virus-like double-stranded RNA (dsRNA) elements (19). Sequence analyses of 306- and 257-bp fragments of the large and small dsRNA elements from human and calf isolates identified both intra- and intergenotypic heterogeneities (19). The diversities between different isolates were 92 to 99% for the large dsRNA and 93 to 99% for the small dsRNA (19). Sequence analyses of both fragments identified two distinct clusters, one with all of the genotype 2 isolates (of human and calf origins) and a second with all of the genotype 1 isolates (19). Sequence analysis of a 173-bp fragment of the small dsRNA from 61 C. parvum isolates (23 from cattle and 38 from humans) identified the presence of 18 distinct nucleotide sequences within genotype 1 and 2 isolates: 8 subgenotypes within genotype 2 and 10 subgenotypes within genotype 1 (50). Isolates from the same outbreak had the same dsRNA subgenotype (50). Unlike the results of the study of Khramtsov et al. (19), clusters corresponding to C. parvum genotypes 1 and 2 were not identified in the 173-bp fragment (50).
The heteroduplex mobility assay (HMA) is a technique used to identify heterogeneity between two segments of DNA, usually PCR amplicons (10). Two segments of DNA, a known sequence (reference material) and a test sample, are mixed together, heated to denaturation into single strands, and then cooled slowly. The single strands reanneal during cooling to reform the original homoduplexes or form heteroduplexes composed of strands from different starting amplicons. The reactions are analyzed by polyacrylamide gel electrophoresis, and heteroduplexes with mismatched bases can be separated from homoduplexes due to slower electrophoretic migration.
We previously developed a reverse transcriptase (RT) PCR-HMA (RT-PCR-HMA) method based on the characterization of the 173-bp fragment of the small dsRNA from C. parvum (21). The 173-bp fragment of the small dsRNA from C. parvum was sequenced from a panel of 18 samples of human origin, previously genotyped by PCR-restriction fragment length polymorphism (RFLP) analysis of the COWP gene (10 isolates of genotype 1 and 8 isolates of genotype 2). A total of 11 distinct nucleotide sequences with 1- to 13-bp differences between them were identified: 5 nucleotide sequences each for genotypes 1 and 2 and a sequence common to the two genotypes (21). Two sequences were selected as reference DNA for the HMA; heteroduplex formation was observed, but for both assays, difficulties were encountered in the characterization of sequence diversity due to the similar electrophoretic mobilities of homo- and heteroduplex bands. Synthetic nucleotides for use as reference material in the HMA were developed by introducing mismatches into a natural sequence by PCR (21). The amplicons were tested by HMA against the parent sequence and, to increase efficiency in generating the novel nucleotides, the heteroduplex bands were excised from the stained polyacrylamide gels and used as templates for PCR of the 173-bp fragment of the small dsRNA. Of nine mutant sequences generated and evaluated as reference material in the HMA, one was selected for improved resolution of heteroduplex bands from homoduplex bands; nine different HMA patterns were identified (21).
The purpose of this study was to apply the HMA method with the artificial reference sequence previously developed plus a second, naturally divergent reference sequence reported in this study as a subgenotyping tool for molecular epidemiological analyses of Cryptosporidium.
MATERIALS AND METHODS
Fecal samples.
A total of 450 specimens were collected as whole feces from humans (395 samples) or livestock animals (55 samples from lambs or calves) between 1995 and 1999 in the United Kingdom, where Cryptosporidium oocysts were identified by using conventional techniques (4). All samples were stored at 4°C without preservatives.
Oocyst disruption, nucleic acid extraction, and PCR-RFLP analysis.
Extraction of nucleic acids from all of the whole-feces samples was performed as previously described (24). C. parvum genotypes 1 and 2 and C. meleagridis were identified by using nested PCR-RFLP analysis for a fragment of the COWP gene as described by Pedraza-Díaz et al. (33). C. meleagridis was further characterized by PCR of an 18S ribosomal DNA (rDNA) fragment (18) and sequence analyses of both this fragment and the COWP gene fragment. C. felis was identified by PCR and sequence analyses of an 18S rDNA fragment (18) and the heat shock protein 70 (HSP70) gene (43) as described by Pedraza-Díaz et al. (31).
RT-PCR for dsRNA elements.
cDNA was generated from all fecal extracts (24) by adding 1 μl of 0.27 mM random hexamers (PdN6; Amersham Biosciences, Little Chalfont, United Kingdom) to 40 μl of nucleic acid extract, denaturing the mixture at 95°C for 5 min, and chilling the mixture on ice. A 13.5-μl volume of 10 mM Tris (pH 8.3)-50 mM KCl-5 mM MgCl2-0.45 mM deoxynucleoside triphosphates (Invitrogen, Paisley, United Kingdom)- 300 U of Moloney murine leukemia virus RTase (Invitrogen) was added to the mixture, which was incubated at 37°C for 60 min. The reaction was terminated by heating to 95°C for 5 min, and the tubes were chilled on ice.
Amplification of the 173-bp fragment of the small dsRNA with primers APBV1 and APBV2, previously described by Xiao et al. (50), was performed by adding 5 μl of cDNA to 45 μl of a PCR mixture containing Expand high-fidelity PCR buffer without MgCl2 (Roche Molecular Biochemicals, Lewes, United Kingdom), 2 mM MgCl2 (Roche), 0.4 mM deoxynucleoside triphosphates (Invitrogen), 20 pmol of each primer, and 0.7 U of Expand high-fidelity Taq polymerase (Roche). PCR conditions were as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 5 min. Fifteen microliters of PCR product was analyzed in 2% agarose (Invitrogen) and stained with ethidium bromide. Amplicons were visualized by UV transillumination.
Cloning and sequencing.
PCR amplicons were cloned into the pCR2.1-TOPO plasmid vector by using a TOPO-TA cloning kit (Invitrogen). A 2-μl aliquot of unpurified PCR product was ligated to the plasmid, and transformed cells were selected on an L agar plate containing ampicillin, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), and isopropyl-β-d-thiogalactopyranoside (IPTG) by following the manufacturer's instructions (Invitrogen). From each transformation, three white colonies were chosen and subcultured on L agar plates at 37°C overnight. A heat denaturation preparation was made for each colony in 50 μl of water, and 1 μl was used as a template for PCR. Clones were screened with PTAG 5′/3′ primers (21) by using an Expand high-fidelity PCR system (Roche). PCR products were purified by using a GeneClean spin kit (Anachem Ltd., Luton, United Kingdom) and sequenced in both directions with PTAG 5′/3′ primers by using a CEQ2000 dye terminator cycle sequencing Quick Start kit (Beckman Coulter, High Wycombe, United Kingdom) according to the manufacturer's instructions and a Beckman Coulter CEQ2000XL automated capillary sequencer. Sequences were analyzed and aligned by using GeneBuilder and Clustal in Bionumerics version 2.5 (Applied Maths, Hortrijk, Belgium). The relationship between isolates was assessed by the unweighted pair-group method with arithmetic means. Multiple alignment analyses were performed with BioEdit Sequence Alignment Editor version 5.0.0 (17).
Preparation of reference strain amplicons.
Plasmid DNAs from reference strain clones 1912 mutant 5C/15C (21) and 1689 (this study) were purified by using a Rapid Pure miniprep kit (Anachem). Multiple PCR mixtures (50-μl volumes each) were prepared by using 1 μl of plasmid DNA diluted 1/15 with water as a template and pooled to provide an adequate quantity of reference strain amplicons for the HMA. Primers, reagents, and PCR conditions were the same as those used for the small-dsRNA PCR. Amplicons were stored at 4°C.
HMA.
Mutation detection enhancement (Biowhittaker Molecular Applications, Wokingham, United Kingdom) gels were prepared as follows. Glass plates (20 by 20 by 1.5 cm) were assembled by using a Hoeffer vertical electrophoresis system (Amersham). Twenty milliliters of mutation detection enhancement gel solution, 2.4 ml of 10× Tris-borate-EDTA (TBE) buffer (Invitrogen), 17.6 ml of water (Sigma, Poole, United Kingdom), 320 μl of freshly prepared 10% ammonium persulfate solution, and 16 μl of TEMED were added to a clean 50-ml conical tube and mixed by inversion; the mixture was poured into plates. A 28-well short-tooth comb was inserted, and the gels were allowed to polymerize for approximately 1.5 h.
HMA reaction mixtures were prepared by adding 5 μl of test amplicon to 5 μl of reference strain amplicon; controls included 10 μl of an individual reference amplicon without a test amplicon. Tubes were briefly centrifuged, and the HMA was performed by using a Perkin-Elmer 9600 thermocycler (PE Applied Biosystems, Warrington, United Kingdom) with initial denaturation at 94°C for 2 min and then cooling to 4°C (slow annealing at 1°C/10 s). Tubes were incubated on ice for 10 min or until loaded onto the gels. A 2-μl aliquot of 6× triple-dye loading buffer (Biowhittaker) containing xylene cyanol, bromophenol blue, and orange G was added to the reaction mixtures. A 1-kb DNA ladder (Invitrogen) was used as a molecular size marker. Vertical electrophoresis was performed with 0.6× TBE running buffer at a constant voltage of 300 V for approximately 3 h, with a water cooling system in a cold room at 4°C, until the bromophenol blue dye band was within 1 cm of the bottom of the gels.
Electrophoresis gels were stained with 100 ml of 0.6× TBE containing a 1/10,000 dilution of Gelstar nucleic acid gel stain (Biowhittaker) and shaken for 1 h. Gels were then rinsed with tap water and photographed by using a Gelstar gel stain photographic yellow filter (Biowhittaker).
HMA pattern identification and HMA type designation.
Heteroduplex bands formed in the areas between the homoduplex and single-stranded DNAs were used for analysis. HMA patterns were identified by comparison of heteroduplex bands to the marker and to controls previously characterized by HMA and sequence analyses. For unequivocal pattern designation, occasional reanalysis was performed by testing samples in adjacent lanes on a single electrophoresis gel. Comparative analysis of different gels and HMA pattern analysis were also performed by using Bionumerics version 2.5.
HMA types were established by combining results obtained with two reference DNAs: a previously described (21) synthetic nucleotide (1912 5C/15C) designated reference sequence I and a sequence from C. meleagridis (isolate 1689) designated reference sequence II. HMA types identified in this study were designated by using a code composed of three letters related to the species (CML for C. meleagridis, CFL for C. felis, and CPV for C. parvum) plus a numerical suffix; e.g., CPV1-1 comprises HMA pattern 1 within C. parvum genotype 1 (for C. parvum isolates, the genotype [1 or 2] was added to the code before the numerical suffix) (Table 1).
TABLE 1.
HMA types identified in this study
| HMA type | Isolate | Species | GenBank accession no. |
|---|---|---|---|
| CML-1 | 1689 | C. meleagridis | AY176647 |
| CFL-1 | 45 | C. felis | AY176650 |
| CFL-2 | 353 | C. felis | AY176648 |
| CFL-3 | 355 | C. felis | AY176649 |
| CFL-4 | 1626 | C. felis | AY176651 |
| CPV1-1 | 1908 | C. parvum genotype 1 | AY135733 |
| CPV1-2 | 1905 | C. parvum genotype 1 | AY135730 |
| CPV1-3 | 1903 | C. parvum genotype 1 | AY135728 |
| CPV1/2-4 | 1911 and 1945 | C. parvum genotypes 1 and 2 | AY135735 |
| CPV1-5 | 1906 | C. parvum genotype 1 | AY135731 |
| CPV1-6 | 1912 | C. parvum genotype 1 | AY135736 |
| CPV1-7 | T89 | C. parvum genotype 1 | AY176652 |
| CPV1-8 | 666 | C. parvum genotype 1 | AY176653 |
| CPV1-9 | 1962 | C. parvum genotype 1 | AY176654 |
| CPV2-1 | 1907 | C. parvum genotype 2 | AY135732 |
| CPV2-2 | 1910 | C. parvum genotype 2 | AY135738 |
| CPV2-3 | 1926 | C. parvum genotype 2 | AY135737 |
| CPV2-5 | 1904 | C. parvum genotype 2 | AY135729 |
| CPV2-6 | 1909 | C. parvum genotype 2 | AY135734 |
| CPV2-7 | 1005 | C. parvum genotype 2 | AY176655 |
| CPV2-8 | 870 | C. parvum genotype 2 | AY176656 |
| CPV2-9 | 1034 | C. parvum genotype 2 | AY176657 |
| CPV2-10 | 1283 | C. parvum genotype 2 | AY176658 |
| CPV2-11 | 1059 | C. parvum genotype 2 | AY176659 |
| CPV2-12 | 340 | C. parvum genotype 2 | AY176660 |
| CPV2-13 | 1876 | C. parvum genotype 2 | AY176661 |
Statistical analysis.
Statistical analysis was performed by using Epi Info 2000 version 1.1.2 (Centers for Disease Control and Prevention, Atlanta, Ga.).
Nucleotide sequence accession numbers.
The nucleotide sequences previously reported for the small dsRNA element in GenBank (19, 20) were as follows: H1, AF169939; H2, AF169940; H3, AF169941; H4, AF169942; H5, AF169943; H6, AF169944; H7, AF169945; C1, AF169934; C2, AF169935; C3, AF169936; C4, AF169937; C5, AF169938; KSU-1, U95996; 1903, AY135728; 1904, AY135729; 1905, AY135730; 1906, AY135731; 1907, AY135732; 1908, AY135733; 1909, AY135734; 1911, AY135735; 1912, AY135736; 1910, AY135738; and 1926, AY135737.
Sequences designated types A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P, Q, and R (50) were obtained from L. Xiao, Division of Parasitic Diseases, Centers for Disease Control and Prevention.
Additional sequences described in this study have been deposited in GenBank under the following accession numbers: 1689, AY176647; 45, AY176650; 353, AY176648; 355, AY176649; 1626, AY176651; T89, AY176652; 666, AY176653; 1962, AY176654; 1005, AY176655; 870, AY176656; 1034, AY176657; 1283, AY176658; 1059, AY176659; 340, AY176660; and 1876, AY176661.
RESULTS
dsRNA fragments from C. felis and C. meleagridis.
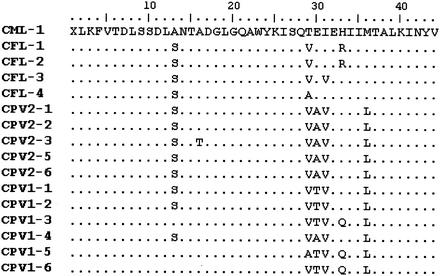
Initial experiments were performed to investigate the presence of the dsRNA fragment in C. meleagridis and C. felis. The RT-PCR procedure for the 173-bp fragment of the small dsRNA was applied to nucleic acid extracts from three fecal samples in which C. meleagridis had been detected and four extracts from fecal samples in which C. felis had been detected. Amplicons of the correct size were obtained for the one sample containing C. meleagridis and for all four samples containing C. felis. Cloning and sequencing of the five fragments showed that the C. felis isolates had differences from each other of between 5 and 7 bp (diversity, 95 to 96%) and differences from the C. meleagridis isolate of between 11 and 13 bp (diversity, 90 to 92%). An alignment of the C. meleagridis and C. felis sequences with sequences of the 173-bp fragment previously obtained from C. parvum (21) is presented in Fig. 1. C. meleagridis showed between 13 and 16 differences from the C. parvum group. C. felis and C. parvum had differences common to each species, i.e., a C at position 114 for all of the C. felis isolates and a C at position 88 for all of the C. parvum isolates (Fig. 1). The C. meleagridis isolate had five nucleotides different from those of all of the other isolates (Fig. 1), while two A's (positions 88 and 105) were seen in the C. felis group. Analysis of the amino acid sequences revealed that the nucleotide change at position 84 reflected an amino acid change from threonine for C. meleagridis to valine or alanine for both C. felis and C. parvum (amino acid 29) (Fig. 2). The two A's shared between C. meleagridis and the C. felis group at positions 88 and 105 of the nucleotide sequence corresponded to amino acids 30 and 36 (Fig. 2), encoded by nucleotide triplets GAG or GAA and ATG, respectively; these triplets encoded glutamic acid and methionine, respectively, in these two species but encoded a lysine in all of the C. parvum isolates (Fig. 2).
FIG. 1.
Sequence diversity in the 173-bp fragment of the small dsRNA element from C. meleagridis (CML), C. felis (CFL), and C. parvum (CPV) isolates. Dots represent identical sequences.
FIG. 2.
Amino acid sequences encoded by the 173-bp fragment of the small dsRNA element from C. meleagridis (CML), C. felis (CFL), and C. parvum (CPV) isolates. Dots represent identical amino acids.
HMA system with mutants and C. meleagridis.
Eighteen human fecal samples containing C. parvum (10 of genotype 1 and 8 of genotype 2) from different laboratories in England were randomly selected from a collection of >3,000 samples, and the 173-bp fragment of the small dsRNA element of C. parvum was generated by RT-PCR from all of them; cloning and sequencing of the fragment identified 11 distinct nucleotide sequences (21). Initial experiments indicated that the natural sequence from C. meleagridis (reference sequence II) and the previously described (21) synthetic sequence (reference sequence I) provided suitable reference material for the HMA to identify diversity in the 173-bp fragments of 11 different sequences from C. parvum (21), 4 from C. felis, and 1 from C. meleagridis. The HMA analysis with reference sequence I identified a total of 14 distinct HMA types: 9 from C. parvum, 4 from C. felis, and a unique profile from C. meleagridis (Fig. 3A). All sequences belonging to the C. felis group formed heteroduplex bands with an electrophoretic mobility slower than that of the C. parvum isolates when the synthetic sequence was used (Fig. 3A). Because the 11 different sequences from C. parvum yielded only nine different HMA patterns (21), HMA analysis with reference sequence II was performed. In the 16 different sequences, 14 HMA patterns (9 from C. parvum, 4 from C. felis, and 1 from C. meleagridis) were recognized (Fig. 3B). With reference sequence II, two pairs of sequences with one or two base mismatches were not differentiated and had identical profiles (i.e., sequences 1907 and 1909, with 14 and 13 mismatches, and sequences 1911 and 1904, with 14 and 16 mismatches, with respect to the reference DNA). However, the combination of results from reference sequences I and II enabled the identification of all 16 sequence types by HMA.
FIG. 3.
HMA analysis of the 173-bp fragment of the small dsRNA element from C. felis and C. parvum isolates with reference sequences I (A) and II (B). Lanes 1 to 16, CFL-4, CFL-2, CFL-1, CFL-3, CPV2-1, CPV2-2, CPV2-2, CPV2-3, CPV2-6, CPV1-1, CPV1/2-4, CPV2-5, CPV1-2, CPV1-3, and CPV1-5, and CPV1-6; lane 17, CML-1 (A) and control reference sequence II (B); lane 18, control reference sequence I (A) and reference sequence II with 1912 mutant 5C/5C (B); lane M, 1-kb DNA ladder.
Reproducibility.
The reproducibility of the HMA technique with the two reference sequences was investigated by comparison of results from the following samples or tests: PCR amplicons obtained by RT-PCR from fecal extracts and from cloned material sharing identical sequences; PCR products amplified from RT-treated fecal extracts and cloned material obtained from the same sample; and multiple tests with different preparations (Table 2). For the C. felis group, original PCR amplicons from only two samples were used for comparative analysis with cloned material, since PCR for the other two samples did not generate a sufficient amount of product suitable for direct HMA analysis. The HMA was reproducible for both reference sequences. However, in one sample, identical HMA profiles were generated with reference DNA preparations from both the original material and the cloned material, but an additional band was present when reference sequence II was used with the original fecal extract.
TABLE 2.
Reproducibility of the HMA technique
| Source of HMA results | Reference sequence I
|
Reference sequence II
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. of:
|
% Repro- ducibility | No. of:
|
% Repro- ducibility | |||||
| Samples | Sequences | HMA patterns | Samples | Sequences | HMA patterns | |||
| Identical sequences from: | ||||||||
| Original PCR ampliconsa from different samples | 12 | 5 | 4 | 100 | 12 | 5 | 5 | 100 |
| Cloned materialb from different samples | 12 | 5 | 4 | 100 | 12 | 5 | 5 | 100 |
| Original PCR ampliconsa and cloned materialb from the same sample | 42 | 14 | 12 | 100 | 42 | 14 | 12 | 95c |
| Multiple tests (five) on the same cloned sample | 23 | 16 | 14 | 100 | 23 | 16 | 14 | 100 |
PCR amplicons obtained by RT-PCR from fecal extracts.
PCR amplicons amplified by PCR from cloned DNA.
For one sample, the initial amplicon gave the pattern of the clone plus an extra band.
Molecular epidemiological analysis by HMA.
To further evaluate the HMA method, samples from epidemiologically related groups (waterborne [drinking water] and intrafamilial outbreaks) and samples from sporadic cases and livestock animals were selected. Amplicons for the HMA analysis were generated by RT-PCR from 303 (68%) of the 443 samples containing C. parvum (Table 3). Categorization of samples into those from waterborne outbreaks or sporadic cases, those containing genotype 1 or 2, and those from livestock showed that there were significant differences between the proportions of samples in which the dsRNA element was amplified (χ2 = 49.15, 4 degrees of freedom; P = <0.0001). The dsRNA element was significantly less amplified in samples collected from livestock than in those collected from humans (χ2 = 26.52; P = <0.0001). This fragment was significantly less commonly amplified in samples collected from sporadic cases than in those collected from outbreaks due to genotype 1 (χ2 = 7.5; P = 0.0062) but not to genotype 2 (χ2 = 2.23; P = 0.14).
TABLE 3.
Summary of RT-PCR amplifications for the 173-bp fragment of the small dsRNA element of Cryptosporidium
| Organism | Source | Case | No. of samples
|
% Ampli- fication | |
|---|---|---|---|---|---|
| Amplified by RT-PCR | Not amplified by RT-PCR | ||||
| C. parvum | |||||
| Genotype 1 | Humans | Outbreaka | 93 | 39 | 70 |
| Genotype 2 | Humans | Outbreaka | 64 | 10 | 86 |
| Genotype 1 | Humans | Sporadic | 30 | 30 | 50 |
| Genotype 2 | Humans | Sporadic | 95 | 27 | 78 |
| Genotype 2 | Livestock animals | Sporadic | 21 | 34 | 38 |
| C. felis | Humans | Sporadic | 4 | 0 | 100 |
| C. meleagridis | Humans | Sporadic | 1 | 2 | 33 |
Waterborne and intrafamilial outbreaks.
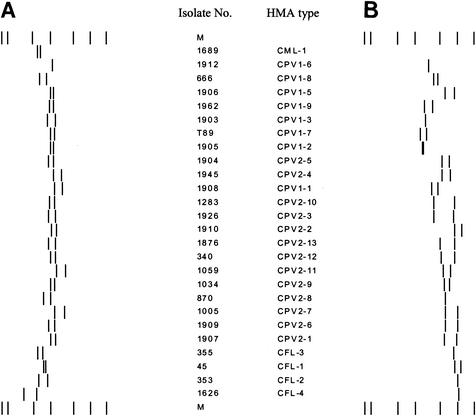
HMA analysis of the 173-bp fragment of the small dsRNA element was performed on amplicons from 260 samples (242 from humans and 18 from livestock) in which C. parvum genotype 1 or 2 had been previously detected. Because selected representatives of each category had already been tested, HMA analysis was not performed on the remaining 43 of the 303 total positive samples. Overall, 21 patterns were distinguished: 8 with genotype 1, 12 with genotype 2, and a pattern common to both genotypes. The C. meleagridis HMA type was designated CML-1, the four C. felis HMA types were designated CFL-1 to CFL-4, and the C. parvum genotype 1 and 2 isolate HMA types were designated CPV1-1 to CPV1-9 and CPV2-1 to CPV2-13, respectively (Table 1). The only common pattern between the two genotypes was named CPV1/2-4. A schematic representation of all HMA profiles identified in this study, generated by Bionumerics, is shown in Fig. 4.
FIG. 4.
Schematic representation of HMA patterns for Cryptosporidium. (A) Reference sequence I. (B) Reference sequence II. The 21 HMA types are identified by the combination of results from both assays. M, marker.
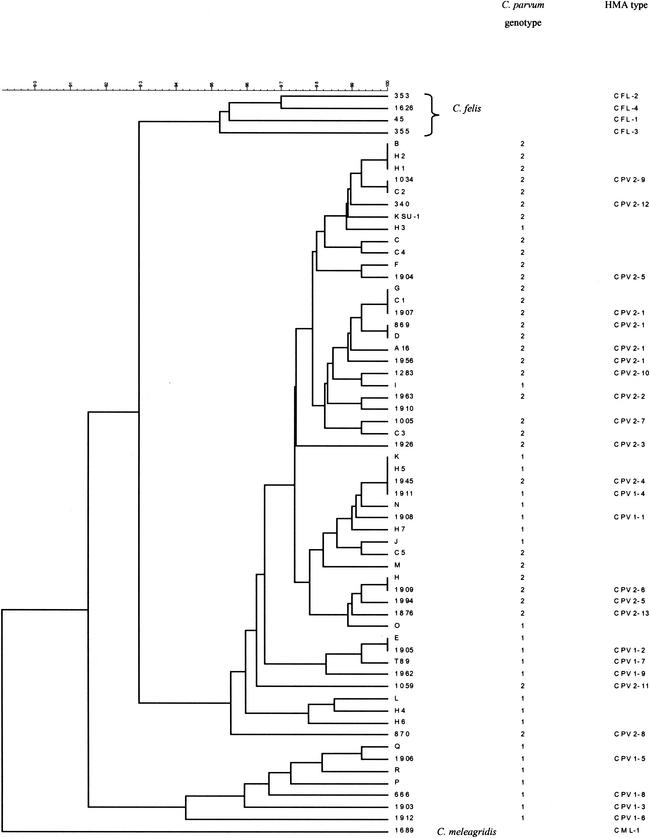
Cloning and sequencing were performed on 106 (41%) of the 260 samples analyzed by HMA; between 27 and 40% of the samples with identical HMA patterns were randomly selected from each HMA type for sequencing, together with all of the samples with unusual HMA types. Sequences corresponding to the 21 HMA patterns showed differences from reference sequence I of between 20 and 31 bp and differences from reference sequence II of between 11 and 17 bp. The 21 HMA types identified all had different sequences. Sequences closely related (Fig. 5) were discriminated by HMA analysis. A single base difference distinguished the following HMA types: CPV2-1 and CPV2-10, CPV2-1 and CPV2-7, CPV1-1 and CPV1-4, CPV2-5 and CPV2-6, and CPV1-2 and CPV1-7. Two base differences distinguished the following HMA types: CPV2-9 and CPV2-12, CPV2-2 and CPV2-7, CPV2-5 and CPV2-13, and CPV1-2 and CPV1-9.
FIG. 5.
Cluster analysis of the 173-bp fragment of the small dsRNA element.
HMA types CPV2-1 and CPV1-2 were found in 206 (79%) of the samples. Of the 109 total samples of genotype 1, 97 (89%) were of HMA type CPV1-2. Of all of the samples generating CPV1-2, 26 (27%) were sequenced and confirmed as a homogeneous group, with a sequence identical to that previously designated type E (50). Of the 26 isolates sequenced, 11, 8, and 7 were from waterborne and intrafamilial outbreaks and sporadic cases, respectively.
Of the 151 samples containing genotype 2, 109 (72%) were of HMA type CPV2-1. Of these, 41 (9 and 4 from waterborne and intrafamilial outbreaks, respectively; 14 from livestock; and 14 from sporadic cases) were cloned and sequenced. Sequencing results showed a homogeneous group for 37 (90%) of the samples, which had a sequence identical to the previously reported (19) C1 sequence (GenBank accession number AF169934); the 4 remaining samples of HMA type CPV2-1 (1 from livestock and 3 from sporadic cases) all had a single base change relative to the C1 sequence, and two had a sequence identical to sequence type D (50).
Of 25 samples of HMA type CPV2-2, 10 (40%) were cloned and sequenced; of these, 2 were from waterborne outbreaks and 8 were from sporadic cases. Sequencing results confirmed a homogeneous group for nine samples, which had a sequence identical to that of 1910, while the sequence of the remaining sample, 1963, had a single base difference.
All of the other samples with unusual HMA types (12 from genotype 1 and 17 from genotype 2) were cloned and sequenced. Of these, all samples with the same sequence (seven HMA types with two or three identical sequences per group) were of the same HMA type, apart from the two samples of HMA type CPV2-5, which had sequences with two base differences.
Waterborne outbreaks.
HMA analysis of the 173-bp fragment was performed with a panel of human samples from five drinking-water-associated outbreaks. Outbreaks 1 (6, 22, 23), 2 (22, 47), and 3 (6, 22) were predominantly caused by C. parvum genotype 1, and contaminated river or borehole waters were implicated in transmission. Outbreaks 4 (1, 22) and 5 (1, 22) were predominantly due to genotype 2, and contamination of tank or reservoir water by livestock animal feces was implicated (1, 2). Sixty-four of 65 samples (98%) from outbreaks caused by genotype 1 (outbreaks 1 to 3) were identified as HMA type CPV1-2; one case due to HMA type CPV1-7 was identified for outbreak 1 (Table 4). For the outbreaks caused by genotype 2, HMA type CPV2-1 was detected in 36 of 43 samples (84%) (Table 5). HMA types CPV2-2 and CPV2-7 were detected in 7 of the 22 patients in outbreak 5 (Table 5).
TABLE 4.
Nine HMA types identified for C. parvum genotype 1
| Case | No. of samples with the following HMA type:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Total | CPV1-1 | CPV1-2 | CPV1-3 | CPV1-4 | CPV1-5 | CPV1-6 | Othera | |
| Waterborne outbreak | 65 | |||||||
| 1 (August-September 1995) | 22 | 21 | 1 | |||||
| 2 (February-April 1997) | 40 | 40 | ||||||
| 3 (January-March 1997) | 3 | 3 | ||||||
| Family outbreak (n = 8)b | 16 | 16 | ||||||
| Sporadic | 28 | 2 | 17 | 3 | 1 | 2 | 1 | 2 |
| Total | 109 | 2 | 97 | 3 | 1 | 2 | 1 | 3 |
| Total sequenced | 38 | 2 | 26 | 3 | 1 | 2 | 1 | 3 |
HMA types CPV1-7, CPV1-8, and CPV1-9.
Two family groups were from waterborne outbreaks (drinking water) 1 and 2, and two other groups were from a swimming pool outbreak.
TABLE 5.
Thirteen HMA types identified for C. parvum genotype 2
| Case | No. of samples with the following HMA type:
|
|||||
|---|---|---|---|---|---|---|
| Total | CPV2-1 | CPV2-2 | CPV2-3 | CPV2-5 | Othera | |
| Waterborne outbreak | 43 | |||||
| 2 (February-April 1997) | 1 | 1 | ||||
| 4 (April 1998) | 20 | 20 | ||||
| 5 (April-May 1999) | 22 | 15 | 5 | 2 | ||
| Family outbreak (n = 7) | 15 | 11 | 4 | |||
| Sporadic | 75 | 44 | 20 | 1 | 2 | 8 |
| Livestock | 18 | 18 | ||||
| Total | 151 | 109 | 25 | 1 | 2 | 14 |
| Total sequenced | 68 | 41 | 10 | 1 | 2 | 14 |
HMA type CPV2-4, CPV2-6, CPV2-7, CPV2-8, CPV2-9, CPV2-10, CPV2-11, CPV2-12, and CPV2-13.
Intrafamilial outbreaks.
Thirty-one individuals in 15 intrafamilial outbreaks (8 of genotype 1 and 7 of genotype 2) were characterized by HMA analysis; 11 family groups (two or three members each) were from sporadic cases which occurred in England between 1998 and 1999, 2 groups (two members each) were from drinking-water-associated outbreaks 1 (6, 22, 23) and 2 (22, 47), and 2 groups (two members each) were from a swimming pool outbreak (October-November 1999) (3). The patients in the two waterborne outbreaks were not included in the above analyses. For all groups, members of the same family had the same HMA pattern. All of the samples associated with genotype 1 were identified as being HMA type CPV1-2 (Table 4). For those associated with genotype 2, HMA type CPV2-1 occurred in 11 of 15 cases (Table 5). Two other HMA types (CPV2-10 and CPV2-12) were detected in two family groups infected with C. parvum genotype 2 (Table 5).
Livestock animal cases.
The RT-PCR-HMA procedure was applied to 18 samples collected from naturally infected livestock between 1996 and 1997, comprising 14 bovine samples from two different regions (8 samples from Scotland and 6 samples from England) and 4 ovine samples (from Scotland). All samples from livestock were of genotype 2, and all yielded HMA type CPV2-1 (Table 5).
Sporadic cases.
A group of 103 samples from sporadic cases diagnosed in humans in England in 1999 were tested with the RT-PCR-HMA procedure. Of these, 28 were of genotype 1 and 75 were of genotype 2. Eight and 10 different HMA types were identified for genotypes 1 and 2, respectively (Tables 4 and 5). HMA types CPV1-2 (17 samples out of 28) in the genotype 1 group and CPV2-1 (44 samples out of 75) in the genotype 2 group were the most common (Tables 4 and 5). Since some of the intrafamilial cases were involved with waterborne (drinking water) outbreaks, a single representative of each family was added to the totals in this group; with these totals, HMA types CPV1-2 (χ2 = 15.2; P < 0.0001) and CPV2-1 (χ2 = 3.98; P = 0.046) were significantly less common in samples collected from sporadic cases than in those collected from waterborne outbreaks.
Since there is a marked seasonality in the distribution of C. parvum genotypes among sporadic cases in England (22), the distributions of HMA types in the samples collected in 1999 were compared for the spring-summer peak (April to June) and the autumn peak (October to December). Among the samples from the spring peak, 1 sample was of genotype 1 (and was identified as HMA type CPV1-8) and 52 samples were of genotype 2; of the latter, 35 were HMA type CPV2-1, 13 were HMA type CPV2-2, and the remaining 4 were HMA types CPV2-8, CPV2-9, CPV2-10, and CPV2-11. Among 50 samples from the autumn peak, 27 were of genotype 1 and 23 were of genotype 2. During this autumn period, among the 27 samples of genotype 1, 17 were HMA type CPV1-2 and the remaining 10 were HMA types CPV1-1, CPV1-3, CPV1-4, CPV1-5, CPV1-6, and CPV1-9. For the 23 samples of genotype 2, 8 (35%) and 7 (32%) were HMA types CPV2-1 and CPV2-2, respectively, and the remaining 8 samples were HMA types CPV2-3, CPV2-4, CPV2-6, CPV2-9, CPV2-12, CPV2-13, and CPV2-5. HMA type CPV2-1 (which was the only type recovered from livestock) was significantly more common among the sporadic cases due to genotype 2 in the spring peak than in the autumn peak (χ2 = 5.22; P = 0.022). HMA types CPV2-1, CPV2-2, and CPV2-9 were present in samples from both the spring and the autumn peaks.
Among all 103 sporadic cases analyzed, 3 were associated with recent foreign travel (1 case due to genotype 2 was associated with travel to Spain, and 2 cases due to genotype 1 were associated with travel to Sudan and Pakistan). The genotype 2 case was HMA type CPV2-1; the two genotype 1 cases were HMA types CPV1-6 and CPV1-8, respectively, which were not detected in any other sample.
DISCUSSION
Previous studies showed the potential for characterization of the dsRNA elements associated with Cryptosporidium as a subgenotyping tool for molecular epidemiological investigations (50). These elements have been detected in C. parvum human and calf isolates in the United States and Australia (19), and the report here of their presence in England confirms their ubiquitous distribution. Analysis of a 173-bp fragment of the small dsRNA element from humans and calves in the United States and humans in Peru and Kenya revealed sequence variations within and between genotypes, although it was not possible to separate isolates into two groups corresponding to the two genotypes (50). Our analysis of human and livestock isolates from England (Fig. 5) by use of the same DNA fragment is consistent with the results reported by others (50) in that samples from England showed variations in the dsRNA fragment which were independent of the genotype. Polymorphisms within a fragment of the 60-kDa glycoprotein (GP60) gene were also independent of C. parvum genotypes (35, 42).
We previously developed an HMA method for rapid characterization of the 173-bp fragment of the small dsRNA element of C. parvum, and a comparison of amplicons with a single artificial reference DNA enabled the identification of nine different types within 11 distinct sequences (21). In this study, we extended the HMA technique by the application of two reference DNAs: either the artificial reference sequence as previously reported (21) or, in a separate reaction, a naturally divergent sequence from C. meleagridis as a second reference preparation.
For the evaluation of the usefulness of epidemiological typing systems, it is important to establish the typeability, discrimination, reproducibility, and ease of application of a technique. We report here the evaluation of an HMA analysis of samples collected in England from sporadic cases, waterborne and intrafamilial outbreaks, and cases of natural infection in livestock. The overall typeability (proportion of samples that could be analyzed by this technique) was 68%. The method described in this study was carried out with total nucleic acid extracted from whole feces, whereas in a previous study, purified oocysts or oocyst-containing fecal material was used and no data about the percentage of success of the amplification were reported (50). A possible reason for the failure of amplification of the dsRNA element from some samples is that the extraction procedure was not suitable for RNA. However, this possibility is unlikely, since the method was originally designed for RNA viral targets (5). A second reason for the absence of amplification may have been degradation of the target prior to extraction. All fecal samples were stored at 4°C on receipt in the laboratory, but some samples (especially those from animals) may have been kept at room temperature for considerable periods prior to dispatch. We are currently investigating field samples collected from livestock animals with a better collection history. Further reasons for the failure of amplification are possible degradation of the target between extraction and random priming for the RT reaction. The RT reaction was performed up to 6 years after the initial nucleic acid extraction, and degradation of the dsRNA during storage might have occurred; this problem might have been especially true for the samples from livestock, which were almost all extracted prior to the samples from humans. A further explanation for the failure of amplification reported here may be the overall sensitivity of the assay. dsRNA elements are estimated to be present at ∼5,1000 molecules of each segment per oocyst (20). Hence, this assay might be expected to be a more sensitive characterization method than, for example, an assay of the COWP gene, which is present at a single copy per genome (four copies per oocyst). However, no formal comparison of the sensitivities of the methods was performed but will be performed in future studies. Differences observed in the ability of the RT-PCR to amplify the 173-bp fragment between genotypes 1 and 2, between samples from sporadic cases and outbreaks, and between humans and livestock may be due to as-yet-uncharacterized variability, e.g., in the number of copies of the dsRNA element or in the sequence at the primer binding regions (data not shown). For the C. meleagridis isolate, one mismatch was identified at both primer annealing regions (data not shown); therefore, variability in target sequences may explain the failure of amplification. We are currently engaged in an ongoing evaluation of the use of specific primers for sequencing of the small dsRNA element in the RT reaction as a more sensitive alternative to the random priming method as well as in an evaluation of different RTs.
Amplicon characterization with two reference DNA preparations enabled the discrimination of all 11 distinct sequences from the panel used in the previous study (21). Further analysis with HMA allowed the identification of 21 HMA types in all 27 different sequences identified in all of the samples analyzed here containing C. parvum (8 in the genotype 1 group, 12 in the genotype 2 group, and a pattern present in both genotypes). Four HMA types were obtained from different sequences identified in samples containing C. felis, and one HMA type was identified in a sample containing C. meleagridis. A limitation of our HMA system is that it did not discriminate small differences (one or two bases) within three groups of sequences characterized as HMA types CPV2-1, CPV2-2, and CPV2-5. An alternative here would be to use another reference DNA to identify these groups, although with the two reference preparations described here, all sequences with divergence outside the major groups were identified. As illustrated by the results of a sequence analysis of 26 samples of HMA type CPV1-2, confirming them all to represent a homogeneous group, the HMA method developed here was a quick and reliable tool which provided an alternative to sequencing.
The HMA system with the two reference DNA preparations was proven to be reproducible, although identical patterns were seen in both the original sample and the cloned material in one instance, except that an additional band was generated from the original fecal sample with one of the reference preparations. A possible explanation for this result is that a heterogeneous mixture of dsRNA sequences was present in the original sample, possibly reflecting a mixed infection with different C. parvum genotype 2 isolates, which would not be identified by a genotyping system based on the conventional analysis of the COWP gene. Mixtures of genotypes 1 and 2 have been reported elsewhere (22). Subsequent experiments identified a second potential mixture within genotype 2 in which a sample yielded HMA type CPV2-11, but the profile generated from the original PCR amplicon had an extra band with one of the reference DNA preparations compared to the profile obtained with the cloned material. Further investigations about the usefulness of this system in identifying mixed infections are ongoing.
dsRNA elements were described by Khramtsov et al. (19) for genotype 1 and 2 isolates of C. parvum but were not detected in other members of the genus (i.e., Cryptosporidium serpentis, Cryptosporidium baileyi, C. muris, and C. meleagridis). We present here the first report of data concerning the amplification of a 173-bp fragment of the small dsRNA element from C. felis (four samples) and C. meleagridis (one sample). The four C. felis isolates all possessed different sequences, and these clustered together in a single group which differed from all of the C. parvum and C. meleagridis sequences. These data are consistent with a phylogenetic analysis of C. felis at other loci (18S rDNA and HSP70) in that they represent a logical grouping which justifies separate species status (31, 43, 51). The sequence diversity of the C. felis dsRNA elements, however, indicates, for the first time, that C. felis infections in humans are due to a heterogeneous group of strains, albeit that these constitute a small proportion of the total identified Cryptosporidium strains associated with human disease. The dsRNA element allowed the separation of C. meleagridis from C. parvum and C. felis. These data are consistent with a phylogenetic analysis at other loci (18S rDNA and HSP70) which justifies separate species status (32, 43, 51). A subsequent sequence analysis of the C. meleagridis dsRNA element showed a single base mismatch at both primer annealing regions (data not shown), and the variability at these primer binding regions may explain the failure of amplification for one other C. meleagridis isolate previously studied (19) and for two other additional samples tested in this study.
This study identified two predominant C. parvum HMA types in samples collected in England: one associated with C. parvum genotype 1 (HMA type CPV1-2) and one associated with C. parvum genotype 2 (HMA type 2-1). HMA type CPV1-2 was identical to sequence type E (50), previously described for a human sample in the United States. This subgenotype was detected in 64 of 65 and in all 16 samples of genotype 1 analyzed from three waterborne outbreaks and intrafamilial outbreaks, respectively; this result suggests that this subgenotype plays a major role in C. parvum genotype 1-associated outbreaks. Glaberman et al. (15) observed similar results for a subgenotyping analysis with the GP60 gene of C. parvum genotype 1 from samples collected in Northern Ireland in that all of the cases in two waterborne outbreaks were of a single subgenotype within genotype 1. An analysis of sequences revealed clustering of a more divergent group of C. parvum isolates (Fig. 5) comprising HMA types CPV1-3, CPC1-5, CPV1-6, and CPV1-8 from this study together with subgenotypes P, Q, and R (50). Of these, subgenotypes Q and R were previously identified in isolates collected from Kenya, and data obtained here showed that patients associated with HMA types CPV1-6 and CPV1-8 had a recent history of foreign travel to Sudan and Pakistan. Our results are consistent with those of Xiao et al. (50) and Feng et al. (13) in that isolates collected from particular geographic areas can have unique subgenotypes not found in other areas.
The predominant HMA type among C. parvum genotype 2 isolates collected from both humans and livestock in the United Kingdom was produced by a sequence identical to that of isolate C1 (GenBank accession number AF169934) and sequence type G (50), previously detected in calves in the United States. Livestock samples tested here were collected from different areas in the United Kingdom and were all of this HMA type. These findings are different from those obtained from calves in the United States, where seven subgenotypes (A, B, D, F, G, H, and M) with differences of between 1 and 5 bp were detected (50), revealing greater variability in livestock infections due to C. parvum genotype 2. An HMA analysis of two waterborne outbreaks associated with genotype 2 detected the presence of HMA type CPV2-1 in 84% of the samples from waterborne outbreaks 4 and 5. These findings are consistent with field epidemiological observations that suggested zoonotic transmission for these outbreaks, where water supplies were contaminated by sheep feces (1, 2, 22). An HMA analysis of samples collected from intrafamilial outbreaks and sporadic cases and a smaller proportion of samples collected from waterborne outbreak 5, all caused by C. parvum genotype 2, revealed the presence of other patterns not detected in samples from livestock. These findings highlight the complexity of transmission cycles for C. parvum genotype 2 and the possibility of human-to-human transmission for some subgenotypes. The greater variability found here with this system for the sporadic cases due to C. parvum genotype 2 in England is similar to the subgenotyping results obtained by Glaberman et al. (15) for sporadic cases in Northern Ireland; in their analysis with the GP60 gene, nine different subgenotypes were described for 14 human clinical samples, while all samples from a waterborne outbreak due to genotype 2 belonged to a predominant subgenotype. The lack of variation at the dsRNA and GP60 loci in C. parvum of both genotype 1 and genotype 2 for samples from the United Kingdom, which is not reflected in the greater variation seen with sporadic cases, is intriguing, especially since neither of these subgenotypes was recognized by the 173-bp dsRNA element in 4 samples from a waterborne outbreak and 13 samples from two food-borne outbreaks investigated in the United States (50). Human volunteer feeding experiments showed that variations in infective doses occur among isolates (29). These subgenotyping data may also reflect biological differences in this parasite, such as variations in infectivity, in survival in the environment, or during water processing. Further work is required to fully characterize these differences.
In summary, the HMA analysis based on the characterization of the 173-bp fragment of the small dsRNA element was confirmed to be a useful subgenotyping tool for the analysis of samples from outbreaks and to be a rapid screening method for distinguishing diversity in samples from sporadic cases of cryptosporidiosis. The HMA method enabled the rapid identification of “unusual” types, which were confirmed by sequencing, and also rapidly provided information on the presence of more common types. The HMA method was proven to be reproducible, easy to perform, applicable to large volumes of samples, and useful for typing Cryptosporidium in epidemiological investigations and provides further insight into the complexity of the transmission cycles of this parasite.
Acknowledgments
We thank clinical and veterinary colleagues for donation of specimens. We also thank D. Brown (Enteric, Respiratory and Neurological Virus Laboratory, Public Health Laboratory Service [PHLS]), F. J. Bolton (Food Safety Microbiology Laboratory, PHLS), and N. Andrews (PHLS Statistics Unit) for helpful discussions.
REFERENCES
- 1.Anonymous. 1999. Surveillance of waterborne disease and water quality. January to June 1999, and summary 1998. Public Health Lab. Serv. Commun. Dis. Rep. 9:305-308. [Google Scholar]
- 2.Anonymous. 1999. Surveillance of waterborne disease and water quality: January to June 1998. Public Health Lab. Serv. Commun. Dis. Rep. 9:73-74. [PubMed] [Google Scholar]
- 3.Anonymous. 2000. Surveillance of waterborne disease and water quality: July to December 1999. Public Health Lab. Serv. Commun. Dis. Rep. 10:65-68. [Google Scholar]
- 4.Arrowood, M. 1997. Diagnosis, p. 43-64. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 5.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. Van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouchier, I. 1998. Cryptosporidium in water supplies: third report of the group of experts. Her Majesty 's Stationery Office, London, United Kingdom.
- 7.Cacció, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed] [Google Scholar]
- 8.Cacció, S., W. Homan, K. van Dijk, and E. Pozio. 1999. Genetic polymorphism at the beta-tubulin locus among human and animal isolates of Cryptosporidium parvum. FEMS Microbiol. Lett. 170:173-179. [DOI] [PubMed] [Google Scholar]
- 9.Cacció, S., F. Spano, and E. Pozio. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082-1086. [DOI] [PubMed] [Google Scholar]
- 10.Delwart, E. L., E. G. Shpaer, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heterduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 11.Fayer, R. 1997. The general biology of Cryptosporidium, p. 1-41. In R. Fayer (ed.), Cryptosporidium and cryptosporidiosis. CRC Press, Inc., Boca Raton, Fla.
- 12.Fayer, R., U. Morgan, and S. J. Upton. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol. 30:1305-1322. [DOI] [PubMed] [Google Scholar]
- 13.Feng, X., S. M. Rich, D. Akiyoshi, J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, S. Tzipori, and G. Widmer. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatei, W., R. W. Ashford, N. J. Beeching, S. K. Kamwati, J. Greensill, and C. A. Hart. 2002. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 8:204-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glaberman, S., J. E. Moore, C. J. Lowery, R. M. Chalmers, I. Sulaiman, K. Elwin, P. J. Rooney, B. C. Millar, J. S. Dooley, A. A. Lal, and L. Xiao. 2002. Three drinking-water-associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 18.Johnson, D. W., N. J. Pieniazek, D. W. Griffin, L. Misener, and J. B. Rose. 1995. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl. Environ. Microbiol. 61:3849-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khramtsov, N. V., P. A. Chung, C. C. Dykstra, J. K. Griffiths, U. M. Morgan, M. J. Arrowood, and S. J. Upton. 2000. Presence of double-stranded RNAs in human and calf isolates of Cryptosporidium parvum. J. Parasitol. 86:275-282. [DOI] [PubMed] [Google Scholar]
- 20.Khramtsov, N. V., K. M. Woods, M. V. Nesterenko, C. C. Dykstra, and S. J. Upton. 1997. Virus-like, double-stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol. Microbiol. 26:289-300. [DOI] [PubMed] [Google Scholar]
- 21.Leoni, F., C. I. Gallimore, J. Green, and J. McLauchlin. A rapid method for identifying diversity within PCR amplicons using a heteroduplex mobility assay and synthetic polynucleotides: application to characterisation of dsRNA elements associated with Cryptosporidium. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 22.McLauchlin, J., C. Amar, S. Pedraza-Díaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLauchlin, J., D. P. Casemore, S. Moran, and S. Patel. 1998. The epidemiology of cryptosporidiosis: application of experimental sub-typing and antibody detection systems to the investigation of water-borne outbreaks. Folia Parasitol. (Prague) 45:83-92. [PubMed] [Google Scholar]
- 24.McLauchlin, J., S. Pedraza-Diaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinhardt, P. L., D. P. Casemore, and K. B. Miller. 1996. Epidemiologic aspects of human cryptosporidiosis and the role of waterborne transmission. Epidemiol. Rev. 18:118-136. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan, U. M., C. C. Constantine, D. A. Forbes, and R. C. Thompson. 1997. Differentiation between human and animal isolates of Cryptosporidium parvum using rDNA sequencing and direct PCR analysis. J. Parasitol. 83:825-830. [PubMed] [Google Scholar]
- 28.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 29.Okhuysen, P. C., C. L. Chappell, J. H. Crabb, C. R. Sterling, and H. L. DuPont. 1999. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J. Infect. Dis. 180:1275-1281. [DOI] [PubMed] [Google Scholar]
- 30.Ong, C. S., D. L. Eisler, A. Alikhani, V. W. Fung, J. Tomblin, W. R. Bowie, and J. L. Isaac-Renton. 2002. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg. Infect. Dis. 8:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedraza-Díaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium ‘dog type’ from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 32.Pedraza-Díaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 33.Pedraza-Díaz, S., C. Amar, G. L. Nichols, and J. McLauchlin. 2001. Nested polymerase chain reaction for amplification of the Cryptosporidium oocyst wall protein gene. Emerg. Infect. Dis. 7:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pedraza-Díaz, S., C. F. Amar, J. McLauchlin, G. L. Nichols, K. M. Cotton, P. Godwin, A. M. Iversen, L. Milne, J. R. Mulla, K. Nye, H. Panigrahl, S. R. Venn, R. Wiggins, M. Williams, and E. R. Youngs. 2001. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J. Infect. 42:243-250. [DOI] [PubMed] [Google Scholar]
- 35.Peng, M. M., O. Matos, W. Gatei, P. Das, M. Stantic-Pavlinic, C. Bern, I. M. Sulaiman, S. Glaberman, A. A. Lal, and L. Xiao. 2001. A comparison of Cryptosporidium subgenotypes from several geographic regions. J. Eukaryot. Microbiol. (2001 Suppl.):28S-31S. [DOI] [PubMed]
- 36.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1998. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed] [Google Scholar]
- 40.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 41.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulaiman, I. M., A. A. Lal, and L. Xiao. 2001. A population genetic study of the Cryptosporidium parvum human genotype parasites. J. Eukaryot. Microbiol. (2001 Suppl.):24S-27S. [DOI] [PubMed]
- 43.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiangtip, R., and S. Jongwutiwes. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health 7:357-364. [DOI] [PubMed] [Google Scholar]
- 46.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 47.Willocks, L., A. Crampin, L. Milne, C. Seng, M. Susman, R. Gair, M. Moulsdale, S. Shafi, R. Wall, R. Wiggins, N. Lightfoot, et al. 1998. A large outbreak of cryptosporidiosis associated with a public water supply from a deep chalk borehole. Commun. Dis. Public Health 1:239-243. [PubMed] [Google Scholar]
- 48.Xiao, L., C. Bern, M. Arrowood, I. Sulaiman, L. Zhou, V. Kawai, A. Vivar, A. A. Lal, and R. H. Gilman. 2002. Identification of the Cryptosporidium pig genotype in a human patient. J. Infect. Dis. 185:1846-1848. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, L., J. Limor, C. Bern, and A. A. Lal. 2001. Tracking Cryptosporidium parvum by sequence analysis of small double-stranded RNA. Emerg. Infect. Dis. 7:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]