Abstract
The phenotypes and genetic determinants for macrolide resistance were determined for 167 erythromycin-resistant Streptococcus pyogenes strains. A cMLS phenotype was shown in 18% of the erythromycin-resistant strains, while inducible resistance was apparent in 31% and the M phenotype was apparent in 50%. The emm gene type of this set of resistant isolates and that of 48 erythromycin-sensitive isolates were determined. emm2 and emm48 were recorded only in the resistant strains of the M phenotype, while approximately all of the strains harboring the emm22 gene had the cMLS phenotype. More than 80% of the emm89-positive strains had the iMLS phenotype, and the same portion of emm4 strains presented the M phenotype. emm3 is recorded only among sensitive strains. The distribution of frequencies of the genetic determinant for the virulence factor M protein was significantly different both among organisms of different types of resistance and between resistant and sensitive populations of S. pyogenes under study.
Group A Streptococcus pyogenes (GAS) is a gram-positive pathogen that causes many infections (pharyngitis, septicemia, toxic shock, and necrotizing fascitis) and postinfectious sequelae (rheumatic heart disease and glomerulonephritis). Humans are the natural host and the sole reservoir of GAS. The organism can survive and replicate in diverse anatomic sites and is provided, like other human pathogenic bacteria, with very smart offensive and defensive molecular weapons. Among them are the virulence factors. The M protein is one of these factors and is surface exposed by means of its amino termini. It has provided the basis for a widely used serological typing scheme (19, 20) and is one of the best-studied proteins in bacteria (13). More recently, a genotypic typing scheme based on the emm genes that encode M and M-like proteins has become widely used, and more than 150 different emm alleles have been characterized (1, 10; http://www.cdc.gov/ncidod/biotech/strep/emmtypes.htm). The antigenic heterogeneity exhibited by this family of genes (and related proteins) reflects the strong impact of host immunity on the generation of diversity within this bacterial species. Numerous genotypic methods other than emm sequence typing have been developed for the genotyping of GAS (9, 12, 14, 21, 24, 26, 31), enabling the determination, to various degrees, of the phylogenetic relationships between isolates and therefore of the level of clonal relatedness in a bacterial population. In this context, it was clearly demonstrated that the emm type appears to correlate closely with clone or clonal complex (9). The present study addressed the possible correlation between emm type (i.e., clonal relatedness) and bacterial resistance to antibiotics. In particular, we subjected to analysis 215 clinical isolates of S. pyogenes, isolated in 1997 from the sore throat of patients with pharyngitis and recovered throughout Italy. Of these, 167 were erythromycin-resistant on the basis of a conventional MIC breakpoint (for resistance, MIC ≥ 1 μg/ml) (23), as determined by using standard methods (22); a set of 48 sensitive strains (MIC ≤ 0.25 mg/liter) has been randomly selected as a comparative control.
On the basis of the triple-disk (erythromycin plus clindamycin and josamycin) assay (15), by which the constitutive resistance phenotype (cMLS), the inducible-resistance phenotypes iMLS-A, iMLS-B, and iMLS-C, and the M phenotype could be discriminated (15), it was shown that the population of erythromycin-resistant isolates under study was strongly characterized (>50%) by the occurrence of the M phenotype, due to the presence of an efflux system. The remaining portion of the population of resistant test strains consists of constitutively resistant (18.1%) and of inducibly resistant (30.7%) isolates. In the latter group, the dominant subpopulation is that of the iMLS-A phenotype, which accounts for 23% of the whole population of resistant isolates. The remaining 8% of iMLS strains is almost equally divided between iMLS-B and iMLS-C phenotypic patterns. This distribution of frequencies is in good agreement with that found in wider Italian population studies (3, 32; G. C. Schito, E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. Tempera, and P. E. Varaldo, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1211, 1999), with the only exception being the proportion represented by the M phenotype strains, which is close to 10% higher. Erythromycin-resistant S. pyogenes isolates were then subjected to PCR analysis to detect erythromycin resistance genetic determinants.
All cMLS isolates harbor the gene coding for the ermB methylase, as does the subfamily of inducible iMLS-A strains. The ermB gene was detected by using the set of primers and the PCR conditions reported by Sutcliffe et al. (30). The ermTR methylase gene was detected in iMLS-B and iMLS-C inducible isolates by using primer TR1 (18) together with a second primer, TR3 (5′-GCTTCAGCACCTGTCTTAATTGAT-3′), designed on the basis of the published sequence (29). The PCR mixture was as described by Seppala et al. (29), and the PCR conditions have been optimized as follows: denaturation at 94°C for 45 s, annealing at 53°C for 45 s, and elongation at 72°C for 45 s for a total of 35 cycles. Amplification of the DNA produced a PCR fragment 550 bp in length. One iMLS-B isolate possesses both ermTR and mef(A) genes; the latter expresses the efflux pump responsible for the transport of erythromycin out of the cell and determines the M phenotypic pattern of resistance. The copresence of both genes has already been described for both S. pyogenes (5; E. Giovanetti, M. P. Montanari, M. Mingoia, S. Bompadre, M. Prenna, S. Ripa, and P. E. Varaldo, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1210, 1999) and S. pneumoniae (11). The mef(A) gene was detected with the primer pair and PCR conditions reported by Clancy et al. (6) and was present in all resistant strains expressing the M phenotype.
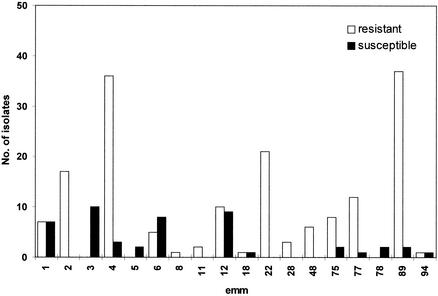
All strains were subjected to PCR M typing to determine specific emm gene types and therefore to infer the corresponding M serotype of the isolate under study. The experiments were basically performed by using the procedure, the emm-specific oligonucleotides (Table 1), and the control tests described by Vitali et al. (33). Subsequently, the PCR M typing on the emm-untypeable strains was performed by using an extra set of 11 emm-specific primers (see Table 1) chosen according to two main criteria: (i) emm11, -22, -28, -29, -77, -87, and -89 were chosen on the basis of their frequency in the Italian GAS population (9), and (ii) the others (emm48, -75, -78, and -94) were chosen on the basis of a sequencing screening (1) conducted on strains randomly sampled from the isolates that could not be typed with the previously used set of primers (33). Detected emm genes belonged to the emm gene families and had the corresponding frequencies listed in Table 2. The most represented emm types in the group of resistant isolates are emm2 (10.2%), emm4 (21.6%), emm12 (6%), emm22 (12.6%), emm77 (7.2%), and emm89 (22.2%). On the other hand, PCR M typing analysis conducted on the susceptible strains revealed an altogether different distribution. In particular, the emm3, emm5, and emm78 types are present, which are missing in the resistant population. In addition, the relative frequencies of emm4 and emm89 in the susceptible isolates group are not as high as those found in the resistant-isolate group (Fig. 1).
TABLE 1.
Oligonucleotide probes used in the present study
| Probe | Nucleotide sequencea | Reference or source |
|---|---|---|
| emm1 | 5′ TTC TAT AAC TTC CCT AGG ATT ACC ATC ACC 3′ | 34 |
| emm2 | 5′ TGC TTC TTT TTT GAC AGG GAC AGG GTT CTT 3′ | 34 |
| emm3 | 5′ CAT GTC TAG GAA ACT CTC CAT TAA CAC TCC 3′ | 34 |
| emm4 | 5′ CCA CGC TGA ATC AGC CTG AGG CTT TTT AAT 3′ | 34 |
| emm5 | 5′ CGG GTC ATT TAT TGT ACC CCT AGT CAC GGC 3′ | 34 |
| emm6 | 5′ TGC TTT GTC CGG GTT TTC TAC CGT CCC CCT 3′ | 34 |
| emm8 | 5′ TCG TTA TTA GAA ATA CTA TGA GAT TTT GGG 3′ | 34 |
| emm11 | 5′ CGC TCA CGT TTG TAC CTT TAG GAG CGC TTT 3′ | This study |
| emm12 | 5′ ACG TTG TTT TTC TGC GAC TAA ATC ACT ATG 3′ | 34 |
| emm18 | 5′ CGT CTT TAT TGT CTG CTG TAG CTC GAG TAA 3′ | 34 |
| emm22 | 5′ CTT GAG AAA TGT TTG ATG ACT CCG CAT TAT 3′ | This study |
| emm24 | 5′ TTC TTG TAC TTT TTC CAG AGT ATC TGT CTG 3′ | 34 |
| emm28 | 5′ CCA TTA GCA GAA GTC TCA GTA CTT TTT GGA 3′ | This study |
| emm29 | 5′ CTA CGT CCT CTT TTG TCA TAC CCC TAG TAA 3′ | This study |
| emm48 | 5′ TGC TTC AGG TGG TAA TTC AGT AAA AGT ACT 3′ | This study |
| emm75 | 5′ GCT TCA TAT GGT AAC TCA GTA AAA GTA CGT 3′ | This study |
| emm77 | 5′ CGG TTA TGT AGT GAT GCA TCT GAA CCT ACA 3′ | This study |
| emm78 | 5′ CTC GTT AGT AAT ACT ACG AGA GTT CTG AGA 3′ | This study |
| emm87 | 5′ GAA GCA GCC AAT TCG TTG GTT ACT TCT CTG 3′ | This study |
| emm89 | 5′ TGA CAG AGA CAG ATC TAT TAA TAT TGT CAC 3′ | This study |
| emm94 | 5′ GTA ATG TGA GTT GCC CAT TAT TTG ATG CTT 3′ | This study |
For emm oligonucleotide probes, the sequences are complementary to the coding strand of the N-terminal region of the corresponding emm gene sequence.
TABLE 2.
Macrolide resistance phenotype and PCR M typing correlation scheme
| PCR M type | Macrolide resistance phenotype (no. of isolates)
|
No. (%) of isolates | |||||
|---|---|---|---|---|---|---|---|
| MLS | iMLS-A | iMLS-B | iMLS-C | M | Susceptible | ||
| emm1 | 3 | 4 | 7 | 14 (6.5) | |||
| emm2 | 17 | 17 (7.9) | |||||
| emm3 | 10 | 10 (4.7) | |||||
| emm4 | 2 | 2 | 1 | 31 | 3 | 39 (18.1) | |
| emm5 | 2 | 2 (0.9) | |||||
| emm6 | 5 | 8 | 13 (6.1) | ||||
| emm8 | 1 | 1 (0.5) | |||||
| emm11 | 2 | 2 (0.9) | |||||
| emm12 | 2 | 8 | 9 | 19 (8.8) | |||
| emm18 | 1 | 1 | 2 (0.9) | ||||
| emm22 | 20 | 1 | 21 (9.8) | ||||
| emm28 | 2 | 1 | 3 (1.4) | ||||
| emm48 | 6 | 6 (2.8) | |||||
| emm75 | 8 | 2 | 10 (4.7) | ||||
| emm77 | 6 | 6 | 1 | 13 (6.1) | |||
| emm78 | 2 | 2 (0.9) | |||||
| emm89 | 2 | 34 | 1 | 2 | 39 (18.1) | ||
| emm94 | 1 | 1 | 2 (0.9) | ||||
| Total no. (%) of isolates | 30 (14) | 39 (18) | 7 (3) | 6 (3) | 85 (40) | 48 (22) | 215 (100) |
FIG. 1.
Number of susceptible and resistant S. pyogenes isolates belonging to different emm types.
The present study was designed to analyze the possible correlation between macrolide resistance (27) and emm gene distribution in Italian GAS isolates. This relationship has never been addressed before, in spite of the fact that virulence and resistance to antibiotics are among the most important factors to be considered when studying a clinically relevant pathogen.
The data presented here are of particular interest if each phenotype (28) or genotype of resistance is related to emm type (Table 2). Fully 66% of cMLS isolates have the emm22 gene, and 87% of the iMLS-A isolates harbor the emm89 gene. Both iMLS-B and iMLS-C phenotypes fit into the emm77 type, with one exception represented by an iMLS-B-emm4 isolate. This isolate, however, shows an extremely important difference with respect to the rest of the iMLS-B-iMLS-C-emm77 group: it is the only one to harbor both ermTR and mefA genetic determinants. By contrast, M phenotype isolates are dispersed in almost all emm type families, with the exception of emm11 and emm77 groups, in which no mefA determined resistance has been recorded. The analysis shows that 100% of emm2 and emm48 isolates (n = 23) and 80% of emm4 and emm75 types (n = 39) fall into the M phenotype subpopulation. A similar correlation was obtained by Brandt et al. (4), who analyzed a limited number of erythromycin-resistant GAS isolates (n = 17) from the region of Aachen, Germany. Homogeneity with respect to resistance phenotype was found in GAS isolates grouped by means of other, more general typing methods (5, 34).
This correlation scheme clearly indicates that antibiotic resistance and the emm gene are associated. In consideration of the fact that emm typing is a good indicator of the clonal complexity of a GAS population, resistance to antibiotic acquisition is (i) nonrandom and (ii) influenced by the genetic background of the cell. This general conclusion might be further extended to the particular case of virulence, since we have analyzed the emm gene, which codes for the M protein. For instance, the M serotype is in good association with many other virulence traits, e.g., opacity factor, T antigen, and other factors belonging to the emm-like gene family (2, 8, 16, 17). Hence, it is possible to formulate the hypothesis that the mode of host invasion and/or colonization is able to positively influence the genetic acquisition of some resistance determinants and to impede the acquisition of some others. Furthermore, this hypothesis seems to be reinforced by the results obtained with sensitive strains: the emm3, emm5, and emm78 (n = 14) families include susceptible isolates only. It seems that these strains are generally not prone to gain macrolide resistance. In this context, it is noteworthy that M3 isolates are characterized as highly invasive and are frequently associated with severe infections (7).
The fact that the environment influences the general biology of a living organism is universally known. Nevertheless, acquisition of foreign DNA by microorganisms is poorly understood if the phenomenon is considered from an ecological point of view. Therefore, at least in S. pyogenes, a specific genetic background, the corresponding phenotype, and the resulting host-parasite interaction could favor acquisition of a particular antibiotic resistance because the microorganism shares a particular environment with bacteria harboring specific transferable resistance genetic determinants (25) and/or because the colonized niches possess especially favorable conditions for competence. Therefore, the general concept in bacterial evolutionary genetics and pathobiology—that horizontal transfer (and recombination) of genes encoding or mediating traits thought to confer adaptive advantage is an important mechanism used by pathogenic microbes to diversify populations and enhance survival—is here extended by the indication that acquisition of specific genetic traits is, to some extent, influenced by the clonal imprinting of the single strain.
Acknowledgments
This work was supported by a grant from the Italian Ministry of the University and Scientific Research (COFIN 2001).
We are grateful to Sheila Beatty for the helpful discussion and the revision of the manuscript.
REFERENCES
- 1.Beall, B., R. Flackman, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beall, B., G. Gherardi, M. Lovgren, R. R. Facklam, B. A. Forwick, and G. J. Tyrrell. 2000. emm and sof gene sequence variation in relation to serological typing of opacity-factor-positive group A streptococci. Microbiology 146:1195-1209. [DOI] [PubMed] [Google Scholar]
- 3.Borzani, M., M. De Luca, and F. Varotto. 1997. A survey of susceptibility to erythromycin amongst Streptococcus pyogenes isolates in Italy. J. Antimicrob. Chemother. 40:457-458. [DOI] [PubMed] [Google Scholar]
- 4.Brandt, C. M., M. Honscha, N. D. Truong, R. Holland, B. Hovener, A. Bryskier, R. Lutticken, and R. R. Reinert. 2001. Macrolide resistance in Streptococcus pyogenes isolates from throat infections in the region of Aachen, Germany. Microb. Drug Resist. 7:165-170. [DOI] [PubMed] [Google Scholar]
- 5.Cascone, C., M. Santagati, S. Noviello, F. Iannelli, S. Esposito, G. Pozzi, and S. Stefani. 2002. Macrolide-resistance genes in clinical isolates of Streptococcus pyogenes. Microb. Drug Resist. 8:129-132. [DOI] [PubMed] [Google Scholar]
- 6.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 7.Cockerill, F. R., III, R. L. Thompson, J. M. Musser, P. M. Schlievert, J. Talbot, K. E. Holley, W. S. Harmsen, D. M. Ilstrup, P. C. Kohner, M. H. Kim, B. Frankfort, J. M. Manahan, J. M. Steckelberg, F. Roberson, W. R. Wilson, et al. 1998. Molecular, serological and clinical features of 16 consecutive cases of invasive streptococcal disease. Clin. Infect. Dis. 26:1448-1458. [DOI] [PubMed] [Google Scholar]
- 8.Dicuonzo, G., G. Gherardi, G. Lorino, S. Angeletti, M. De Cesaris, E. Fiscarelli, D. E. Bessen, and B. Beall. 2001. Group A streptococcal genotypes from pediatric throat isolates in Rome, Italy. J. Clin. Microbiol. 39:1687-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facklam, R., B. Beall, A. Efstratiou, V. Fischetti, D. Johnson, E. Kaplan, P. Kriz, M. Lovgren, D. Martin, B. Schwartz, A. Totolian, D. Bessen, S. Hollingshead, F. Rubin, J. Scott, and G. Tyrrell. 1999. emm typing and validation of provisional M types for group A streptococci. Emerg. Infect. Dis. 5:247-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrell, D. J., I. Morrissey, S. Bakker, and D. Felmingham. 2001. Detection of macrolide resistance mechanisms in Streptococcus pneumoniae and Streptococcus pyogenes using a multiplex rapid cycle PCR with microwell-format probe hybridization. J. Antimicrob. Chemother. 48:541-544. [DOI] [PubMed] [Google Scholar]
- 12.Feil, E. J., and B. J. Spratt. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561-590. [DOI] [PubMed] [Google Scholar]
- 13.Fischetti, V. A. 1989. Streptococcal M protein: molecular design and biological behavior. Clin. Microbiol. Rev. 2:285-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner, D. L., J. Hartas, B. Currie, J. D. Mathews, D. J. Kempt, and K. S. Sriprakash. 1995. Vir typing: a long-PCR typing method for group A streptococci. PCR Methods Appl. 4:288-293. [DOI] [PubMed] [Google Scholar]
- 15.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gooder, H. 1961. Association of a serum opacity reaction with serological type in Streptococcus pyogenes. J. Gen. Microbiol. 25:347-352. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, D. R., and E. L. Kaplan. 1993. A review of the correlation of T-agglutination patterns and M-protein typing and opacity factor production in the identification of group A streptococci. J. Med. Microbiol. 38:311-315. [DOI] [PubMed] [Google Scholar]
- 18.Kataja, J., P. Huovinen, A. Muotiala, J. Vuopio-Varkila, A. Efstratiou, G. Hallas, H. Seppälä, et al. 1998. Clonal spread of group A streptococcus with the new type of erythromycin resistance. J. Infect. Dis. 177:786-789. [DOI] [PubMed] [Google Scholar]
- 19.Lancefield, R. C. 1928. The antigenic complex of Streptococcus hemolyticus. I. Demonstration of the type-specific substance in extracts of Streptococcus hemolyticus. J. Exp. Med. 47:91-103. [DOI] [PMC free article] [PubMed]
- 20.Lancefield, R. C. 1962. Current knowledge of the type specific M antigens of group A streptococci. J. Immunol. 89:307-313. [PubMed] [Google Scholar]
- 21.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement M100-S9. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 23.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 5th ed. NCCLS document M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 24.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:55-61. [DOI] [PubMed] [Google Scholar]
- 25.Santagati, M., F. Iannelli, M. R. Oggioni, S. Stefani, and G. Pozzi. 2000. Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2585-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selander, R. K., D. A. Caugant, H. Ochman, J. M. Musser, M. N. Gilmour, and T. S. Whittam. 1986. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl. Environ. Microbiol. 51:873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seppälä, H., A. Nissinen, H. Järvinen, S. Huovinen, T. Henriksson, E. Herva, S. E. Holm, M. Jahkola, M. L. Katila, T. Klaukka, S. Kontiainen, O. Liimatainen, S. Oinonen, L. Passi-Metsomaa, and P. Huovinen. 1992. Resistance to erythromycin in group A streptococci. N. Engl. J. Med. 326:292-297. [DOI] [PubMed] [Google Scholar]
- 28.Seppälä, H., A. Nissinen, Q. Yu, and P. Huovinen. 1993. Three different phenotypes of erythromycin-resistant Streptococcus pyogenes in Finland. J. Antimicrob. Chemother. 32:885-891. [DOI] [PubMed] [Google Scholar]
- 29.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upton, M., P. E. Carter, G. Orange, and T. H. Pennington. 1996. Genetic heterogeneity of M type 3 group A streptococci causing severe infections in Tayside, Scotland. J. Clin. Microbiol. 34:196-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varaldo, P. E., E. A. Debbia, G. Nicoletti, D. Pavesio, S. Ripa, G. C. Schito, G. Tempera, and the Artemis-Italy Study Group. 1999. Nationwide survey in Italy of treatment of Streptococcus pyogenes pharyngitis in children: influence of macrolide resistance on clinical and microbiological outcomes. Clin. Infect. Dis. 29:869-873. [DOI] [PubMed] [Google Scholar]
- 33.Vitali, L. A., C. Zampaloni, M. Prenna, and S. Ripa. 2002. PCR M typing: a new method for rapid typing of group A streptococci. J. Clin. Microbiol. 40:679-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zampaloni, C., L. A. Vitali, M. Prenna, M. A. Toscano, G. Tempera, and S. Ripa. 2002. Erythromycin resistance in Italian isolates of Streptococcus pyogenes and correlations with pulsed-field gel electrophoresis analysis. Microb. Drug. Resist. 8:39-44. [DOI] [PubMed] [Google Scholar]