Abstract
The colonization by the probiotic Lactobacillus casei subsp. rhamnosus Lcr35 of the gastrointestinal tracts of mice and humans was studied. The mice were orally given 109 CFU of Lcr35 either once or three times at 24-h intervals. A 16S ribosomal nucleic probe used in hybridization assays detected Lcr35 in the feces of mice for up to 3 days after the feeding, at a level of 108 to 109 CFU/g of feces. In the human assay, 12 healthy volunteers were enrolled in a randomized trial and ingested Lcr35 at a dosage of 108 or 1010 or 1012 CFU every day for 7 days. Then, after a 3-week posttreatment period, there was a second intake period similar to the first one. Analysis of fecal samples showed significant increases in the number of lactobacilli during the first intake period, whatever the dose given. The greatest increases were observed in subjects harboring the lowest indigenous population of Lcr35-like bacteria. During the 3-week posttreatment period, the number of CFU slightly decreased over time, and an increase, although not a statistically significant one, was observed during the second test period. These findings suggest that Lcr35 is able to survive within the gastrointestinal tract.
Probiotics have been defined as viable microorganisms that have a beneficial effect on health (3). Oral consumption of probiotics has been associated with the prevention or cure of diverse intestinal disorders such as antibiotic-induced diarrheal disease, viral and bacterial diarrhea, lactose intolerance, and inflammatory bowel diseases (10). Much of the early evidence on the actual health effects of probiotics was anecdotal, but in recent years there has been accumulating evidence from rigorous clinical studies that certain well-characterized strains have real health-promoting properties (7). Many mechanisms by which probiotics may protect the host from intestinal disorders have been proposed, including production of inhibitory substances, blockage of adhesion sites, competition for nutrients, and stimulation of immunity. Whatever the underlying mechanism, in order to produce their health effects, the probiotic microorganisms must be able to survive within the gastrointestinal (GI) tract (5) and persist at high levels in the intestine. The minimum effective dose is not precisely known, but the usual recommended oral administration is in excess of 109 CFU/day. Several in vitro assays have been used to demonstrate the survival of probiotic strains inside the GI tract, including bile and acid resistance assays (1). However, only human trials can provide evidence of the survival of the probiotic strains in vivo and therefore are required as a basis of a credible claim.
Lactobacillus casei subsp. rhamnosus Lcr35 has been successfully exploited commercially as a pharmaceutical product for more than 20 years. Its beneficial effects include treatment and prevention of nonorganic diarrhea. We recently showed in vitro that this strain has probiotic activities such as the ability to adhere to intestinal cells and antibacterial activity against several pathogens (2). The aim of the present study was to determine if Lcr35 is able to survive passage through the GI tract and to evaluate the persistence of the strain after discontinuation of its administration.
Design of a nucleic probe specific for Lcr35.
A specific DNA probe was designed on the basis of the Lcr35 16S ribosomal DNA (rDNA) sequence. A 219-bp fragment internal to the 16S sequence was amplified with primers Lcr1 (5′-ATTTTGAACGAGTGGCGGAC-3′) and Lcr2 (5′-AACCTCTCAGTTCGGCTACG-3′) at a concentration of 0.5 μM each. Taq DNA polymerase and PCR buffer (final concentrations of 20 mM Tris-HCl, 1.5 mM MgCl2, 0.2 mg of bovine serum albumin/ml, and 100 mM KCl [pH 8.0]) and deoxynucleotides were purchased from Boehringer. The amount of Taq DNA polymerase used was 1.0 U in a total reaction volume of 50 μl. A PCR System 2400 apparatus (Perkin-Elmer Cetus) was used for PCR cycling. The genomic DNA from Lcr35 was obtained by an isolation protocol based on the ultrasonic lysis of cells developed by Müller et al. (9). Initial denaturation was carried out at 94°C for 5 min and was followed by 25 amplification cycles (annealing for 30 s at 94°C, hybridization for 60 s at 56°C, and extension at 72°C for 40 s). The specificity of the resulting DNA probe was confirmed by hybridization assay with strains of five different Lactobacillus species (L. rhamnosus A157T, L. casei 103.137T, L. brevis 102.806T, L. acidophilus 76.13T, and L. paracasei subsp paracasei 103.918T) and 10 other bacterial genera, including Listeria, Enterococcus, Streptococcus, Staphylococcus, Bacillus, Clostridium, Escherichia, Klebsiella, Enterobacter, and Pseudomonas. Colony hybridization assays were performed with an [α-32P]dATP-labeled DNA probe with rapid hybridization buffer (Amersham) under the conditions specified by the manufacturer. No aspecific hybridization signal was observed with any of the bacteria tested, and this DNA probe was therefore selected for further experiments.
Detection of Lcr35 in murine fecal samples.
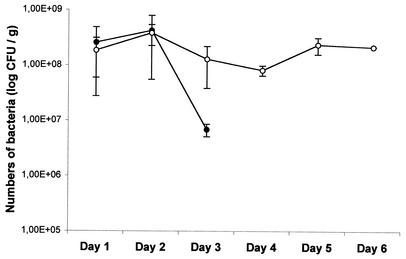
Detection of Lcr35 in feces after oral administration was initially performed in a murine model. Lcr35 was administered (inoculum of 109 CFU) to BALB/c mice in drinking water. Three mice received the inoculum once, and three others received it three times at 24-h intervals. Two other animals were used as controls and did not receive any bacteria. Mouse feces were collected and weighed, and samples were homogenized in saline and diluted 10-fold to 10−8 in MRS broth. Aliquots of the suspensions were plated onto MRS agar plates and incubated for 48 h at 37°C under a CO2 atmosphere. Samples from the colonies were then hybridized with the nucleic probe specific for the 16S rDNA gene of Lcr35. The results were expressed as the numbers of Lcr35 CFU per gram of feces (Fig. 1). Lcr35 was recovered from feces for up to 3 days in the mice inoculated only once, at levels greater than 8 log10 CFU/g for the first 2 days and then lower thereafter (Fig. 1). The counts of Lcr35 in the feces of mice inoculated with three doses were similar to those obtained after only one administration (between 8 log10 and 9 log10 CFU/g) and were detected at the same level even 3 days after termination of feeding (Fig. 1). No Lcr35 or Lcr35-like colonies were detected in the control mice. These results suggest that daily administration is necessary for the maintenance of high probiotic levels in mice.
FIG. 1.
Detection of Lcr35-like lactobacilli in mouse feces after oral administration of 109 CFU once (•) or three times at intervals of 24 h (○).
Human volunteers, Lcr35 administration, and detection of Lcr35 in fecal samples.
A total of 12 healthy volunteers, 5 women and 7 men, with a median age of 23 years (range, 18 to 30 years) were enrolled in a randomized trial. Informed consent from all volunteers was obtained before the experiment, and the protocol was approved by the Regional Health Authority Ethics Committee. No antibiotic had been taken by any volunteer during the 2 weeks before the study, and no antibiotics were administered during the investigation period. No particular diet was required during the test period.
The preparations of Lcr35 were manufactured by the Lyocentre Pharmaceutical Company (Aurillac, France). All aliquots were lyophilized and stored at room temperature until reconstitution and administration in water. Lcr35 was administered at a daily dosage of 108 or 1010 or 1012 CFU for two periods of 7 days with an intervening gap of 3 weeks. The dose administered to each volunteer was randomly chosen.
Fecal samples from volunteers were collected on day 3 before intake (D−3), every day during the 7-day intake period (D0 to D6), and on D7, D8, D10, D12, and D14. On D0, Lcr35 was given after collection of feces, and so results from D0 fecal analysis were included in those for the pretreatment period. A weighted sample of feces was homogenized in saline and processed as described above. The results were expressed as the numbers of CFU per gram of feces, and the statistical evaluation of the significance of the differences in the numbers of bacteria was performed by the Wilcoxon signed-rank test and the Friedman test for matched-pair studies with SPSS, version 10.0, software (12). The results were compared sample by sample and then with a mobile average on a 3-day test.
During the pretreatment period (D−3 and D0), hybridization-positive colonies were detected in the feces of all subjects (median, 4.30 log10 CFU/g; range, 3.25 to 5.74 log10 CFU/g) (Table 1). This indicates that Lcr35-like bacteria, unlike the Lactobacillus species tested during the DNA probe design process, were present in the volunteers' GI tracts. They probably belonged to the L. casei group, which includes bacterial species (L. casei, L. paracasei, L. rhamnosus, and L. zeae) that have highly similar 16S rDNA sequences. No such cross-reaction was detected in the feces of mice collected during the prefeeding period or from the controls, probably due to an absence of members of the L. casei group in these animals' intestinal ecosystems. In humans, these strains are likely to represent part of the Lactobacillus autochthonous flora, i.e., strains of stable and durable residence. Previous studies have shown that some Lactobacillus strains are long-term residents of the intestinal tracts of humans (6, 8). Using ribotyping and pulsed-field electrophoresis, the authors showed that most of the individuals tested harbored a unique collection of lactobacilli (6, 8). They also reported that Lactobacillus numbers vary greatly among subjects and even among samples collected from the same individual (6).
TABLE 1.
Populations of Lcr35-like bacteria in fecal samples
| Subject | First period
|
Second period
|
||||||
|---|---|---|---|---|---|---|---|---|
| Dose of bacteria daily ingested (log10 CFU) | Mean Lactobacillus population (log10 CFU/g) (SD) at:
|
Dose of bacteria daily ingested (log10 CFU) | Mean Lactobacillus population (log10 CFU/g) (SD) at:
|
|||||
| D−3, D0 (control) | D1-D8 (test) | D10, D12, D14 (posteriori) | D32, D35 (control) | D36-D43 (test) | D45, D47, D49 (posteriori) | |||
| 1 | 10 | 5.74 (0.19) | 4.90 (0.56) | 4.25 (1.69) | 8 | 5.02 (1.25) | 5.28 (0.77) | 6.21 (0.18) |
| 2 | 8 | 4.97 (0.58) | 5.27 (0.77) | 6.21 (0.18) | 12 | 5.08 (0.40) | 4.81 (0.76) | 4.70 (0.45) |
| 3 | 12 | 5.50 (0.14) | 4.48 (0.81) | 5.08 (1.25) | 10 | 5.15 (0.21) | 5.39 (0.34) | 4.92 (0.52) |
| 4 | 8 | 4.27 (1.04) | 4.72 (0.77) | 5.35 (0.46) | 12 | 3.80 (2.12) | 4.37 (0.92) | 5.17 (0.67) |
| 5 | 12 | 3.59 (1.14) | 4.34 (1.39) | 4.23 (0.41) | 10 | 4.48 (0.12) | 4.21 (0.68) | 3.79 (0.10) |
| 6 | 10 | 4.42 (0.60) | 4.49 (0.60) | 4.03 (0.53) | 8 | 5.14 (0.06) | 4.93 (0.57) | 5.67 (1.17) |
| 7 | 10 | 3.25 (0.79) | 4.25 (2.12) | 3.82 (1.24) | 8 | 4.78 | 4.22 (0.77) | 5.41 (0.02) |
| 8 | 8 | 3.29 (0.66) | 4.45 (0.92) | 4.36 (0.41) | 12 | 4.68 (1.13) | 5.06 (0.92) | 5.36 (1.44) |
| 9 | 12 | 3.48 (0.50) | 5.00 (1.00) | 4.58 (0.93) | 10 | 4.14 (1.74) | 5.49 (0.95) | 5.40 (0.28) |
| 10 | 8 | 4.21 (2.39) | 4.67 (0.51) | 5.10 (0.53) | 12 | 5.60 (0.48) | 5.24 (0.26) | 6.27 (0.34) |
| 11 | 12 | 4.28 (1.02) | 4.05 (0.90) | 4.40 (0.52) | 10 | 4.57 (0.35) | 5.35 (0.38) | 5.51 (0.15) |
| 12 | 10 | 4.56 (1.60) | 4.05 (0.53) | 5.02 (2.09) | 8 | 4.80 (2.12) | 5.55 (0.66) | 4.73 (0.57) |
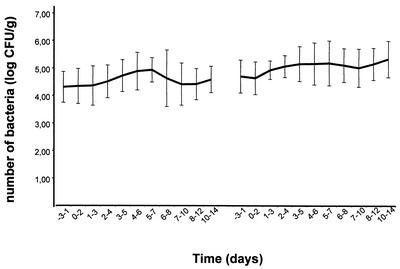
The mobile-average test showed significant increases in the average number of Lcr35-like bacteria in the feces during the intake periods, whatever the dose ingested by the subjects (108 or 1010 or 1012 CFU) (Fig. 2), and there was no relation between the average number of CFU in the feces and the doses ingested by the subjects. During the first treatment period, the mobile-average data indicated 4.30 log10 CFU/g between D−3 and D1 as against 4.92 log10 CFU/g between D3 and D5 (P < 0.01). The highest increase during the treatment period was obtained in subjects harboring the lowest initial colonization levels (<4 log10 CFU/g) (Table 1). In contrast, in individuals harboring the highest colonization levels in the pretreatment period (>5 log10 CFU/g; subjects 1 and 3), the numbers of Lcr35-like bacteria detected slightly decreased during the treatment period. Last, no major variation in individuals (subjects 4, 6, 10, 11, and 12) with initial colonization levels between 4 and 5 log10 CFU/g was observed. In a recent study performed with L. rhamnosus strain DR20, Tannock et al. showed that consumption of this probiotic results in greater frequency of detection of lactobacilli in fecal samples from human volunteers, with major differences among individuals (11). Moreover, the establishment of strain DR20 in the subjects' intestinal microflora was inversely related to the presence of a stable indigenous population of lactobacilli (11). Thus there seem to be great differences in the composition of endogenous Lactobacillus microflora among individuals, and these differences considerably influence the implantation of new Lactobacillus members.
FIG. 2.
Detection of Lcr35-like lactobacilli in the human feces of 12 volunteers during the control, test, and postadministration periods. The numbers of CFU are expressed as the mobile averages of the CFU obtained within a 3-day period, whatever the dose of Lcr35 ingested by the subjects.
During the posttreatment period, the number of CFU detected decreased as a function of time after Lcr35 administration was discontinued (median, 4.8 log10 CFU/g; range, 3.5 to 6.8 log10 CFU/g at day 14, i.e., 7 days after the last intake). After a 3-week period without any intake, hybridization-positive CFU were still detected in the subjects' feces, at levels similar to those observed at the end of period 1. This suggests that the proliferation of Lactobacillus populations induced by the oral absorption of Lcr35 had a prolonged effect on the level of the bacteria within the GI tracts of the volunteers. During test period 2, the number increased, but not to a significant degree, and the levels of CFU per gram of feces remained high (Fig. 2).
Lcr35 has been shown to adhere in vitro to the Caco-2 and Int-407 human intestinal cell lines (2). The finding reported here is that Lcr35 can survive in the GI tracts of humans after oral administration, regardless of the dietary and physiological differences among individuals. As described by Jacobsen et al., Lactobacillus strains with adhesion properties survive passage through the intestinal tract at higher rates than those without adhesion properties (4). The fact that the concentrations of these bacteria were still high after discontinuation of administration indicated that they were able to persist inside the intestine despite rapid turnover and/or to stimulate the proliferation of Lcr35-like lactobacilli. Recent studies performed with L. rhamnosus strain GG showed that this probiotic was able to attach in vivo to colonic mucosae and probably multiplied on the colonic surface at high rates (1). If Lcr35 behaves the same way, its persistence in fecal samples for prolonged periods after discontinuation of administration of the probiotic could be explained.
Acknowledgments
This work was supported by l'Agence Nationale de Valorisation de la Recherche (ANVAR Auvergne) and Lyocentre S.A.
REFERENCES
- 1.Alander, M., R. Satokari, R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forestier, C., C. De Champs, C. Vatoux, and B. Joly. 2001. Probiotic activities of Lactobacillus casei rhamnosus: in vitro adherence to intestinal cells and antimicrobial properties. Res. Microbiol. 152:167-173. [DOI] [PubMed] [Google Scholar]
- 3.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 4.Jacobsen, C. N., V. R. Nielsen, A. E. Hayford, P. L. Møller, K. F. Michaelsen, A. Pœrregaard, B. Sandström, M. Tvede, and M. Jakobsen. 1999. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaila, M., E. Isolauri, M. Saxelin, H. Arvilommi, and T. Vesikari. 1995. Viable versus inactivated Lactobacillus strain GG in acute rotavirus diarrhoea. Arch. Dis. Child. 72:51-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 63:3394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, Y. K., and S. Salminen. 1995. The coming age of probiotics. Trends Food Sci. Technol. 6:241-245. [Google Scholar]
- 8.McCartney, A. L., W. Wenzhi, and G. W. Tannock. 1996. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 62:4608-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Müller, M. R., M. A. Ehrmann, and R. F. Vogel. 2000. Multiplex PCR for the detection of Lactobacillus pontis and two related species in a sourdough fermentation. Appl. Environ. Microbiol. 66:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rolfe, R. D. 2000. The role of probiotic cultures in the control of gastrointestinal health. J. Nutr. 130:396S-402S. [DOI] [PubMed]
- 11.Tannock, G. W., K. Munro, H. J. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolson, R. F. 1987. Comparison of two groups: t-tests and rank tests, p. 145-203. In R. F. Woolson (ed.), Statistical methods for the analysis of biomedical data. John Wiley & Sons, New York, N.Y.