Abstract
Comparative studies on the ABC model of floral development have revealed extensive conservation of B and C class genes, but have failed to identify similar conservation for A class genes. Using a reverse genetic approach, we show that the previous inability to obtain Antirrhinum mutants corresponding to the A class gene AP2 of Arabidopsis reflects greater genetic redundancy in Antirrhinum. Antirrhinum has two genes corresponding to AP2, termed LIP1 and LIP2, both of which need to be inactivated to give a mutant phenotype. Analysis of interactions between LIP and class B/C genes shows that unlike AP2 in Arabidopsis, LIP genes are not required for repression of C in outer whorls of the flower. However, like AP2, LIP genes play a role in sepal, petal and ovule development, although some of their detailed effects are different, reflecting the diverse morphologies of Antirrhinum and Arabidopsis flowers. The dual functions for which AP2 is required in Arabidopsis are therefore separate in Antirrhinum, showing that the genetic basis of some aspects of organ identity have undergone major evolutionary change.
Keywords: ABC model/Antirrhinum/apetala2/Arabidopsis/perianth
Introduction
Studies of parallel mutations in a variety of plant species, most notably Arabidopsis and Antirrhinum, have led to a general model for the control of floral organ identity based on three genetic functions A, B and C (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). The model has been elaborated more recently by the discovery of additional genes that act in combination with the ABC genes (Davies et al., 1996; Egea-Cortines et al., 1999; Pelaz et al., 2000; Honma and Goto, 2001). The ABC model was proposed originally on the basis of mutant phenotypes, raising the question of whether there is also a molecular correspondence between the relevant genes in different species. In the case of B and C class genes, comparative molecular studies have revealed striking conservation between species, although some differences in expression and function have also been observed (Pnueli et al., 1994; Samach et al., 1997; Kramer and Irish, 1999; Ambrose et al., 2000). In contrast, it has been difficult to establish any clear structural and functional relationships for class A genes across species (Egea-Cortines and Davies, 2000). Here we address this issue by using a reverse genetic approach to compare the function of orthologous class A genes from Arabidopsis and Antirrhinum.
Both Arabidopsis and Antirrhinum flowers have a concentric arrangement of four types of organs: sepals in whorl 1, petals in whorl 2, stamens in whorl 3 and carpels in whorl 4. According to the ABC model, the identity of these organs depends on three functions, which act in the combinations A, AB, BC and C in whorls 1–4, respectively (Carpenter and Coen, 1990; Bowman et al., 1991). A key feature of the model is that the A and C functions are antagonistic and thus occupy exclusive domains. The ABC functions also act in combination with those of the SEPALLATA genes to specify organ identity in whorls 2–4 (Pelaz et al., 2000; Honma and Goto, 2001).
Although Arabidopsis and Antirrhinum flowers have a similar pattern of organ types, they differ in the number and detailed morphology of organs. Arabidopsis flowers are organized according to a basic number of four organs per whorl (tetramerous flower) whereas Antirrhinum has a basic number of five organs per whorl (pentamerous flower). In addition, Antirrhinum flowers are formed in the axil of a small leaf-like organ, termed a bract, while Arabidopsis flowers do not have a subtending bract. The morphology of the floral organs, most notably the petals, is also different. In Arabidopsis, the petals are separate and have a relatively simple outline that is similar for all petals within a flower. In contrast, the proximal regions of Antirrhinum petals are united to form a corolla tube (comprising a throat and palate), while the more distal regions comprise lobes which are partially united to form lips. The petals differ in shape and size along the dorsoventral axis of the flower: there are two dorsal petals, two lateral petals and one ventral petal. The more complex morphology of Antirrhinum petals is thought to be an evolutionarily derived condition that arose as a specialization for insect pollination.
The primary class A gene in Arabidopsis is APETALA2 (AP2; Bowman et al., 1991; Meyerowitz et al., 1991). Mutations in AP2 lead to the development of reproductive or leaf-like organs in the outer whorls (perianth), where sepals and petals normally would form. In weak ap2 mutants, such as ap2-1, sepals are replaced by leaf-like organs, and petals by stamenoid organs (Bowman et al., 1989, 1991; Kunst et al., 1989). In strong ap2 mutants, the flower comprises two whorls of carpels with occasional stamens lying between these whorls (Bowman et al., 1991). AP2 is the founding member of a plant-specific transcription factor family and is expressed at a low level in all floral whorls (Jofuku et al., 1994; Okamuro et al., 1997; Riechmann and Meyerowitz, 1998). Molecular and genetic studies have shown that AP2 has two roles in the control of organ identity (Pruitt et al., 1987; Komaki et al., 1988; Kunst et al., 1989; Bowman et al., 1991; Drews et al., 1991). First, AP2 prevents expression of the C class gene AGAMOUS (AG) in whorls 1 and 2, ensuring that C activity and hence reproductive organ identity is restricted to whorls 3 and 4 of the flower. Secondly, AP2 has a role in sepal and petal development that can be separated from its role in repressing C. This is demonstrated by the observation that ap2 mutations still confer a phenotype in the absence of C activity: ap2 ag double mutants have leaf-like organs in whorl 1 and modified petals in whorls 2 and 3 (Bowman et al., 1991).
The A function in Antirrhinum was postulated originally on the basis of a semi-dominant mutation which resulted in reproductive organs growing in place of sepals and petals. This mutation was later shown to be a gain-of-function mutation in the C class gene PLENA (PLE), resulting in its ectopic expression in outer whorls (Bradley et al., 1993). This has raised the question of what genes are involved normally in repressing C and controlling organ identity in outer whorls of Antirrhinum flowers.
Although several genes in Antirrhinum have been shown to be involved in repressing C (McSteen et al., 1998; Motte et al., 1998; Wilkinson et al., 2000), screens so far have not revealed recessive mutants with phenotypes comparable with those of ap2 mutants in Arabidopsis. One possibility is that the screens have not been extensive enough. However, this seems unlikely as mutations in class B and C genes have been recovered multiple times. Another possibility is that AP2-like genes do not play a role in either floral organ identity or repression of C in Antirrhinum. Some support for this has come from the observation that inactivation of an AP2-like gene from Petunia, a near relative of Antirrhinum, had no effect on floral development (Maes et al., 2001). A third possibility is that there is greater genetic redundancy in Antirrhinum so that single mutations in AP2-like genes give no clear mutant phenotype.
To distinguish these possibilities, we have characterized AP2-like genes from Antirrhinum and inactivated them using a reverse genetic approach. We show that there are two likely orthologues of AP2, termed LIP1 and LIP2, that are expressed in floral tissue. Inactivation of either gene alone does not give a mutant phenotype, whereas inactivation of both genes gives bract/leaf-like organs in whorl 1 and reduced petals without lips or palate in whorl 2. This indicates that orthologues of AP2 play a role in development of outer floral whorls of Antirrhinum, and the inability to recover the mutants reflects genetic redundancy. However, unlike the situation in Arabidopsis, inactivation of both LIP genes does not result in ectopic expression of the C class gene PLE in outer whorls, indicating that repression of C is dependent on other genes in Antirrhinum.
Results
Isolation and inactivation of AP2-like genes of Antirrhinum
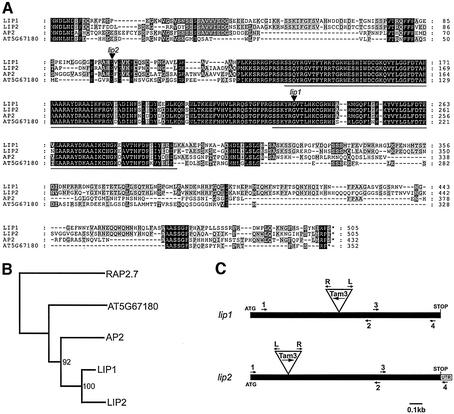
To isolate AP2-like genes from Antirrhinum, an inflorescence cDNA library was probed at low stringency with AP2 from Arabidopsis. This yielded 10 clones, eight of which were of one type, termed LIPLESS1 or LIP1 (for reasons that will become clear later), and two clones of another type, named LIPLESS2 (LIP2). The proteins encoded by LIP1 and LIP2 showed ∼65% identity with AP2 and 72% identity with each other (Figure 1A). To determine whether these genes represented likely orthologues of AP2, the predicted proteins were compared with other members of the AP2 gene family from Arabidopsis using PHYLO_WIN (Galtier et al., 1996). This showed that LIP1 and LIP2 were more closely related to AP2 than to any other genes of the family (Figure 1B). The simplest interpretation is that LIP1 and LIP2 in Antirrhinum arose by duplication of an ancestral AP2-like gene since divergence of the Antirrhinum and Arabidopsis lineages. Independent screens of an Antirrhinum cDNA library also yielded only LIP1 and LIP2 as the closest relatives of AP2 (H.Sommer, personal communication). The putative AP2 orthologue from Petunia, Phap2A, falls within the LIP/AP2 clade, although the LIP genes of Antirrhinum are more closely related to each other than to Phap2A (not shown).
Fig. 1. LIP1 and LIP2 genes. (A) Sequence of LIP genes. The predicted amino acid sequence for the LIP1 and LIP2 products compared with those of AP2 and AT5G67180, another AP2-like gene from Arabidopsis. Amino acid identities are in black, and conservative amino acid changes in grey. Black triangles mark the transposon (Tam3) insertion sites for lip1 and lip2 mutant alleles. The AP2 domains are underlined. (B) Neighbour joining tree showing the phylogenetic relationship, based on amino acid sequence comparisons, between LIP genes and other members of the AP2 gene family. Bootstrap values are shown (500 replicates). (C) Schematic diagram of LIP1 and LIP2. The solid black box represents the coding region. Primers used in PCR screening experiments are shown as numbered arrows. Tam3 is represented as a triangle, with transposon primers, designed to the left (L) and right (R) ends of Tam3, shown as arrows.
To investigate the function of LIP1 and LIP2, the genes were inactivated using a PCR-based screening method. Seed and leaf material was harvested from 27 000 transposon-mutagenized Antirrhinum plants and pooled in batches (for details see Materials and methods). DNA was extracted from the leaves and screened by PCR using transposon-specific primers in combination with primers matching various regions of the LIP genes (Figure 1C). Seeds corresponding to the positives were sown, and individuals carrying the transposon insertions identified by PCR. The genotype of progeny from these individuals was determined by Southern blotting to identify plants homozygous for the insertion.
For LIP1, a Tam3 insertion was obtained within the coding region, after amino acid 233 (Figure 1A). Plants homozygous for this mutation were indistinguishable from wild type. To rule out the possibility that this was because the mutation was leaky, a deletion of LIP1 was obtained. This was achieved by growing plants homozygous for the Tam3 insertion at 15°C, a temperature that increases the activity of this transposon (Harrison and Fincham, 1964; Carpenter et al., 1987). Plants carrying deletions of the gene, caused by imprecise Tam3 excision, were screened for by PCR using primers either side of Tam3. Out of 480 plants screened, two yielded shorter PCR products and contained deletions of 1.3 kb or 900 bp. Plants homozygous for either deletion again were indistinguishable from wild type.
For LIP2, three independent Tam3 insertions were obtained. However, only one was located within the coding region (Figure 1A). No apparent mutant phenotype was observed in plants homozygous for this event.
The lack of a clear mutant phenotype with any of these plants suggested that the functions of LIP1 and LIP2 might be genetically redundant. To test this, plants carrying mutations in both genes were obtained by crossing the LIP1 deletion line with the line carrying Tam3 in the LIP2 coding region. Twelve out of 82 plants in the resulting F2 progeny showed a novel phenotype. Southern blots on the F2 population showed that all plants with the novel phenotype were homozygous for both mutant alleles.
The double mutant phenotype was shown to be the null phenotype by generation of a line carrying deletions in both genes. A deletion of LIP2 was found fortuitously in the progeny of the F2 population as follows. One F3 family, thought to carry deletions in only LIP1, contained plants which showed the double mutant phenotype, and gave no signal on Southern blots when probed with LIP2, indicating that the gene had been deleted. These plants therefore carried deletions in both LIP1 and LIP2. The phenotype was the same as that observed in the original double mutant.
Characterization of the double mutant phenotype
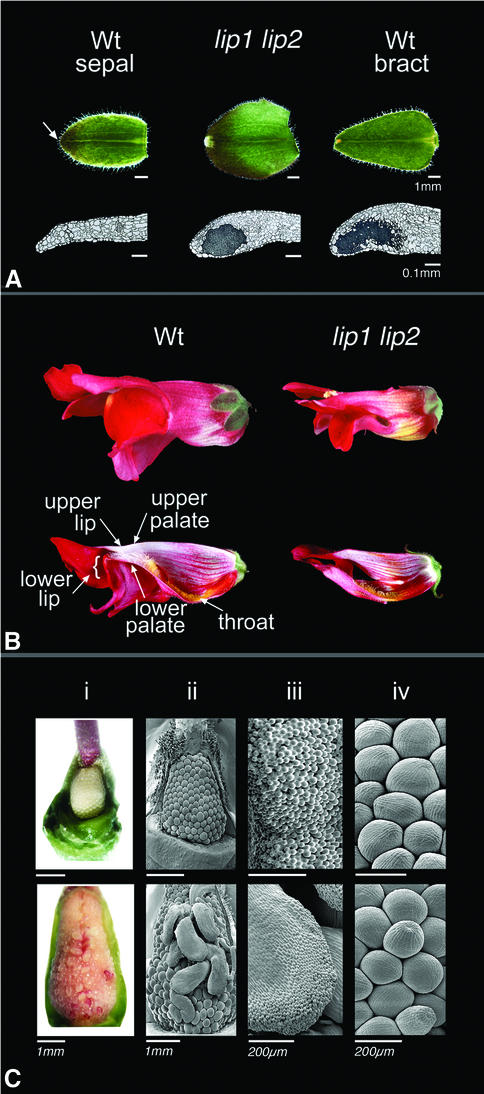
The lip1 lip2 double mutants showed defects in all four whorls of the flower. Starting with the outermost whorl, the mutant organs had about twice the area of wild-type sepals. They also had glandular structures at their tips, resembling those seen in bracts and leaves (Munz, 1926). Histological sections confirmed that these glands had a similar morphology to those of bracts and that they were absent in wild-type sepals (Figure 2A).
Fig. 2. Phenotype of the lip1 lip2 double mutant. (A) Comparison of wild-type sepals, bracts and whorl 1 organs of the lip1 lip2 double mutant. Longitudinal sections of the tips show that lip1 lip2 sepals have glands (dark grey areas) normally found in the tips of bracts and leaves, but not in wild-type sepals. The first whorl organs are also larger in lip1 lip2 than in wild type. The upper scale bar = 1 mm and the lower scale bar = 0.1 mm. The arrow points to tip of wild-type sepal. (B) Lip and palate growth is affected in the lip1 lip2 double mutant. Entire flowers (upper) or flowers that have been cut open longitudinally are shown in side view (stamens and carpels removed from cut flowers). Regions that make up the upper and lower palate and lips, as seen in the wild type, are either missing or very much reduced in lip1 lip2. (C) Carpel development is altered in the lip1 lip2 double mutant. Compared with the wild-type ovary (upper panel), mutant ovules (lower panel) often display style-like outgrowths (i and ii). These are tipped with cells that resemble stigmatic cells of wild type (iii). Not all ovules are transformed into stylar projections, but may still appear abnormal in shape (iv). Scale bars = 1 mm for live tissue in (i). In all cases, the ovary walls have been removed. Scale bars = 1 mm in (ii) and 200 µm in (iii and iv) for SEMs.
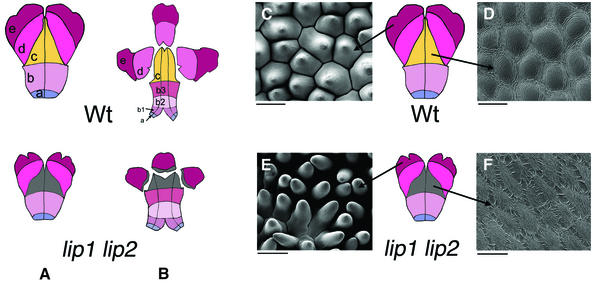
The most striking defect in whorl 2 was the loss or reduction of the upper and lower lips of the petal, hence the name of the genes (Figure 2B). To study the differences between wild type and the double mutant, the petals were flattened and various zones roughly demarcated according to the colour and cell types within them (Figure 3A and B). This showed that the regions forming the palate and lips of wild type (zones c and d) were greatly reduced in the double mutant and replaced by a region with no easily identified cell types, shown as a grey area in Figure 3A and B. Cells in this region were smaller than those of wild type and were surrounded by small ridges on the surface (compare Figure 3D and F).
Fig. 3. Petal domains in wild type and lip1 lip2 mutant. The petal shape is altered in the lip1 lip2 double mutant when compared with wild type. Dorsal (A) and lateral/ventral (B) petals were flattened and divided into several regions according to cell types: zones a and b (b1–b3 for ventral petals) form the throat; zone c forms the upper and lower palate; zone d forms the lips; and zones e and d comprise the petal lobes. In lip1 lip2, zones c and d are very much reduced or absent and are represented as a grey area. Cells in this region, as shown by SEM, appear to be of a morphology different from those in the corresponding region of wild-type petals (D and F). In addition, cells from the distal petal lobes of lip1 lip2 (zone e) are reduced in size and have a changed morphology (C and E). Scale bars = 50 µm.
The distal region of the lobes (zone e) was also reduced in the double mutant. Cells in this region were smaller and appeared narrower than those of the corresponding region in wild type (Figure 3C and E). Cell counts per unit area revealed that the mutant cells were ∼77% of wild type in area, accounting for some, but not all, of the reduction in petal lobe size (the lobes of the double mutant were 35–40% of wild type in area).
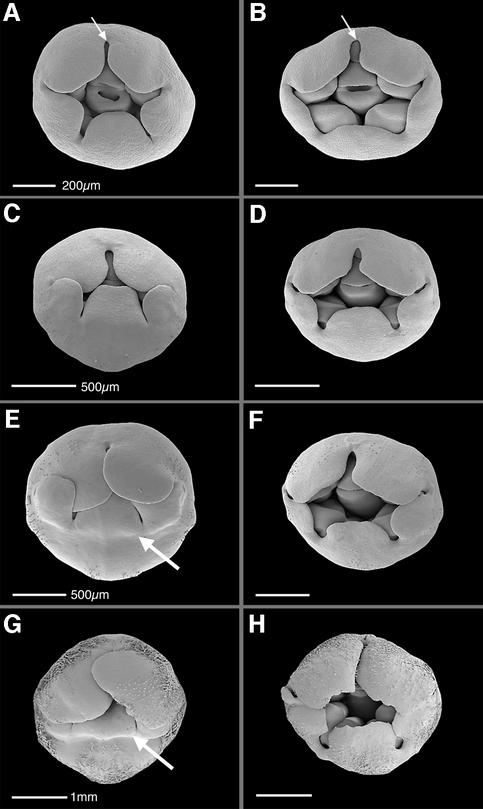
The development of the double mutant was also compared with wild type by scanning electron microscopy (SEM) analysis. Floral meristems arise sequentially on the periphery of the inflorescence apex, and their development can be divided into 15 stages, each stage lasting ∼40 h when the plants are grown at 25°C (Carpenter et al., 1995; C.Vincent and E.Coen, unpublished results). Development of the double mutant was indistinguishable from wild type until stage 7, when the five petal lobes normally cover the developing anthers (Figure 4A). In the double mutant at this stage, the lobes were smaller and only partially covered the anthers (Figure 4B). In addition, the gap between the petal junctions appeared slightly wider than in wild type. These differences became more apparent during stage 8, when the petal lobes normally cover the carpel (Figure 4C and D). Towards the end of stage 8, a furrow developed between the ventral tube and lobes of wild type (Figure 4E, arrow). This furrow is destined to form the lower palate and lips of the flower, but was missing in the double mutant (Figure 4F). This difference was maintained through later stages (Figure 4G and H), consistent with the reduced lip and palate of the mature double mutant flower.
Fig. 4. Floral development in wild type and lip1 lip2. SEMs of wild-type and lip1 lip2 flower buds at various stages of development. Differences in morphology can be detected by stage 7 (A and B), when petals in the lip1 lip2 double mutant are smaller than wild-type petals. Gaps between the petal lobes also appear to be more marked at this stage (arrows). At early stage 8 (C and D), these differences become more apparent. Towards the end of stage 8 (E and F), the furrow destined to become the lower palate and lips (arrowed) is missing in lip1 lip2. This defect is maintained throughout later stages of development (G and H, stage 10). Scale bars in (A) and (B) = 200 µm (C–F) = 500 µm and in (G) and (H) = 1 mm. Sepals have been removed in all cases.
Stamens and carpels were also affected in the double mutant. Stamens were ∼85% of the length of wild-type stamens. In the fourth whorl, the style was ∼65% the length of a wild-type style, but the ovary was about twice the length of wild type. The ovules within the double mutant ovary often displayed outgrowths of style-like tissue tipped with stigmatic cells, suggesting that they had acquired partial carpel identity (Figure 2C). The plants were female sterile, but male fertile.
Expression pattern of LIP genes
RNA in situ hybridizations showed that LIP1 and LIP2 were expressed in a similar pattern. Expression was first seen transiently in young bracts (stage 1) flanking the inflorescence apex (Figure 5A). Expression was next detected at stages 3 and 4 of flower development, in the emerging sepal primordia surrounding the central meristematic dome (Figure 5B). At stage 6, when carpel primoridia had emerged, expression was no longer seen in sepals but was observed in the distal part of the petal primordia (Figure 5C) and in the carpels, although the signal was weak. Weak expression was also sometimes seen in stamens. This pattern of expression was maintained during later stages of development. At stage 10, expression in the ventral petals was concentrated around the furrow destined to form the lower palate and lips (Figure 5D, arrowed), correlating with the region that is reduced in the double mutant.
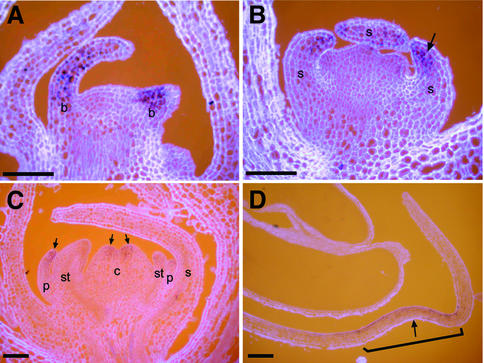
Fig. 5. Expression of LIP genes in wild type. RNA in situ hybridization with a LIP1 probe on wild-type longitudinal sections. (A) Young bracts of an inflorescence apex at stage 1 showing expression of LIP1. Young floral bud at stage 4 (B) and stage 6 (C) showing LIP1 expression in floral tissues (arrows). (D) Ventral petal of a stage 10 floral bud showing localized expression (marked with bracket) of LIP1 within the furrow (arrow) destined to form the lips and lower palate. B, bracts; s, sepal; p, petal; st, stamen; and c, carpel. Scale bar = 100 µm.
Interactions with organ identity genes
Unlike the Arabidopsis ap2 mutant, the lip1 lip2 double mutant showed no evidence of carpel or stamen transformations in the outer two whorls of the flower. This indicated that the LIP genes were not required for repressing the C class gene PLE. However, it was possible that some aspects of the double mutant phenotype were caused by weak ectopic expression of PLE. If this were true, then removing PLE activity should have an effect on the outer whorls of the lip1 lip2 double mutant. To test this, we constructed the lip1 lip2 ple triple mutant. The outer whorls of this mutant resembled those of the lip1 lip2 double mutant (Figures 6A and B, and 2B), indicating that ectopic PLE was not contributing to the phenotype. This was confirmed further by in situ hybridizations on the lip1 lip2 double mutant, which showed that PLE expression was restricted to whorls 3 and 4, as seen in wild type (Figure 7A and B; Bradley et al., 1993). These experiments also demonstrated that expression of PLE in developing ovules was not greatly affected, although a quantitative effect cannot be ruled out.
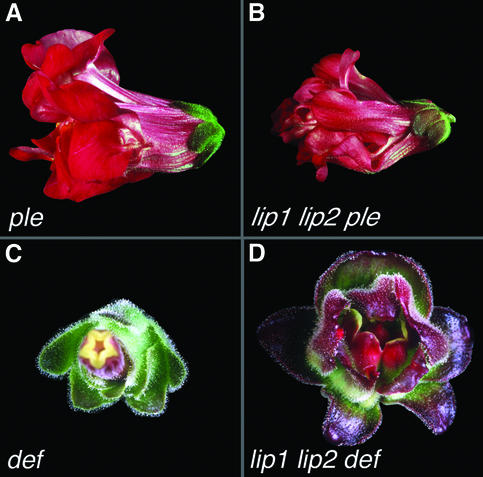
Fig. 6. Interactions with organ identity genes. Flowers of ple (A) and the lip1 lip2 ple triple mutant shown in side view (B). Flowers of def (C) and the lip1 lip2 def triple mutant (D) shown in face view.
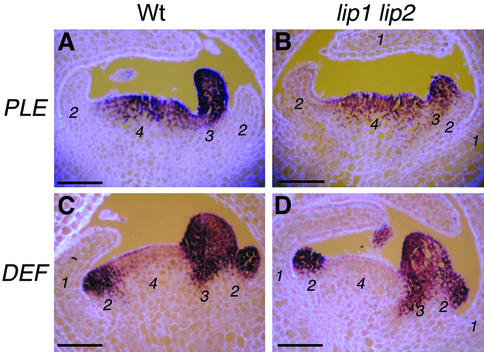
Fig. 7. Organ identity gene expression in wild type and lip1 lip2 double mutant. RNA in situ hybidizations on wild-type (A and C) and lip1 lip2 (B and D) sections of floral buds at stage 6, probed with PLE (A and B) or DEF (C and D). Expression patterns for PLE and DEF in the double mutant sections are similar to those observed in wild type. In particular, no ectopic expression of PLE is seen in whorl 2. Floral whorls labelled as sepal (1), petal (2), stamen (3) and carpel (4). Scale bars = 100 µm.
A further possibility was that other genes might be masking the effect of LIP genes on PLE expression. For example, the FISTULATA (FIS) gene normally has a role in repressing PLE expression in outer whorls (McSteen et al., 1998; Motte et al., 1998; Wilkinson et al., 2000). The lack of ectopic PLE expression in the lip1 lip2 double mutant might therefore be because FIS can substitute for the LIP genes. To test this, the lip1 lip2 fis mutant was constructed. This triple mutant showed no increase in stamen or carpel characters in the outer whorls compared with fis single mutants, indicating that the LIP genes had little effect on PLE even in a fis mutant background. However, this does not rule out the possibility that genes other than FIS are masking a repressive effect of LIP genes on PLE.
It was also possible that LIP genes exerted some of their effects on organ development by influencing other types of organ identity gene, such as DEFICIENS (DEF) belonging to the B class. To test this, the def lip1 lip2 triple mutant was constructed. The phenotype was essentially additive: the outer two whorls comprised bract-like sepals resembling those of the lip1 lip2 double mutant, while the third whorl comprised carpels with short styles and containing ovules bearing outgrowths (Figure 6C and D). Unlike the def single mutant, carpels sometimes developed in whorl 4 of the triple mutant, indicating that there may be an interaction between the genes with respect to whorl number. The overall lack of interaction between DEF and LIP genes was confirmed by in situ hybridizations, which showed that DEF was expressed normally in whorls 2 and 3 of the lip1 lip2 double mutant (Figure 7C and D).
Discussion
We have shown that the inability to obtain mutations in counterparts of AP2 through phenotypic screens in Antirrhinum is a consequence of genetic redundancy. Antirrhinum contains two genes, LIP1 and LIP2, that are more similar to each other and to AP2 than to any other genes of the AP2 family in Arabidopsis. The high degree of similarity between LIP1 and LIP2 indicates that they most probably arose by duplication since the divergence of Antirrhinum and Arabidopsis from their common ancestor. Like AP2 of Arabidopsis, LIP genes are expressed in floral meristems. LIP expression appears to be sequential, first appearing in bract primordia, then in sepal primordia and finally in petal and carpel primordia.
Mutation of either LIP gene alone does not produce a mutant phenotype. However, plants carrying null mutations in both genes have flowers in which the identity and development of organs are altered. A previous study in which a single LIP/AP2-like gene from Petunia was inactivated also revealed no phenotype (Maes et al., 2001). This may be because, as in Antirrhinum, there is a duplicate gene in Petunia.
The lip1 lip2 double mutant of Antirrhinum shows both differences from and similarities to ap2 mutants of Arabidopsis. Unlike ap2 mutants, there is no evidence for ectopic expression of C class genes in the outer whorls of the double mutant. RNA in situ hybridizations show that expression of the C class gene, PLE, is still restricted to whorls 3 and 4 in lip1 lip2 double mutants. Moreover, the phenotype of whorls 1 and 2 is not influenced by introduction of a plena mutation into the lip1 lip2 mutant background. This indicates that unlike ap2, the LIP genes are not needed to repress C in the outer whorls of Antirrhinum, and that other genes, such as STYLOSA (STY), FLORICAULA (FLO), FISTULATA (FIS) and CHORIPETALA (CHO), perform this role instead (McSteen et al., 1998; Motte et al., 1998; Wilkinson et al., 2000).
Although LIP genes are not required for repression of C, the ap2-1 mutant of Arabidopsis can be complemented by introduction of the Petunia AP2/LIP-like gene under the control of the 35S promoter (Maes et al., 2001). Because Petunia is more closely related to Antirrhinum than Arabidopsis, this suggests that LIP genes may be able to repress the C gene, AGAMOUS, in the context of Arabidopsis. The differences between Antirrhinum and Arabidopsis with respect to C regulation therefore most probably reflects changes in genes downstream of LIP/AP2, such as modifications in the cis-regulatory sequences of C class genes.
There are several evolutionary scenarios that might be envisaged. One is that the common ancestor of AP2 and LIP was required to repress C in outer whorls but that this requirement subsequently was lost in the lineage leading to Antirrhinum. This could have happened if, for example, PLE came under the control of other repressor genes, rendering the contribution of LIP genes redundant. PLE could then have either retained or lost its ability to interact with LIP genes. Another scenario is that the common ancestor of AP2/LIP did not repress C in outer whorls and that this role subsequently arose in the lineage leading to Arabidopsis. This might have happened if AG acquired an ability to interact with AP2 in outer whorls. However, this scenario also requires a second event in which the genes that originally repressed AG/PLE in the common ancestor adopted other roles so that AP2 became a requirement for C repression.
Although LIP genes differ from AP2 with respect to their requirement for regulation of C in outer whorls, their effects on floral organ development, as revealed by the lip1 lip2 double mutant, show some similarities. In this regard, it is most useful to compare the outer whorls of the lip1 lip2 double mutant with those of the ap2 ag double mutant of Arabidopsis, as this reveals the role of AP2 in organ development as distinct from its role in regulating C.
The whorl 1 organs of lip1 lip2 double mutants have glands at their tips that normally are found in leaves or bracts but not in sepals, indicating that LIP genes have a role in promoting sepal versus leaf/bract identity. This is comparable with the role of AP2 in Arabidopsis, as ap2 ag double mutants have organs with leaf-like characteristics in whorl 1 instead of sepals (Bowman et al., 1991). The whorl 1 organs of lip1 lip2 double mutants are also larger than wild-type sepals or bracts. It is not clear whether this reflects a more leaf-like state or whether it is a separate effect on organ growth. The effect of LIP genes on whorl 1 is not a consequence of ectopic B gene expression, as DEF interacts additively with the LIP genes. Moreover, in situ hybridizations revealed no ectopic expression of DEF in whorl 1 of the lip1 lip2 double mutant. Thus, the organ identity effects of the LIP genes in whorl 1 are not mediated by class B or C organ identity genes, but reflect a separate role in distinguishing sepals from leaves/bracts.
It is unlikely that expression of the LIP genes alone is sufficient to specify sepal identity, as LIP genes are also expressed in developing bracts. This suggests that other factors, which are expressed preferentially in floral meristems, such as SQUAMOSA (Huijser et al., 1992; Carpenter et al., 1995), may be needed together with LIP activity to distinguish sepal from bract identity. This would be similar to the proposed involvement in Arabidopsis of both APETALA1 (a likely orthologue of SQUAMOSA) and AP2 in conferring sepal identity (Irish and Sussex, 1990; Mandel et al., 1992a; Bowman et al., 1993).
The whorl 2 organs of the lip1 lip2 double mutant have reduced lip and palate regions. These defects are first apparent about half way through floral development, when whorl 2 organ primordia start to cover the developing stamens. The results suggest that the LIP genes play an important role in the growth and patterning of petal development. The AP2 gene of Arabidopsis also plays a role in whorl 2 growth and development, as evidenced by the modified shape and cell types of these organs in ag ap2 double mutants compared with wild-type petals (Bowman et al., 1991). However, the effects of AP2 in Arabidopsis are not precisely the same as those seen for LIP genes in Antirrhinum. This most probably reflects evolutionary divergence underlying the distinctive petal morphologies in these two species. For example, Arabidopsis petals have no obvious counterparts to lip or palate regions, raising the possibility that LIP genes have been involved in the evolution of these features in the Antirrhinum lineage. Such divergence in function may have been accelerated if the requirement for LIP genes for repressing C was lost in the Antirrhinum lineage, allowing their function to evolve in a less constrained manner.
The whorl 2 organs of lip1 lip2 double mutants are smaller than those of wild type, reflecting a reduction in both the size and number of cells. A more distantly related gene belonging to the AP2 family, AINTEGUMENTA, also influences organ size, although in this case the major contribution to final size is through effects on cell proliferation rather than cell size (Mizukami and Fischer 2000).
In addition to their effects on the outer two whorls, LIP genes also affect development of whorls 3 and 4. In whorl 3, the stamens of lip1 lip2 double mutants have slightly shorter filaments than wild type. In whorl 4, the style is shorter but the ovary is larger than wild type. The ovary also contains outgrowths tipped with stylar tissue, suggesting that carpel-like organs have grown in place of ovules. Such carpeloid structures have also been observed in strong ap2 mutants, where they have been proposed to reflect ectopic AG expression (Modrusan et al., 1994), and in plants overexpressing C class genes (Mandel et al., 1992b; Ray et al., 1994). By analogy, it is possible that the carpeloid ovules in lip1 lip2 double mutants result from ectopic PLE expression. In situ hybridizations did not reveal obvious ectopic expression of PLE in ovules of lip1 lip2 double mutants, although it is difficult to rule out the possibility that some ectopic activity is involved, particularly as the carpeloidy is variable. If ectopic PLE expression is responsible for the phenotype, it would imply that LIP genes play a role in repressing C class genes in ovules. As carpeloid ovules are observed in both Arabidopsis and Antirrhinum mutants, this might represent the most ancestral role of LIP/AP2 in regulation of class C genes. It is also possible that the carpeloid ovules reflect interactions with other genes, such as BEL1 in Arabidopsis or FBP 7 and FBP 11 in Petunia, which also give mutants with carpeloid ovules (Modrusan et al., 1994; Ray et al., 1994; Angenent et al., 1995).
In summary, the molecular basis of the A function in Antirrhinum differs to some extent from that in Arabidopsis. In Arabidopsis, AP2 has a dual role as a class A gene: establishment of normal sepal and petal development and repression of C. In Antirrhinum, these two roles are carried out more separately. LIP genes play a role in establishing sepal and petal development, but are not required for repression of C, which depends on other genes such as FIS, FLO, CHO and STY. Perhaps this divergence in the A function reflects greater evolutionary flexibility for genes controlling perianth as compared with reproductive organ development, and may have been enhanced by gene duplication within the Antirrhinum lineage.
Materials and methods
Isolation and sequencing of cDNA clones
Both LIP genes were obtained by screening an Antirrhinum λgt10 inflorescence cDNA library previously constructed by R.Simon. About 1 × 106 phage were screened at low stringency (55°C). Two LIP1 clones, each with an insert size of ∼1.85 kb, were subcloned into the KpnI site of pBluescript KS(+) to yield plasmid pJAM 2005. Two LIP2 clones with inserts of 1.1 kb were subcloned into the KpnI site of pBluescript SK(+) to yield pJAM 2006. Clones were sequenced using the automated ABI PRISM™ Big Dye Terminator Kit (Perkin Elmer), with additional sequence information being provided by NinoVillaroel, Alec Forsyth and Sandra Doyle. Predicted amino acid sequences were compared using Wisconsin GCG and GeneDoc programs.
Gene inactivation
Plants carrying transposon insertions in LIP1 and LIP2 were identified using a PCR-based reverse genetic screen. Several lines containing active transposons (JI2, 98, 75) were grown and self-pollinated at 15°C, a temperature which increases the rate of transposition (Harrison and Fincham, 1964; Carpenter et al., 1987).
A total of 1800 seed capsules from these plants individually were sown to give 1800 families with 15 plants each, giving a total of 27 000 M1 plants. The M1 plants were self-pollinated, and the resulting seed from each family pooled to give 1800 family bags of seed. Samples of seed from each bag were sown to generate leaf material for DNA extraction. Leaves were pooled in batches of 10 families to generate 180 DNA pools, each representing 150 M1 plants. To reduce the number of PCRs in the initial screen, the DNA pools were combined in groups of three to give 60 superpools. In parallel with this, pooled leaf material from each family was collected and stored separately at –80°C. Once a hit was detected in one of the 60 superpools in the first round of PCR screening, it was then traced to one of the corresponding three pools. DNA was then extracted from the 10 corresponding family samples stored at –80°C, and PCR carried out to identify the particular family containing the insertion. Seed from the corresponding family bag was sown, and the hit was then traced to individual progeny.
PCR primers at various sites within the LIP1 and LIP2 genes were used in various combinations with primers designed to either end of the transposon, Tam3 (Figure 1C). A 10 µl aliquot of each reaction was blotted onto Hybond N+ (Amersham International, UK), cross-linked and probed with the corresponding gene at 60°C. PCR was carried out in 50 µl reactions at 94°C for 1 min, then 94°C for 1 min, 55°C for 2 min and 72°C for 3 min, 35 cycles, plus 72°C for 10 min. Gene-specific and Tam3 primer sequences shown in Figure 1C are: LIP1 (1) 5′-GTGGTTGTCGAGGATGGTTC-3′, (2) 5′-CCGAGTTACCCAAACTCAGGTCC-3′, (3) 5′-GTTTGTAGACCAGCCTTCTTCG-3′, (4) 5′-AGTCACGTGGTCGAGAACTAC-3′; LIP2 (1) 5′-GTGGGATCTCAACGATTCGC-3′, (2) 5′-CAGGGTTACCGACGTTGTTT-3′, (3) 5′-CAATACGGATACAACGAGACG-3′, (4) 5′-TTCTTTCTTTGATATTGGCCC-3′; Tam3L, 5′-CACGGCCCAATTCACATCTTTA-3′; and Tam3R, 5′-CTCGGCACGTTTCACATCTTTA-3′.
Phenotypic analysis
Histological analysis was performed on 0.5 µm sections on the tips of bracts and sepals stained with aniline blue and photographed under a light microscope. For SEMs, floral buds at different stages of development were fixed in 3% (v/v) glutaraldehyde, dehydrated through an acetone series, critical point dried (Polaron, UK) and sputter coated with gold before being viewed and photographed. SEMs were also taken from plastic casts of buds as previously described by Carpenter et al. (1995). SEMs of wild-type and mutant ovaries used partly dissected material and the CT1500 HF cryo-system. Petal lobe areas were calculated using a MatLab program on dissected petals that had been flattened and scanned into a computer. Petal cell counts and cell size per unit area were determined by SEM, using the CT1500 HF cryo-system.
Antirrhinum stocks
The lip1 lip2 double mutant was crossed to several lines carrying def-621, ple-625 or fis alleles. The origins of these alleles have been been described previously by Carpenter and Coen (1990) and McSteen et al. (1998). Stock JI 75 was used as a wild type for comparison with lip1 lip2. Plants were grown as described by Carpenter et al. (1987).
In situ hybridization
RNA in situ hybridization and preparation of the DEF, FLO and PLE probes were carried out according to Bradley et al. (1993). LIP1 and LIP2 probes were prepared by subcloning PCR fragments, amplified from the 5′ region of the open reading frame, into the pGEM-T easy vector (Promega) in both orientations. Templates were linearized with SalI and transcribed by T7 polymerase to generate sense and antisense RNA probes for both LIP1 and LIP2.
Accession numbers
The DDBJ/EMBL/GenBank accession Nos for the LIP1 and LIP2 sequences are AY223518 and AY223519, respectively.
Acknowledgments
Acknowledgements
We thank Jack Okamuro for providing the AP2 clone, John Bowman for seed of the ap2-2 ag-1 mutant, Hans Sommer for sharing information on AP2-like genes in Antirrhinum, Nino Villaroel, Alec Forsyth and Sandra Doyle for providing sequence information, Desmond Bradley, Zsuzsanna Schwarz-Sommer and Karen Lee for helpful comments on the manuscript, Kim Findlay for help with sectioning and light microscopy, and Peter Walker for growing the plants.
References
- Ambrose B.A., Lerner,D.R., Ciceri,P., Padilla,C.M., Yanofsky,M.F. and Schmidt,R.J. (2000) Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell, 5, 569–579. [DOI] [PubMed] [Google Scholar]
- Angenent G.C., Franken,J., Busscher,M., van Dijken,A., van Went,J.L., Dons,H.J. and van Tunen,A.J. (1995) A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell, 7, 1569–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth,D.R. and Meyerowitz,E.M. (1989) Genes directing flower development in Arabidopsis. Plant Cell, 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth,D.R. and Meyerowitz,E.M. (1991) Genetic interactions among floral homeotic genes of Arabidopsis. Development, 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Alvarez,J., Weigel,D., Meyerowitz,E.M. and Smyth,D.R. (1993) Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development, 119, 721–743. [Google Scholar]
- Bradley D., Carpenter,R., Sommer,H., Hartley,N. and Coen,E. (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell, 72, 85–95. [DOI] [PubMed] [Google Scholar]
- Carpenter R. and Coen,E.S. (1990) Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev., 4, 1483–1493. [DOI] [PubMed] [Google Scholar]
- Carpenter R., Martin,C. and Coen,E.S. (1987) Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol. Gen. Genet., 207, 82–89. [Google Scholar]
- Carpenter R., Copsey,L., Vincent,C., Doyle,S., Magrath,R. and Coen,E. (1995) Control of flower development and phyllotaxy by meristem identity genes in Antirrhinum. Plant Cell, 7, 2001–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen E.S. and Meyerowitz,E.M. (1991) The war of the whorls: genetic interactions controlling flower development. Nature, 353, 31–37. [DOI] [PubMed] [Google Scholar]
- Davies B., Egea-Cortines,M., de Andrade Silva,E., Saedler,H. and Sommer,H. (1996) Multiple interactions amongst floral homeotic MADS box proteins. EMBO J., 15, 4330–4343. [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Bowman,J.L. and Meyerowitz,E.M. (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell, 65, 991–1002. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M. and Davies,B. (2000) Beyond the ABCs: ternary complex formation in the control of floral organ identity. Trends Plant Sci., 5, 471–476. [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler,H. and Sommer,H. (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J., 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- Harrison B.J. and Fincham,J.R.S. (1964) Instability at the PAL locus in Antirrhinum majus. 3. A gene controlling mutation frequency. Heredity, 23, 112–115. [Google Scholar]
- Honma T. and Goto,K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature, 409, 525–529. [DOI] [PubMed] [Google Scholar]
- Huijser P., Klein,J., Lonnig,W.E., Meijer,H., Saedler,H. and Sommer,H. (1992) Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J., 11, 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V.F. and Sussex,I.M. (1990) Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell, 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K.D., den Boer,B.G., Van Montagu,M. and Okamuro,J.K. (1994) Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell, 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki M.K., Okada,K., Nishino,E. and Shimura,Y. (1988) Isolation and characterization of novel mutants of Arabidopsis thaliana defective in flower development. Development, 104, 195–203. [Google Scholar]
- Kramer E.M. and Irish,V.F. (1999) Evolution of genetic mechanisms controlling petal development. Nature, 399, 144–148. [DOI] [PubMed] [Google Scholar]
- Kunst L., Klenz,J.E., Martinez-Zapater,J. and Haughn,G.W. (1989) AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell, 1, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes T., Van De Steene,N., Zethof,J., Karimi,M., D’Hauw,M., Mares,G., Van Montagu,M. and Gerats,T. (2001) Petunia Ap2-like genes and their role in flower and seed development. Plant Cell, 13, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel M.A., Gustafson-Brown,C., Savidge,B. and Yanofsky,M.F. (1992a) Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature, 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Mandel M.A., Bowman,J.L., Kempin,S.A., Ma,H., Meyerowitz,E.M. and Yanofsky,M.F. (1992b) Manipulation of flower structure in transgenic tobacco. Cell, 71, 133–143. [DOI] [PubMed] [Google Scholar]
- McSteen P.C., Vincent,C.A., Doyle,S., Carpenter,R. and Coen,E.S. (1998) Control of floral homeotic gene expression and organ morphogenesis in Antirrhinum. Development, 125, 2359–2369. [DOI] [PubMed] [Google Scholar]
- Meyerowitz E.M., Bowman,J.L., Brockman,L.L., Drews,G.N., Jack,T., Sieburth,L.E. and Weigel,D. (1991) A genetic and molecular model for flower development in Arabidopsis thaliana. Development, Suppl. 1, 157–167. [PubMed] [Google Scholar]
- Mizukami Y. and Fischer,R.L. (2000) Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl Acad. Sci. USA, 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrusan Z., Reiser,L., Feldmnn,A., Fischer,R.L. and Haughn,G.W. (1994) Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell, 6, 333–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte P., Saedler,H. and Schwarz-Sommer,Z. (1998) STYLOSA and FISTULATA: regulatory components of the homeotic control of Antirrhinum floral organogenesis. Development, 125, 71–84. [DOI] [PubMed] [Google Scholar]
- Munz P.A. (1926) The Antirrhinoideae—Antirrhineae of the new world. Proc. Calif. Acad. Sci., 15, 323–397. [Google Scholar]
- Okamuro J.K., Caster,B.,Villarroel,R.,Van Montagu,M. and Jofuku,K.D. (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl Acad. Sci. USA, 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta,G.S., Baumann,E., Wisman,E. and Yanofsky,M.F. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature, 405, 200–203. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Hareven,D., Rounsley,S.D., Yanofsky,M.F. and Lifschitz,E. (1994) Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell, 6, 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R.E., Chang,C., Pang,P.P.Y. and Meyerowitz,E.M. (1987) Molecular genetics and development of Arabidopsis. In Loomis,W.R. (ed.), Genetic Regulation of Development. A.R.Liss, New York, NY, pp. 327–338.
- Ray A., Robinson-Beers,K., Ray,S., Baker,S.C., Lang,J.D., Preuss,D., Milligan,S.B. and Gasser,C.S. (1994) Arabidopsis floral homeotic gene BELL (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl Acad. Sci. USA, 91, 5761–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J.L. and Meyerowitz,E.M. (1998) The AP2/EREBP family of plant transcription factors. Biol. Chem., 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Samach A., Kohalmi,S.E., Motte,P., Datla,R. and Haughn,G.W. (1997) Divergence of function and regulation of class B floral organ identity genes. Plant Cell, 9, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. and Meyerowitz,E.M. (1994) The ABCs of floral homeotic genes. Cell, 78, 203–209. [DOI] [PubMed] [Google Scholar]
- Wilkinson M., de Andrade Silva,E., Zachgo,S., Saedler,H. and Schwarz-Sommer,Z. (2000) CHORIPETALA and DESPENTEADO: general regulators during plant development and potential floral targets of FIMBRIATA-mediated degradation. Development, 127, 3725–3734. [DOI] [PubMed] [Google Scholar]