Abstract
Cdc2p is a cyclin-dependent kinase (CDK) essential for both mitotic and meiotic cell cycle progression in fission yeast. We have found that the spindle checkpoint kinase Bub1p becomes phosphorylated by Cdc2p during spindle damage in mitotic cells. Cdc2p directly phosphorylates Bub1p in vitro at the CDK consensus sites. A Bub1p mutant that cannot be phosphorylated by Cdc2p is checkpoint defective, indicating that Cdc2p-dependent Bub1p phosphorylation is required to activate the checkpoint after spindle damage. The kinase activity of Bub1p is required, but is not sufficient, for complete spindle checkpoint function. The role of Bub1p in maintaining centromeric localization of Rec8p during meiosis I is entirely dependent upon its kinase activity, suggesting that Bub1p kinase activity is essential for establishing proper kinetochore function. Finally, we show that there is a Bub1p-dependent meiotic checkpoint, which is activated in recombination mutants.
Keywords: Bub1p kinase/Cdc2p/meiotic checkpoint/Rec8p/spindle checkpoint
Introduction
The spindle checkpoint plays an essential role in eukaryotic cells to inhibit onset of anaphase until proper attachment of chromosomes to the spindle (reviewed by Amon, 1999; Millband et al., 2002). One of the proteins involved in this checkpoint is the conserved Bub1p protein kinase that localizes to kinetochores during mitosis (reviewed by Straight, 1997). In this paper, we investigate the roles that Bub1p phosphorylation and kinase activity play in spindle checkpoints during the mitotic and meiotic cell cycles of fission yeast.
Bub1p was initially identified in budding yeast as being required for pre-anaphase delay in response to spindle damage (Hoyt et al., 1991; Roberts et al., 1994). It plays a role in maintaining chromosomal stability in a number of eukaryotes, and mutations in the bub1 gene have been found in some cancers (reviewed by Cahill et al., 1998; Jallepalli and Lengauer, 2001). Bub1p localizes at the kinetochores during prophase and prometaphase of mitosis (Taylor and McKeon, 1997; Basu et al., 1999) and is thought to be recruited to unattached kinetochores. Bub1p physically associates with Bub3p (Roberts et al., 1994) and Mad1p (Brady and Hardwick, 2000), and is required for the kinetochore localization of Bub3p (Basu et al., 1999; Sharp-Baker and Chen, 2001). Bub1, Bub3p, Mad1p, Mad2p and Mad3p probably form a complex at the unattached kinetochore (reviewed by Amon, 1999). Bub1p has kinase activity in vitro and can phosphorylate itself as well as Bub3p (Roberts et al., 1994) and Mad1p (Seeley et al., 1999) in vitro. Other spindle checkpoint components such as Mad1p, Mad2p and Mad3p interact with Cdc20–APC (the anaphase-promoting complex/cyclosome), which inhibits the metaphase–anaphase transition (reviewed by Amon, 1999; Millband et al., 2002). In budding yeast, a point mutation in the conserved lysine residue of the kinase domain abolished in vitro kinase activity, and such mutants appeared to be checkpoint defective (Roberts et al., 1994). In Xenopus laevis, Bub1p recruits other spindle checkpoint components (Mad1p, Mad2p and Bub3p) to kinetochores independently of its kinase activity (Sharp-Baker and Chen, 2001).
The role of Bub1p during meiosis I (MI) differs from mitosis and meiosis II (MII) because chromosomal segregation is reductional, with sister chromatids remaining attached and moving together to one pole (reviewed by Kleckner, 1996). The spindle checkpoint proteins are recruited to kinetochores during meiosis (Yu et al., 1999; Kallio et al., 2000), and both Bub1p and Mad2p prevent non-disjunction events during MI in yeast (Shonn et al., 2000; Bernard et al., 2001). In fission yeast, Bub1p has been reported to play a role in ensuring reductional segregation during MI (Bernard et al., 2001). The meiotic cohesin Rec8p also plays an essential role in establishing reductional segregation (Watanabe and Nurse, 1999; Buonomo et al., 2000), and its association with the inner centromeres is important to establish monopolar orientation (Watanabe et al., 2001). Fission yeast Bub1p is associated with the kinetochores during both meiosis and mitosis (Bernard et al., 1998, 2001), and is required for the centromeric localization of Rec8p during anaphase I.
How Bub1p is regulated is not well understood. Sequence analysis of Bub1p reveals four putative CDK phosphorylation sites in the amino acid sequence of Bub1p. Cdc2p is a cyclin-dependent kinase that plays multiple roles during both mitosis and meiosis (reviewed by Nigg, 1995; Nurse, 2000). Cdc2p has been shown to be localized at centromeres during meiosis in fission yeast (Decottignies et al., 2001), and so we have investigated whether Cdc2p plays a role in regulating Bub1p during the cell cycle. We show that at least one of the four putative CDK sites in Bub1p is phosphorylated by Cdc2p, and have investigated the roles this phosphorylation and the Bub1p kinase domain play in the spindle checkpoint during mitosis and the first meiotic division.
Results
Phosphorylation of Bub1p during mitosis
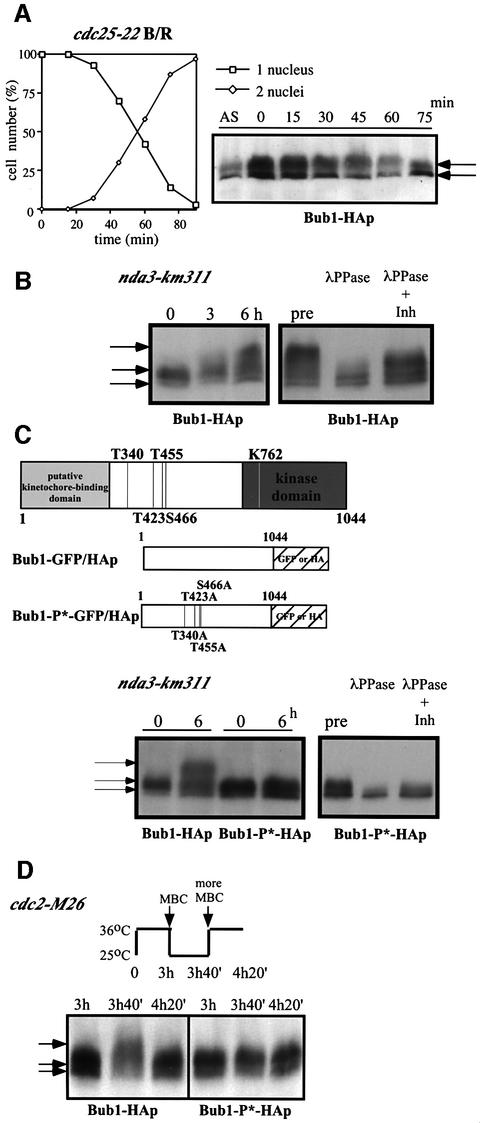
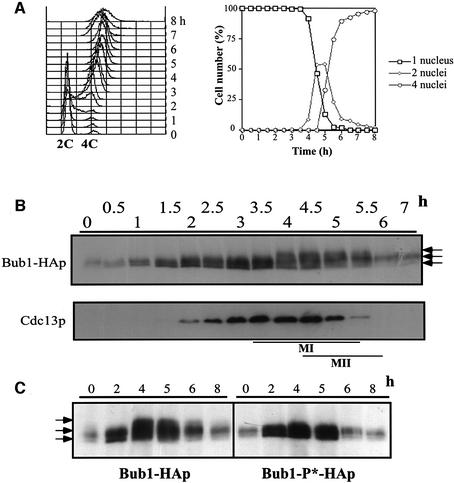
To determine whether Bub1p phosphorylation changes during the mitotic cell cycle, cells were synchronized in G2 by incubating a cdc25–22 mutant strain in which the endogenous bub1+ was tagged with HA at 36°C for 4 h before being released into mitosis at 25°C (Figure 1A). Western blot analysis using anti-HA antibodies revealed multiple bands of Bub1-HAp in exponentially growing cells, but these remained unchanged throughout the cell cycle (Figure 1A). Next, we investigated Bub1p phosphorylation during spindle damage, which is known to involve Bub1p function (reviewed by Amon, 1999). To disrupt the mitotic spindle, cold-sensitive β-tubulin mutant nda3-km311 cells were incubated at 18°C (Hiraoka et al., 1984). Upon shifting the cells to 18°C, slower migrating forms of Bub1-HAp appeared on the gel in addition to the multiple bands present in asynchronously growing cells (Figure 1B). All the bands that appeared in the nda3 block were sensitive to phosphatase treatment (Figure 1B). These results indicate that Bub1p becomes hyperphosphorylated in an nda3 block.
Fig. 1. Phosphorylation of Bub1p during mitosis. (A) Percentage of cells with 1 or 2 nuclei (left panel) and western blotting (right panel) of G2 block and release experiments. Cdc25-22 bub1-HA cells were incubated at 36°C for 4 h, then released at 25°C for the indicated time to proceed through synchronous mitosis. Anti-HA antibody was used to detect Bub1-HAp. (B) Bub1p became hyperphosphorylated in an nda3 block. Nda3-km311 cells were incubated at 18°C for the indicated time in hours (left panel). Extracts were prepared from nda3-km311 cells blocked at 18°C for 6 h (pre) and incubated with λ phosphatase in the absence (λPP) or presence (λPP+Inh) of phosphatase inhibitors (right panel). (C) Hyperphosphorylation was abolished in nda3-km311 bub1-P*-HA strain. The four CDK consensus sites were mutated to alanine in the bub1-P* mutant (left panel). Hyperphosphorylation in an nda3 block was eliminated in bub1-P*-HA strain (centre panel). Phosphatase treatment of extracts from bub1-P*-HA cells (right panel). (D) Hyperphosphorylation of Bub1p was dependent on Cdc2p kinase activity. Cdc2-M26 bub1-HA and cdc2-M26 bub1-P*-HA cells were incubated at 36°C for 3 h, released to 25°C in the presence of 50 µg/ml MBC for 40 min until cells blocked in mitosis, and then shifted back to 36°C in the presence of 100 µg/ml MBC.
Sequence analysis revealed four putative CDK consensus sites (S/T-P-X-K/R) (Maller et al., 1989) in the middle region of Bub1p (Figure 1C). To investigate whether hyperphosphorylation of Bub1p in a mitotic block occurs at these CDK sites, we constructed an ‘unphosphorylatable’ mutant (bub1-P*) of Bub1p by changing the four S/T residues T340, T423, T455 and S466 to alanine (Figure 1C). Bub1p hyperphosphorylation was eliminated in this bub1-P*-HA strain in an nda3 block (Figure 1C). Phosphatase treatment revealed that the other bands observed in the bub1-P*-HA strain were also due to phosphorylation (Figure 1C), suggesting that Bub1p may be additionally phosphorylated by other kinases. These results demonstrate that Bub1p becomes hyperphosphorylated at the CDK consensus sites when spindle formation is disrupted.
To determine whether Cdc2p kinase activity is required for Bub1p hyperphosphorylation, we destabilized microtubules (MTs) with drugs in a thermosensitive (ts) cdc2 mutant strain (Figure 1D). Cdc2-M26 cells were incubated for 3 h at 36°C, arresting most cells in late G2 (Carr et al., 1989). The cells were released to 25°C in the presence of 50 µg/ml carbendazim (MBC), and after 40 min, both bub1-HA and bub1-P*-HA cells were analysed by western blotting (Figure 1D). Slower migrating forms of Bub1-HAp were detected in addition to the normal multiple bands (Figure 1D, left panel, 3h40). In contrast, hyperphosphorylation of Bub1-P*-HAp was not observed in the bub1-P*-HA cells (Figure 1D, right panel, 3h40). The upper bands of Bub1-HAp disappeared when cells were shifted back to 36°C (Figure 1D, left panel, 4h20), establishing that Cdc2p kinase activity is continuously required to maintain the MBC-induced hyperphosphorylation of Bub1p.
Cdc2p can phosphorylate Bub1p in vitro
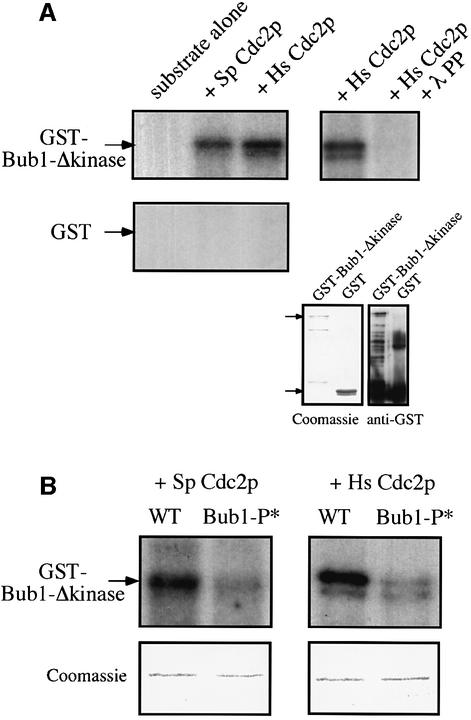
To investigate whether this Cdc2p-dependent phosphorylation of Bub1p is direct or not, we performed an in vitro kinase assay. Since Bub1p is able to autophosphorylate (Roberts et al., 1994), bacterially-produced glutathione S-transferase (GST) fusion protein of Bub1p lacking the kinase domain was used as a substrate for Cdc2p. When GST–Bub1-Δkinasep was incubated with either Schizosaccharomyces pombe or Homo sapiens Cdc2p, we detected the phosphorylation of a 115 kDa band (Figure 2A). This was not due to autophosphorylation of Bub1p, as GST–Bub1-Δkinasep alone did not give any signal (Figure 2A). We concluded that Cdc2p can phosphorylate Bub1p in vitro.
Fig. 2. Cdc2p phosphorylates Bub1p at CDK consensus sites in vitro. (A) Left panel: GST–Bub1-Δkinasep or GST was incubated either alone (substrate alone) or with S.pombe Cdc2p (Sp Cdc2p) or H.sapiens Cdc2p (Hs Cdc2p). The band just below GST–Bub1-Δkinasep is a degradation product, as shown by the western blotting. Top right panel: GST–Bub1-Δkinasep was incubated either with Hs Cdc2p alone or with Hs Cdc2p and λ phosphatase (λPP). Bottom right panel: Coomassie Blue staining and anti-GST western blotting of GST–Bub1-Δkinase and GST. (B) GST–Bub1-Δkinasep (WT) or GST–Bub1-P*-Δkinasep (Bub1-P*) was incubated with either Sp Cdc2p (left panel) or Hs Cdc2p (right panel). Coomassie Blue staining is shown as loading control.
To investigate whether this phosphorylation by Cdc2p occurs at the CDK consensus sites, we purified unphosphorylatable mutant of Bub1p from bacteria, in which four CDK consensus sites are mutated (GST–Bub1-P*-Δkinasep). Incubation with either S.pombe or H.sapiens Cdc2p revealed that the [32P]Bub1p signal was much reduced in the GST–Bub1-P*-Δkinasep mutant protein (Figure 2B), indicating that Cdc2p phosphorylates Bub1p at the CDK consensus sites in vitro.
Cdc2p-dependent hyperphosphorylation of Bub1p and the spindle checkpoint
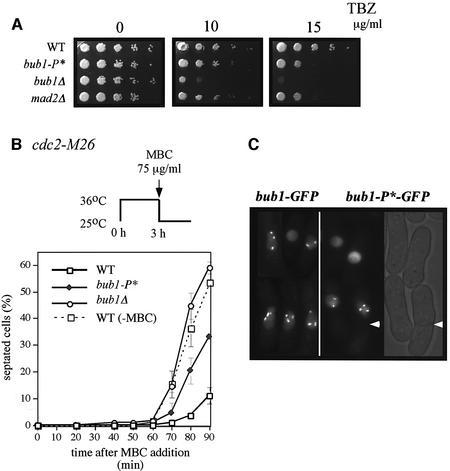
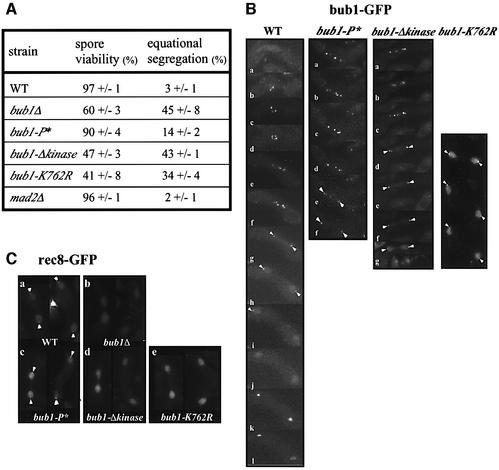
We next tested whether Cdc2p-mediated hyperphosphorylation of Bub1p plays a role in the spindle checkpoint. Bub1-P* cells were more sensitive to thiabendazole (TBZ) than wild-type (WT) cells, but more resistant than bub1Δ cells (Figure 3A). The TBZ sensitivity of bub1-P* cells was comparable with that of mad2Δ cells (Figure 3A), which have been shown to be sensitive to MT drugs (Li and Murray, 1991; Kim et al., 1998). To quantify these effects, the number of cells that could proceed through mitosis to septation in the presence of MBC was scored. Cdc2-M26 cells were synchronized in late G2 by incubation at 36°C and subsequently released to 25°C in the presence of 75 µg/ml MBC. Bub1+ cells became blocked in mitosis before septation, and 80 min after release only 3% of the cells were found to be septated (Figure 3B). In contrast, the spindle checkpoint is defective in bub1Δ cells and 45% of the cells were septated at 80 min. In the bub1-P* mutant, 20% of the cells were septated, suggesting that phosphorylation is required to completely block cells in mitosis. The inability of Bub1-P*p to completely activate the checkpoint was not due to a defect in kinetochore localization because Bub1–GFPp and Bub1-P*–GFPp both localized similarly to kinetochores in the presence of MBC (Figure 3C).
Fig. 3. Bub1-P* mutant cells are checkpoint defective. (A) WT, bub1-P*, bub1Δ and mad2Δ cells were grown to exponential phase, spotted onto YE5S plates containing 0, 10 or 15 µg/ml TBZ and incubated for 2–3 days. (B) Cdc2-M26 (WT), cdc2-M26 bub1-P* (bub1-P*), cdc2-M26 bub1Δ (bub1Δ) strains were incubated at 36°C for 3 h, released to 25°C in the presence of 75µg/ml MBC and incubated for the indicated time in minutes. Cdc2-M26 strain released in the absence of MBC (WT, –MBC) is indicated as a control. Septated cells were scored by calcofluor staining. (C) Bub1–GFPp and Bub1-P*–GFPp were observed by live fluorescence microscopy after 40 min of release at 25°C in the presence of MBC.
Bub1p kinase domain and the spindle checkpoint
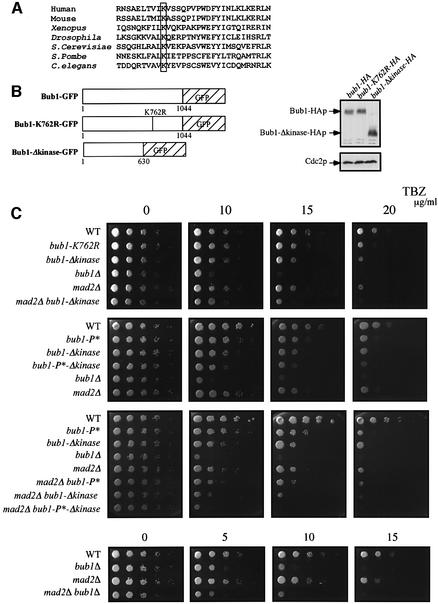
The kinase domain is at the C-terminus of the Bub1p protein (Figure 1C) and includes the conserved K762 residue in the ATP-binding site (Figure 4A). To investigate the role of Bub1p kinase activity in the spindle checkpoint, we constructed two different mutants, one with the K762R mutation, which abolishes the kinase activity of Saccharomyces cerevisiae Bub1p (Roberts et al., 1994), and a second that lacked the entire kinase domain (bub1-Δkinase) (Figure 4B). Both mutants were more sensitive to TBZ than WT cells, but not as sensitive as bub1Δ cells (Figure 4C). We quantified these effects using synchronized cdc2-M26 cells. Eighty minutes after release from G2 in the presence of MBC, 21% of the bub1-Δkinase cells and 22% of the bub1-K762R cells were septated compared with 3% of WT cells and 45% of bub1Δ cells (Figure 4D). These observations suggest that the kinase activity of Bub1p is necessary for complete spindle checkpoint function. However, there must also be an additional role for Bub1p that is independent of its kinase activity and is necessary for full operation of the checkpoint. Fusion of the Bub1-Δkinase protein to green fluorescent protein (GFP) revealed that the mutant protein could still localize to the kinetochores of MBC-treated cells (Figure 4E). The defective spindle checkpoint in kinase mutants was not due to a reduction in protein level, as the protein amount of Bub1p in bub1-K762R, bub1-Δkinase and WT cells was comparable (Figure 4B, right panel). Because the bub1-P* and bub1-Δkinase mutants are both partially defective in the spindle checkpoint response, we investigated the phenotype of the bub1-P*-Δkinase double mutant. TBZ resistance tests revealed that the bub1-P*-Δkinase mutant was more sensitive to the drug than the single mutants, but was still less sensitive than a bub1Δ mutant (Figure 4C). Quantitative experiments in liquid medium gave similar results (Figure 4D). Therefore, we conclude that although both CDK phosphorylation of Bub1p and the kinase domain of Bub1p contribute to the spindle checkpoint, other aspects of Bub1p function must also be required for a full response.
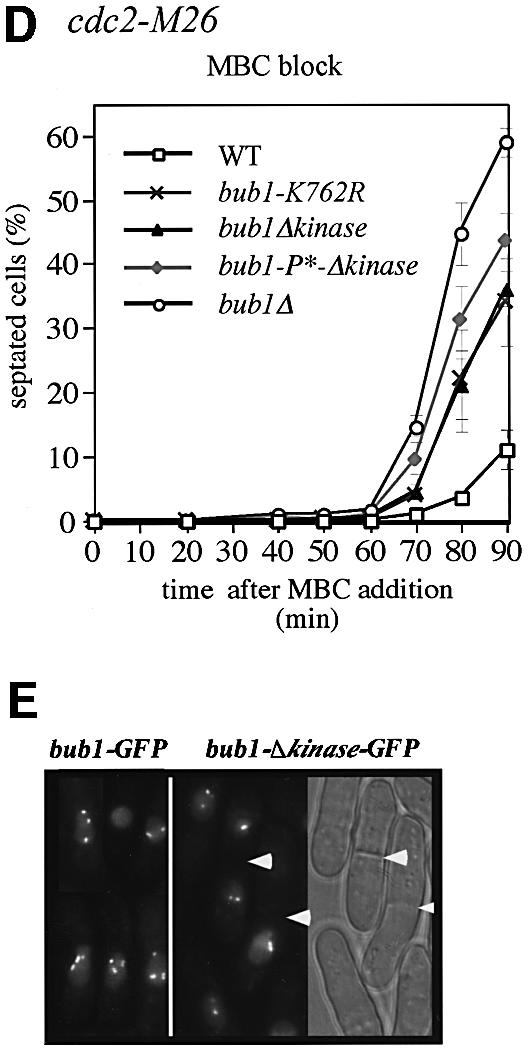
Fig. 4. Kinase activity of Bub1p is required but not sufficient for spindle checkpoint. (A) Conservation of K762 residue in the kinase domain of Bub1p. (B) Left panel: kinase mutants. bub1-K762R carries a point mutation and bub1-Δkinase lacks the entire kinase domain. Right panel: cell extracts from exponentially growing bub1-HA, bub1-K762R-HA and bub1-Δkinase-HA strains were analysed by western blotting using anti-HA antibody. Cdc2p is shown as loading control. (C) See Figure 33 legend for details. The concentrations of TBZ used are indicated in µg/ml. (D) Cdc2-M26 (WT), cdc2-M26 bub1-K762R (bub1-K762R), cdc2-M26 bub1Δkinase (bub1Δkinase), cdc2-M26 bub1-P*-Δkinase (bub1-P*-Δkinase) and cdc2-M26 bub1Δ (bub1Δ) strains were used for the experiment described in Figure 33B. (E) The experiment described in Figure 33C was performed using kinase mutants.
We next investigated the relationship between Mad2p and Bub1p kinase activity, by testing to see if the double mutant of bub1-Δkinase and mad2Δ would give the same phenotype as the single mutants. This was not the case: bub1-Δkinase mad2Δ double mutant cells were more sensitive to TBZ than the single mutants (Figure 4C), indicating that Mad2p is not the only target of Bub1p kinase activity. In a mad2Δ bub1-P*-Δ kinase mutant, TBZ sensitivity was similar to bub1Δ cells (Figure 4C), establishing that CDK phosphorylation, Bub1p kinase activity and the Mad2p-dependent pathway have additive effects. The mad2Δ bub1Δ double mutant showed similar TBZ sensitivity to that of bub1Δ single mutant (Figure 4C). However, we found that Mad2p is still recruited to the kinetochores during spindle damage in the absence of Bub1p (data not shown), suggesting that Bub1p is not required for Mad2p localization at the kinetochore.
Bub1p becomes hyperphosphorylated at CDK sites during meiosis
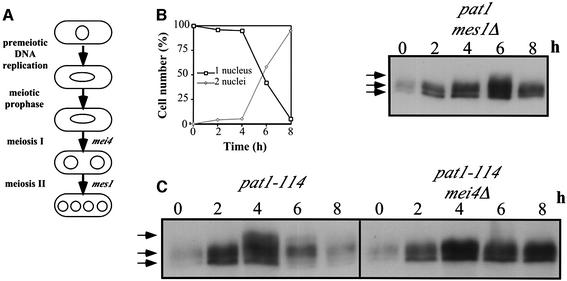
Bub1p is required for reductional segregation of sister chromatids during MI (Bernard et al., 2001), and both Cdc2p and Bub1p localize to the centromeres during meiosis (Bernard et al., 2001; Decottignies et al., 2001). Therefore we examined the phosphorylation state of Bub1p in meiotic cells and the relevance of the CDK consensus sites for this phosphorylation. Pat1-114/pat1-114 mutant cells were used to induce synchronous meiotic divisions (Bähler et al., 1991). Pre-meiotic DNA synthesis occurred 2 h after the shift (Figure 5A, left panel), followed by MI and MII (Figure 5A, right panel). Western blot analysis revealed that Bub1-HAp becomes hyperphosphorylated 4 h after the shift, corresponding to the time of the meiotic nuclear divisions (Figure 5B). This hyperphosphorylation is maintained for 1 h and was not present when the Cdc2p consensus sites were mutated (Figure 5C). In mes1 mutant cells, which block between MI and MII (Figure 6A; Shimoda et al, 1985), Bub1-HAp became hyperphosphorylated at the first meiotic division (Figure 6B) and dephosphorylated at MI exit (Figure 6B). In mei4 mutant cells, which arrest at meiotic prophase (Figure 6A; Shimoda et al., 1985; Bähler et al., 1993; Horie et al., 1998), Bub1-HAp did not become hyperphosphorylated (Figure 6C). These data show that Bub1p hyperphosphorylation at Cdc2p consensus sites occurs during MI, after meiotic prophase.
Fig. 5. Bub1p becomes hyperphosphorylated at CDK consensus sites during meiotic divisions. (A) FACS analysis (left panel) and percentage of cells with 1, 2 or 4 nuclei (right panel) of pat1-induced synchronous meiosis using pat1-114/pat1-114 bub1-HA/bub1-HA strain (pat1-114 bub1-HA). Time after induction is indicated in hours. (B) Western blotting of cell extracts from the time-course experiment described in (A) using anti-HA and anti-Cdc13p antibodies. Cdc13p level is shown as an indication of Cdc13p-associated Cdc2p kinase activity. (C) Western blotting of cell extracts from synchronous meiosis using pat1-114 bub1-HA and pat1-114 bub1-P*-HA strains.
Fig. 6. Bub1p becomes phosphorylated during meiosis I. (A) mei4 is required for exit from prophase I, and mes1 is required for entry into meiosis II. (B) Percentage of cells with 1 or 2 nuclei at each time point as scored by DAPI staining (left panel) and western blotting of cell extracts (right panel) from synchronous meiosis using pat1-114 mes1Δ strain. (C) Western blotting of extracts from pat1-114 and pat1-114 mei4Δ strains. Time after induction is indicated in hours.
Bub1p phosphorylation and sister chromatid segregation
To determine whether Cdc2p-dependent phosphorylation of Bub1p is required for reductional segregation, we scored cen1-GFP distribution during MI (Nabeshima et al., 1998; Watanabe and Nurse, 1999). In bub1+ cells, only 3% of the cells showed equational distribution. In contrast, in the absence of Bub1p, cells underwent an equational division in 45% of MI events (Figure 6A), in agreement with data published previously (Bernard et al., 2001). Spore viability was also reduced to 60% (Figure 7A). In the bub1-P* mutant (90% spore viability), equational divisions occurred in 14% of MI events (Figure 7A). The Bub1-P*–GFP protein had a localization similar to WT Bub1–GFPp (Figure 7B). We conclude that although phosphorylation has little effect on spore viability, it does have some role in ensuring reductional division during MI.
Fig. 7. Bub1p kinase activity is required for maintenance of centromeric Rec8p during anaphase I and reductional segregation. (A) Spore viability (%) and equational segregation frequency (%) of WT, bub1Δ, bub1-P*, bub1-Δkinase, bub1-K762R and mad2Δ cells. (B) In vivo localization of Bub1–GFPp (WT), Bub1-P*–GFPp (bub1-P*), Bub1-Δkinase–GFPp (bub1-Δkinase) and Bub1-K762R–GFPp (bub1-K762R) during meiosis. (C) Rec8–GFPp localization during anaphase I in WT, bub1Δ, bub1-P*, bub1-Δkinase and bub1-K762R cells.
Reductional segregation requires Bub1p kinase activity
To examine the role of the Bub1p kinase domain in sister chromatid cohesion during MI, cen1-GFP distribution in both the bub1Δkinase and the bub1-K762R mutants was examined. In the bub1Δkinase mutant, equational segregation of cen1-GFP occurred in 43% of MI events, a value similar to the 45% observed in bub1Δ cells (Figure 7A). In the bub1-K762R mutant, 34% of the MI cells underwent equational segregation, whilst in the mad2Δ cells, equational segregation was observed in only 2% of MI cells (Figure 7A; Bernard et al., 2001). These results show that the kinase activity of Bub1p is required for complete reductional segregation of sister chromatids during MI. The failure of Bub1p kinase mutants to maintain efficient sister chromatid cohesion during MI may be due to mislocalization of the mutant protein. We examined the localization of Bub1p kinase mutants during meiotic divisions (Figure 7B). In zygotes undergoing meiotic divisions, WT Bub1–GFPp became associated with the centromeres after meiotic prophase and remained associated until anaphase I, confirming previous reports using fixed cells (Bernard et al., 2001; Figure 7B, WT, f–h). Both the Bub1Δkinase–GFPp and Bub1-K762R–GFPp proteins were associated with centromeres during anaphase I (Figure 7B, bub1-Δkinase, d–g and bub1-K762R). These data suggest that the effect of Bub1p kinase domain on sister chromatid segregation in MI is not due to a failure of mutant proteins to localize to the centromeres, although the localization of kinase mutant proteins within the centromeric region may be modified. Watanabe and Nurse (1999) have shown that maintenance of Rec8p at the centromeric regions during anaphase I is required for reductional segregation of sister chromatids. In MII, centromeric Rec8p is degraded, allowing separation of sister chromatids. In vivo centromeric staining of Rec8–GFPp was detected as bright dots in dividing nuclei (Figure 7Ca, arrows), but in the absence of Bub1p, centromeric Rec8–GFPp was no longer maintained during anaphase I (Figure 7Cb), in agreement with the results using fixed cells (Bernard et al., 2001). As expected from cen1–GFP segregation in MI (Figure 7A), the centromeric localization of Rec8–GFPp was lost during anaphase I in the majority of bub1Δkinase and bub1-K762R cells (Figure 7Cd and e). These results show that even though Bub1p kinase mutants localize at the centromeric region during anaphase I, they are no longer capable of maintaining Rec8p at this location.
A Bub1p-dependent meiotic spindle checkpoint
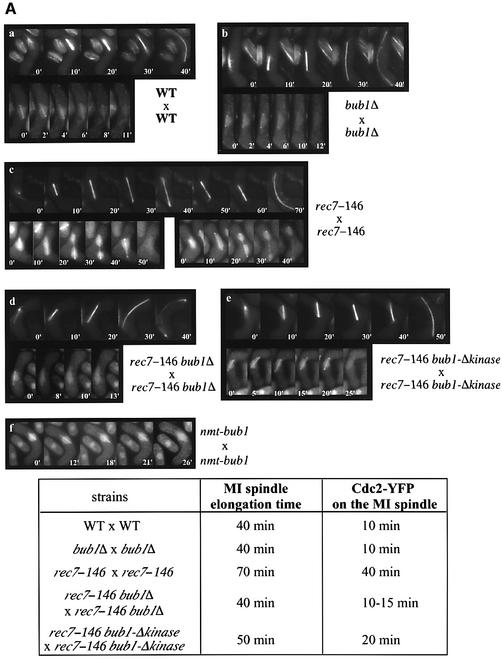
To investigate the existence of a meiotic spindle checkpoint in fission yeast, we used recombination-deficient (rec) mutants, which exhibit delays in MI (Molnar et al., 2001). We used the GFP–α2-tubulin allele (Yamamoto et al., 1999) to measure the length of MI. In WT cells, the time-course experiment shows that MI spindle elongation occurs in three steps: the spindle elongates slowly between 0 and 10 min (phase I), there is no elongation between 10 and 20 min (phase II) and, finally, the spindle elongates rapidly between 20 and 40 min (phase III) (Figure 8Aa, upper panel; Mallavarapu et al., 1999). Rec7+ is an early meiotic recombination gene, deletion of which leads to non-disjunction of homologous chromosomes and low spore viability (Ponticelli and Smith, 1989; Lin et al., 1992; Molnar et al., 2001). In rec7-146 cells, we found that, after the first step of slow elongation between 0 and 10 min (phase I), the spindle continues to elongate slowly and then undergoes regression for ∼60 min (phase II) before it starts to elongate rapidly until 70 min (phase III) (Figure 8Ac, upper panel). We found that this MI delay is dependent upon Bub1p because MI spindle elongation time is reduced to 40 min in rec7-146 bub1Δ cells (Figure 8Ad, upper panel). In this case, however, cells seem to lack phase II and spindle elongates continuously between 0 and 40 min (Figure 8Ad, upper panel). Next, we tested whether the delay is dependent on Bub1p kinase activity. In a rec7-146 bub1Δkinase mutant, the MI delay occurs, although reduced by ∼20 min compared with rec7-146 bub1+ cells; no spindle regression was observed during phase II (Figure 8Ae, upper panel). This Bub1p-dependent delay was also observed in a rec14Δ mutant (data not shown; Evans et al., 1997; Cervantes et al., 2000).
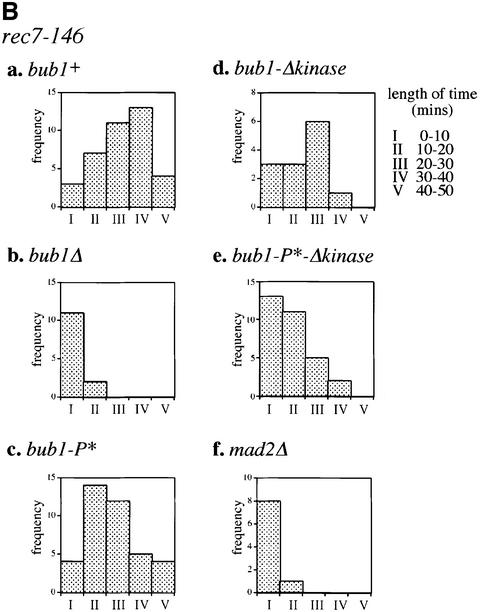
Fig. 8. Bub1p-dependent delay during meiosis I in rec7 mutants. (A) α-tubulin–GFPp (top panel) and Cdc2–YFPp on MI spindles (bottom panel) were observed in the following strains: (a) WT, (b) bub1Δ, (c) rec7-146, (d) rec7-146 bub1Δ, and (e) rec7-146 bub1Δkinase. (f) Cdc2–YFP was observed on MI spindle in bub1+-overexpressing cells (nmt1-bub1). MI spindle elongation time and the time of Cdc2–YFPp fluorescence association with MI spindle (Cdc2–YFP on the MI spindle) in each strain are shown in the table. (B) Association time of Cdc2–YFPp fluorescence with MI spindle was measured in rec7-146 cells combined with (a) bub1+, (b) bub1Δ, (c) bub1-P*, (d) bub1-Δkinase, (e) bub1-P*-Δkinase and (f) mad2Δ. The association times were grouped into five categories (I: 0–10, II: 10–20, III: 20–30, IV: 30–40, V: 40–50 mins). Histograms show the number of cells falling into each category.
Our data suggest the existence of a Bub1p-dependent spindle checkpoint in MI. To investigate whether CDK activity would be a target of Bub1p-dependent meiotic spindle checkpoint activation, we looked at the association of Cdc2–YFPp (Decottignies et al., 2001) with MI spindle since CDKs from different species are known to regulate MT dynamics in mitosis by direct interaction with MT-associated proteins (reviewed by Andersen, 2000). In WT cells, Cdc2–YFPp fluorescence stays on the MI spindle for <10 min (Figure 8Aa, lower panel). In rec7-146 cells, we found that Cdc2–YFPp remained at the spindle for ∼40 min (Figure 8Ac, lower panel and Ba), persisting on the elongated spindle after separation of the nucleus into two DNA masses (Figure 8Ac, lower panel). This is never observed in WT cells. In agreement with GFP–α2-tubulin data, the association of Cdc2–YFPp with the MI spindle is reduced to <10 min in rec7-146 bub1Δ cells (Figure 8Ad, lower panel and Bb). The same reduction was observed in rec7-146 mad2Δ cells (Figure 8Bf). In rec7-146 bub1Δkinase cells, the MI delay was reduced by ∼20 min (Figure 8Ae, lower panel and Bd) and Cdc2–YFPp was never found on the elongated spindles. As CDK-dependent phosphorylation of Bub1p occurs during the first meiotic division (Figures 5 and 6), we tested whether this phosphorylation plays any role in the meiotic checkpoint. However, in a rec7-146 bub1-P* mutant, the delay in MI was still present (Figure 8Bc), indicating that CDK-dependent phosphorylation does not play a significant role in this checkpoint. These observations suggest that the Bub1p-dependent delay in MI acts on Cdc2p association with the spindle and occurs at two distinct stages, an early stage with one nuclear mass, and a later stage with separated nuclei which requires kinase activity. We also observed a similar early MI delay in cells overexpressing bub1+ (Figure 8Af).
Discussion
We have found that Bub1p becomes hyperphosphorylated at CDK consensus sites in a Cdc2p-dependent manner when the fission yeast mitotic spindle is destabilized using drugs or a β-tubulin mutant. Our in vitro results have shown that Cdc2p directly phosphorylates Bub1p at these CDK consensus sites. A Bub1-P*p mutant, which cannot be phosphorylated on the four CDK consensus sites, has a spindle checkpoint defect, although the defect is not as pronounced as in the bub1Δ mutant. In vivo observation of Bub1-P*–GFPp revealed that Cdc2p-dependent phosphorylation of Bub1p is not required to maintain the kinetochore localization of the enzyme during spindle damage. It has also been shown that human Bub1p is phosphorylated when the spindle is disrupted, but not during normal mitosis (Taylor et al., 2001). There are putative CDK sites present in Bub1p from human, mouse, fly, budding yeast and Xenopus laevis, although the position of the sites is not conserved. Therefore it is possible that there may be a role for CDK phosphorylation of Bub1p in the mitotic spindle of eukaryotes generally. In this respect, Xenopus Bub1p has been shown to be directly phosphorylated by Cdc2p in vitro (Schwab et al., 2001). However, the function of this phosphorylation is still unclear, as the kinase activity of Bub1p was not affected by this phosphorylation (Schwab et al., 2001). In fission yeast, the main function affected by Cdc2p-dependent phosphorylation is unlikely to be Bub1p kinase activity, as the effects of mutations in phosphorylation sites and kinase activity regarding sensitivity to MT-destabilizing drugs are additive. One hypothesis would be that Cdc2p-dependent phosphorylation of Bub1p regulates signal transmission to downstream effectors like Mad3p (Millband and Hardwick, 2002). Alternatively, CDK may regulate Bub1p interaction with other spindle checkpoint components at the kinetochore, as previously shown for the budding yeast Mps1p kinase, which is required for formation of the Mad1p/Bub1p/Bub3p complex (Brady and Hardwick, 2000).
Strains lacking the kinase domain of Bub1p (bub1-Δkinase) or with a point mutation in the kinase domain (bub1-K762R) are partially checkpoint defective. In Xenopus, it has been reported that Bub1p kinase activity is required for metaphase arrest (Tunquist et al., 2002), although recruitment of other spindle checkpoint proteins does not require Bub1p kinase activity (Sharp-Baker and Chen, 2001). Bub1-Δkinase–GFPp and Bub1-K762R– GFPp are able to localize to kinetochores during spindle damage, indicating that Bub1p kinase activity is not required for Bub1p recruitment to the kinetochores. When both the CDK sites and the kinase domain of Bub1p are mutated, cells are more sensitive to MT-destabilizing drugs, but are less sensitive than bub1Δ cells. We conclude that both CDK phosphorylation and kinase activity are important for Bub1p function, and that Bub1p may also have a role in the structural maintenance of the mitotic kinetochore.
Unlike the situation in mitosis, we found that Bub1p becomes hyperphosphorylated at CDK sites during meiosis. This hyperphosphorylation occurs during MI, after meiotic prophase. Sister chromatids of chromosome I separate in 14% of the bub1-P* cells during MI compared with 3% in WT cells, indicating that Cdc2p-dependent phosphorylation of Bub1p plays some role in chromosome segregation during MI. Xenopus Bub1p has also been reported to be regulated by phosphorylation through the MAPK pathway during MI and MII, and it has been suggested that phosphorylation may be required for metaphase arrest in oocytes (Schwab et al., 2001). Deletion of the kinase domain of Bub1p has a greater effect on chromosome segregation in MI, with equational segregation of chromosome I occurring in 43% of the bub1-Δkinase cells. There is also a premature loss of centromeric Rec8–GFPp in this mutant, although the Bub1-Δkinase–GFPp protein is still localized at the kinetochores. Bub1Δ cells show 45% equational segregation, indicating that Bub1p function in MI is largely dependent on its kinase domain. The inability of Bub1-Δkinasep to maintain centromeric Rec8p suggests that the role of Bub1p in meiosis may be to phosphorylate kinetochore components that specifically protect centromeric cohesin from being degraded or delocalized. The maintenance of centromeric cohesion may reflect a ‘local’ effect of Bub1p kinase activity since the majority of Rec8p, found along the chromatid arms, is degraded in anaphase I (Watanabe and Nurse, 1999).
Live observation of GFP–α-tubulin in rec7-146 mutants revealed that progression through MI was delayed (Molnar et al., 2001). We found that MI spindle elongation time was increased by ∼30 min compared with WT cells, with the spindle undergoing slow elongation followed by regression before the final elongation step. Deletion of the bub1 gene abolishes the MI delay in rec7 mutants, demonstrating that there is a bub1-dependent delay during meiotic progression. The delay in meiotic progression may be analogous to the spindle checkpoint during mitosis because defects in homologous chromosome pairing are likely to reduce the efficiency of kinetochore capture by MTs. Observation of CDK Cdc2–YFPp in rec7-146 and rec14Δ diploids revealed that, like in mitosis, CDK is likely to be a target of the spindle checkpoint in meiosis, since Cdc2–YFPp remains associated with the spindle of separating nuclei, in contrast to WT cells, where the CDK leaves the metaphase spindle before anaphase I (Decottignies et al., 2001). The increased association time of Cdc2–YFPp with the spindle was no longer observed in a rec7-146 bub1Δ (or rec14Δ bub1Δ) diploid, suggesting that, in recombination-deficient mutants, a bub1- dependent checkpoint signal is sent, resulting in Cdc2p remaining on the spindle. Like the mitotic spindle checkpoint, some checkpoint function remains when the kinase domain of Bub1p is deleted. On the other hand, Cdc2p-dependent phosphorylation of Bub1p plays only a minor role in MI delay.
We have identified a new role for Cdc2p in the Bub1p-dependent spindle checkpoint control in fission yeast. Phosphorylation by a CDK is required for complete function of Bub1p during spindle damage, and may also have a role during meiosis I. Cdc2p-dependent regulation of Bub1p and the kinase activity of Bub1p appear to act in different pathways, and Bub1p kinase activity is required to establish proper kinetochore function. The conservation of Bub1p kinases from yeast to human suggests that similar controls may operate in higher eukaryotes.
Materials and methods
Fission yeast strains and methods
All experiments were carried out in Edinburgh minimal media (EMM2) or rich media (YE5S), and growth conditions are as described previously (Moreno et al., 1991). Strains used in this study are shown in Table I. Temperature-sensitive strains were grown exponentially at 25°C and shifted to 36°C, the nda3-km311 strain was grown at 32°C and shifted to 18°C, and other strains were grown at 32°C. Yeast transformation was performed as described previously (Bähler et al., 1998). Septation was scored by calcofluor staining and nuclear division by DAPI staining, after fixing cells in 70% ethanol.
Table I. Schizosaccharomyces pombe strains used in this study.
| PN4252 | bub1::bub1-HA[kanR] leu1-32 h- <bub1-HA> | |
| PN4262 | cdc25–22 bub1-HA leu1-32 h- | |
| PN3822 | nda3-km311 leu1– his2– h+ | T.Toda |
| PN4270 | bub1::bub1-P*-HA[kanR] leu1-32 ura4-D18 h+ <bub1-P*-HA> | |
| PN4465 | nda3-km311 bub1-HA his2– leu1-32 h+ | |
| PN4163 | nda3-km311 bub1-P*-HA leu1-32 h+ | |
| PN72 | cdc2-M26 leu1-32 h- | |
| PN4253 | bub1-HA cdc2-M26 leu1-32 h+ | |
| PN4273 | bub1-P*-HA cdc2-M26 leu1-32 h+ | |
| PN4255 | bub1Δ::ura4+ ura4-D18 leu1-32 h- | Bernard et al. (1998) |
| PN1 | 972 h- | |
| PN4320 | bub1::bub1-P*[ura4+] ura4-D18 leu1-32 h+ | |
| PN2647 | mad2Δ::ura4+ ura4-D18 leu1-32 h- | Kim et al. (1998) |
| PN4294 | bub1Δ::ura4+ cdc2-M26 leu1-32 ura4-D18 h- | |
| PN4329 | cdc2-M26 bub1-P*[ura4+] ura4-D18 leu1-32 h- | |
| PN4091 | bub1::bub1-GFP[kanR] leu1-32 ura4-D18 ade6-704 h- | |
| PN4464 | bub1::bub1-P*-GFP[kanR] ura4-D18 leu1-32 h- | |
| PN4157 | bub1::bub1Δkinase-GFP[kanR] ura4-D18 leu1-32 ade6-704 h- | |
| PN4298 | bub1::bub1Δkinase[kanR] leu1-32 h- | |
| PN4260 | bub1Δ::ura4+ mad2Δ::ura4+ ura4-D18 leu1-32 h+ | |
| PN4472 | bub1::bub1Δkinase[kanR] mad2Δ::ura4+ ura4-D18 leu1-32 h- | |
| PN4506 | mad2Δ::ura4+ ura4-D18 bub1-P*-Δkinase[kanR] leu1-32 | |
| PN4353 | bub1::bub1-K762R leu1-32 h- | |
| PN4628 | bub1::bub1-K762R-HA[kanR] ura4-D18 leu1-32 | |
| PN4156 | bub1::bub1Δkinase-HA[kanR] ura4-D18 leu1-32 ade6-704 h- | |
| PN4347 | bub1::bub1-K762R-GFP[kanR] ura4-D18 leu1-32 h- | |
| PN4466 | bub1::bub1-K762R cdc2-M26 leu1-32 h+ | |
| PN4330 | bub1::bub1Δkinase[kanR] cdc2-M26 leu1-32 h- | |
| PN4340 | cdc2-M26 bub1-P*-Δkinase[kanR] leu1-32 | |
| PN4259 | bub1-HA/bub1-HA pat1-114/pat1-114 leu1-32/leu1-32 ade6-M210/ade6-M216 | |
| PN4297 | pat1-114/pat1-114 bub1-P*-HA/bub1-P*-HA leu1-32/leu1-32 | |
| PN4295 | pat1-114/pat1-114 bub1-HA/bub1-HA mei4Δ::ura4+/mei4Δ::ura4+ leu1-32/leu1-32 ura4-D18/ura4-D18 ade6-M210/ade6-M216 | |
| PN4274 | pat1-114/pat1-114 bub1-HA/bub1-HA mes1Δ::LEU2/ mes1Δ::LEU2 leu1-32/ leu1-32 ade6-M210/ade6-M216 | |
| MKY7A-4 | LacO[lys1+] GFP-LacI-NLS[his7+] ura4– leu1– h+ <cen1-GFP> | Nabeshima et al. (1998) |
| PN4095 | bub1Δ::ura4+ ura4-D18 leu1-32 ade6-M210 cen1-GFP h+ | |
| PN4468 | bub1-P*[ura4+] cen1-GFP leu1-32 ura4-D18 h+ | |
| PN4303 | bub1Δkinase[kanR] cen1-GFP leu1-32 h+ | |
| PN4354 | bub1-K762R cen1-GFP ura4-D18 leu1-32 h+ | |
| PN4144 | mad2Δ::ura4+ ura4-D18 leu1-32 cen1-GFP ade6– h+ | |
| PN2377 | rec8::rec8-GFP[kanR] h- <rec8-GFP> | Watanabe and Nurse (1999) |
| PN4106 | rec8-GFP bub1Δ::ura4+ ura4-D18 ade6-M210 h+ | |
| PN4309 | rec8-GFP bub1Δkinase[kanR] h+ | |
| PN4467 | rec8-GFP bub1-K762R h+ | |
| PN4469 | rec8-GFP bub1-P*[ura4+] ura4– h+ | |
| PN4145 | rec8Δ::kanR bub1-GFP[kanR] ura4-D18 h+ | |
| GP985 | rec7-146::Tn10LLK ade6-M26 leu1-32 ura4-294 h+ <rec7-146> | G.Smith |
| PN4068 | cdc2-YFP[sup3-5] Δnmt1-cdc2::kanR leu1-32 ura4-D18 ade6-704 h-<cdc2-YFP> | Decottignies et al. (2001) |
| PN4470 | rec7-146 cdc2-YFP ura4– leu1-32 h- | |
| PN4474 | rec7-146 cdc2-YFP bub1Δ::ura4+ ura4– leu1-32 h- | |
| PN4473 | rec7-146 cdc2-YFP bub1Δkinase[kanR] ura4– leu1-32 h- | |
| PN4476 | rec7-146 cdc2-YFP bub1-P*[ura4+] ura4– leu1-32 h- | |
| PN4357 | bub1::nmt1-bub1[kanR] cdc2-YFP leu1-32 ura4-D18 h- | |
| PN4617 | rec7-146::Tn10LLK cdc2-YFP bub1-P*-Δkinase[kanR] leu1-32 ura4-D18 | |
| PN4502 | rec7-146::Tn10LLK cdc2-YFP mad2Δ::ura4+ ura4-D18 leu1-32 | |
| GP2004 | rec14Δ::LEU2 leu1-32 ade6-52 h- | G.Smith |
| PN3774 | leu1-32 ura4D18 nmt1::GFP-α-tubulin integrant h- <GFP-tub> | Yamamoto et al. (1999) |
| PN4622 | GFP-tub bub1Δ::ura4+ ura4-D18 leu1-32 | |
| PN4623 | GFP-tub rec7-146::Tn10LLK leu1-32 | |
| PN4624 | GFP-tub rec7-146::Tn10LLK bub1Δ::ura4+ leu1-32 ura4-D18 | |
| PN4626 | GFP-tub rec7-146::Tn10LLK bub1-Δkinase[kanR] leu1-32 ura4-D18 | |
| MAG071 | mad2::mad2-GFP[kanR] leu1-32 ura4– h- | Garcia et al. (2001) |
| PN4619 | mad2-GFP[kanR] cdc2-M26 leu1-32 | |
| PN4620 | mad2-GFP[kanR] bub1Δ::ura4+ cdc2-M26 ura4-D18 leu1-32 |
Western blotting and phosphatase treatment
Cells were boiled for 6 min after washing with STOP buffer (150 mM NaCl, 50 mM NaF, 1 mM NaN3, 10 mM EDTA pH 8.0). Protein extracts were made by vortexing with glass beads in HB buffer (Moreno et al., 1991) and protein concentration was determined with the BCA assay kit (Sigma). Cell extracts were boiled in 2× sample buffer for 3 min and 50 µg of each sample were loaded onto an 8% SDS–polyacrylamide gel (Laemmli, 1970). Visualization of Bub1p phosphorylation was achieved using acrylamide containing low bis concentration (200:1) (Lindsay et al., 1998). Proteins were transferred onto Immobilon™-P membrane (Millipore) and detected using ECL (Amersham). The antibodies used were anti-HA monoclonal antibody 12CA5 (1:1000; Babco), anti-Cdc13p monoclonal antibody 6F (1:500; Hayles and Steel), anti-Cdc2p monoclonal antibody Y63 (1:200; Yamano et al., 1996) and anti-GST polyclonal antibody GST1 (1:2000; Sigma). λ phosphatase treatments were performed as described previously (Yamaguchi et al., 2000), using λ phosphatase (New England Biolab).
Site-directed mutagenesis of CDK phosphorylation consensus sites and kinase domain
The serine and threonine residues of the four CDK phosphorylation consensus sites S/T-P-X-K/R (T340, T423, T455 and S466) and K762 were changed to alanine and arginine respectively, using the Quikchange site-directed mutagenesis kit (Stratagene). The mutated clones were confirmed by sequencing of the open reading frame. The genomic bub1 fragment carrying the mutations was then transformed into bub1Δ::ura4+ cells and integrants were selected for 5-FOA resistance and confirmed by colony PCR.
Construction of GST–Bub1-Δkinasep
The 4.5 kb KpnI/DraIII fragment containing bub1+ ORF was isolated from cosmid SPCC1322 (kindly provided by the Sanger Center). The purified 4.5 kb was subsequently digested by BamHI and cloned into BamHI/KpnI-cleaved pSK. The bub1Δkinase gene was isolated from EcoRI/BamHI-cleaved pSK::bub1+and cloned into EcoRI/BamHI-cleaved pGEX-KT (Hakes and Dixon, 1992). The 1.2 kb BamHI/SacI fragment of pGEX-KT::bub1Δkinase was replaced with a 0.8 kb PCR fragment amplified on S.pombe genomic DNA using the 5′-CGGGATCCATGTCCGATTGGCGG and 5′-CGAGCTCTTATCATAGAGCAGG primers. The resulting pGEX-KT::short bub1Δkinase plasmid allows bacterial expression of GST–Bub1Δkinasep. The same strategy was used to get pGEX-KT::short bub1-P*-Δkinase.
Purification of GST–Bub1-Δkinasep
Bacteria cells (JM109) carrying pGEX-KT::short bub1Δkinase or pGEX-KT::short bub1-P*-Δkinase plasmid were grown to OD595 = 0.3 and protein expression was induced by adding 1 mM IPTG for 4 h at 25°C. Extracts were purified using glutathione–Sepharose 4 fast flow (Amersham Bioscience). Protein concentration was estimated on gel by Coomassie Blue staining.
In vitro phosphorylation
Five microliters of anti-Cdc2p polyclonal antibody C2 (Simanis and Nurse, 1986) were incubated in the presence of Sepharose bead-coupled protein A in HB buffer (Moreno et al., 1991) for 30 min at room temperature. After three washes with HB buffer, 5 mg of S.pombe cell extracts were added to the beads and rotated for 3 h at 4°C. Immunoprecipitates (S.pombe Cdc2p) were washed three times with HB and incubated with 200 µM ATP, 40 µCi/ml [γ-32P]ATP, and GST–Bub1-Δkinasep, GST–Bub1-P*-Δkinasep or GST (Sigma) at 30°C for 30 min. In vitro phosphorylation by H.sapiens Cdc2p (New England Biolab) was performed in the same conditions. The reactions were stopped by boiling for 3 min in the presence of 2× SDS sample buffer. Samples were analysed by SDS–polyacrylamide gel.
pat1ts-induced meiosis
To induce meiosis, pat1-114/pat1-114 ade6-M216/M210 leu1-32/leu1-32 cells were grown to mid-exponential phase at 25°C in minimal medium containing 1% glucose, 5 g/l NH4Cl and 0.1 g/l leucine. Cells were washed three times, shifted to minimal medium containing 1% glucose and 0.05 g/l leucine and incubated for 16 h at 25°C to arrest the cells in G1. Cells were then shifted to 34°C to inactivate Pat1p and at the same time 0.5 g/l NH4Cl and 0.05 g/l leucine were added. Samples were taken every 30 min for 8 h and processed for western blot analysis, FACS analysis and DAPI staining.
Cell methods
FACS analysis was performed as described previously (Sazer and Sherwood, 1990) (Becton Dickinson).
The sister chromatid segregation pattern during MI was scored by using cen1–GFP strains (Nabeshima et al., 1998). Cells of opposite mating types, one of which carried cen1–GFP, were crossed and cells in anaphase I were scored.
Spore viability was scored using tetrad analysis.
Live fluorescence microscopy was performed as described in Decottignies et al. (2001). For Figure 8B, cells with MI spindles were observed every 10 min for 50 min.
GFP- and HA-C-terminus tagging of bub1 was perfomed as described previously (Bähler et al., 1998).
For mitotic block experiments, cdc2-M26 strains were grown to exponential phase in rich medium at 25°C, shifted to 36°C and incubated for 3 h. Cells were then released to 25°C in the presence of 50 µg/ml MBC for 40 min. When the cells were shifted back to 36°C, additional MBC was added to bring the final concentration to 100 µg/ml.
For the TBZ sensitivity test, TBZ was dissolved in DMSO at 10 mg/ml and added to YE5S plates at a final concentration of 0 (DMSO only), 10, 15 or 20 µg/ml. Cells were grown to 1 × 107 cells/ml in rich medium and serial dilutions (1:10) of the cells were spotted onto the plates.
Acknowledgments
Acknowledgements
We would like to thank Drs Jean-Paul Javerzat, Tomohiro Matsumoto, Gerald Smith, Yasushi Hiraoka, Takashi Toda, Hiroyuki Yamano and Mitsuhiro Yanagida for providing us with strains and antibodies. We would like to thank Drs Heidi Browning, Rafael Carazo-Salas, Naomi Fersht, Damien Hermand, Klaus Leonhard, Frank Uhlmann and Takashi Toda for critical reading of the manuscript. We would like to thank Drs Annie Borgne, Yoshinori Watanabe and Stefan Weitzer for technical advice. We would like to thank all the people in the Cell Cycle Laboratory for their help and support. This work was supported by Cancer Research UK. S.Y. was supported by a Japan Society for the Promotion of Science predoctoral fellowship. A.D. was supported by the European Commission, Biotechnology Programme and Cancer Research UK.
References
- Amon A. (1999) The spindle checkpoint. Curr. Opin. Genet. Dev., 9, 69–75. [DOI] [PubMed] [Google Scholar]
- Anderson S.S. (2000) Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol., 10, 261–267. [DOI] [PubMed] [Google Scholar]
- Bähler J., Schuchert,P., Grimm,C. and Kohli,J. (1991) Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr. Genet., 19, 445–451. [DOI] [PubMed] [Google Scholar]
- Bähler J., Wyler,T., Loidl,J. and Kohli,J. (1993) Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J. Cell Biol., 121, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J., Wu,J.Q., Longtine,M.S., Shah,N.G., McKenzie,A.,III, Steever,A.B., Wach,A., Philippsen,P. and Pringle,J.R. (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast, 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa,H., Logarinho,E., Li,Z., Williams,B.C., Lopes,C., Sunkel,C.E. and Goldberg,M.L. (1999) Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol., 146, 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Hardwick,K. and Javerzat,J.P. (1998) Fission yeast bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol., 143, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P., Maure,J.F. and Javerzat,J.P. (2001) Fission yeast Bub1 is essential in setting up the meiotic pattern of chromosome segregation. Nat. Cell Biol., 3, 522–526. [DOI] [PubMed] [Google Scholar]
- Brady D.M. and Hardwick,K.G. (2000) Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr. Biol., 10, 675–678. [DOI] [PubMed] [Google Scholar]
- Buonomo S.B., Clyne,R.K., Fuchs,J., Loidl,J., Uhlmann,F. and Nasmyth,K. (2000) Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell, 103, 387–398. [DOI] [PubMed] [Google Scholar]
- Cahill D.P., Lengauer,C., Yu,J., Riggins,G.J., Willson,J.K., Markowitz,S.D., Kinzler,K.W. and Vogelstein,B. (1998) Mutations of mitotic checkpoint genes in human cancers. Nature, 392, 300–303. [DOI] [PubMed] [Google Scholar]
- Carr A.M., MacNeill,S.A., Hayles,J. and Nurse,P. (1989) Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 protein kinase gene: implications for cdc2+ protein structure and function. Mol. Gen. Genet., 218, 41–49. [DOI] [PubMed] [Google Scholar]
- Cervantes M.D., Farah,J.A. and Smith,G.R. (2000) Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell, 5, 883–888. [DOI] [PubMed] [Google Scholar]
- Decottignies A., Zarzov,P. and Nurse,P. (2001) In vivo localisation of fission yeast cyclin-dependent kinase cdc2p and cyclin B cdc13p during mitosis and meiosis. J. Cell Sci., 114, 2627–2640. [DOI] [PubMed] [Google Scholar]
- Evans D.H., Li,Y.F., Fox,M.E. and Smith,G.R. (1997) A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics, 146, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M.A., Vardy,L., Koonrugsa,N. and Toda,T. (2001) Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J., 20, 3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakes D.J. and Dixon,J.E. (1992) New vectors for high level expression of recombinant proteins in bacteria. Anal. Biochem., 202, 293–298. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y., Toda,T. and Yanagida,M. (1984) The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell, 39, 349–358. [DOI] [PubMed] [Google Scholar]
- Horie S., Watanabe,Y., Tanaka,K., Nishiwaki,S., Fujioka,H., Abe,H., Yamamoto,M. and Shimoda,C. (1998) The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol., 18, 2118–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis,L. and Roberts,B.T. (1991) S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell, 66, 507–517. [DOI] [PubMed] [Google Scholar]
- Jallepalli P.V. and Lengauer,C. (2001) Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer, 1, 109–117. [DOI] [PubMed] [Google Scholar]
- Kallio M., Eriksson,J.E. and Gorbsky,G.J. (2000) Differences in spindle association of the mitotic checkpoint protein Mad2 in mammalian spermatogenesis and oogenesis. Dev. Biol., 225, 112–123. [DOI] [PubMed] [Google Scholar]
- Kim S.H., Lin,D.P., Matsumoto,S., Kitazono,A. and Matsumoto,T. (1998) Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science, 279, 1045–1047. [DOI] [PubMed] [Google Scholar]
- Kleckner N. (1996) Meiosis: how could it work? Proc. Natl Acad. Sci. USA, 93, 8167–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li R. and Murray,A.W. (1991) Feedback control of mitosis in budding yeast. Cell, 66, 519–531. [DOI] [PubMed] [Google Scholar]
- Lin Y., Larson,K.L., Dorer,R. and Smith,G.R. (1992) Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics, 132, 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H.D., Griffiths,D.J., Edwards,R.J., Christensen,P.U., Murray,J.M., Osman,F., Walworth,N. and Carr,A.M. (1998) S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev., 12, 382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A., Sawin,K. and Mitchison,T. (1999) A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr. Biol., 9, 1423–1426. [DOI] [PubMed] [Google Scholar]
- Maller J., Gautier,J., Langan,T.A., Lohka,M.J., Shenoy,S., Shalloway,D. and Nurse,P. (1989) Maturation-promoting factor and the regulation of the cell cycle. J. Cell Sci. (Suppl.), 12, 53–63. [DOI] [PubMed] [Google Scholar]
- Millband D.N. and Hardwick,K.G. (2002) Fission yeast Mad3p is required for Mad2p to inhibit the anaphase-promoting complex and localizes to kinetochores in a Bub1p-, Bub3p- and Mph1p-dependent manner. Mol. Cell. Biol., 22, 2728–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millband D.N., Campbell,L. and Hardwick,K.G. (2002) The awesome power of multiple model systems: interpreting the complex nature of spindle checkpoint signaling. Trends Cell Biol., 12, 205–209. [DOI] [PubMed] [Google Scholar]
- Molnar M., Parisi,S., Kakihara,Y., Nojima,H., Yamamoto,A., Hiraoka,Y., Bozsik,A., Sipiczki,M. and Kohli,J. (2001) Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics, 157, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. (1995) Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays, 17, 471–480. [DOI] [PubMed] [Google Scholar]
- Nurse P. (2000) A long twentieth century of the cell cycle and beyond. Cell, 100, 71–78. [DOI] [PubMed] [Google Scholar]
- Ponticelli A.S. and Smith,G.R. (1989) Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics, 123, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B.T., Farr,K.A. and Hoyt,M.A. (1994) The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol. Cell. Biol., 14, 8282–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S. and Sherwood,S.W. (1990) Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci., 97, 509–516. [DOI] [PubMed] [Google Scholar]
- Schwab M.S., Roberts,B.T., Gross,S.D., Tunquist,B.J., Taieb,F.E., Lewellyn,A.L. and Maller,J.L. (2001) Bub1 is activated by the protein kinase p90(Rsk) during Xenopus oocyte maturation. Curr. Biol., 11, 141–150. [DOI] [PubMed] [Google Scholar]
- Seeley T.W., Wang,L. and Zhen,J.Y. (1999) Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem. Biophys. Res. Commun., 257, 589–595. [DOI] [PubMed] [Google Scholar]
- Sharp-Baker H. and Chen,R.H. (2001) Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3 and CENP-E, independently of its kinase activity. J. Cell Biol., 153, 1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda C., Hirata,A., Kishida,M., Hashida,T. and Tanaka,K. (1985) Characterization of meiosis-deficient mutants by electron microscopy and mapping of four essential genes in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 200, 252–257. [DOI] [PubMed] [Google Scholar]
- Shonn M.A., McCarroll,R. and Murray,A.W. (2000) Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science, 289, 300–303. [DOI] [PubMed] [Google Scholar]
- Simanis V. and Nurse,P. (1986) The cell cycle control gene cdc2+ of fission yeast encodes a protein kinase potentially regulated by phosphorylation. Cell, 45, 261–268. [DOI] [PubMed] [Google Scholar]
- Straight A.F. (1997) Cell cycle: checkpoint proteins and kinetochores. Curr. Biol., 7, R613–R616. [DOI] [PubMed] [Google Scholar]
- Taylor S.S., Hussein,D., Wang,Y., Elderkin,S. and Morrow,C.J. (2001) Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci., 114, 4385–4395. [DOI] [PubMed] [Google Scholar]
- Taylor S.S. and McKeon,F. (1997) Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell, 89, 727–735. [DOI] [PubMed] [Google Scholar]
- Tunquist B.J., Schwab,M.S., Chen,L.G. and Mallar,J.L. (2002) The spindle checkpoint kinase Bub1 and Cyclin E/Cdk2 both contribute to the establishment of meitoc metaphase arrest by cytostatic factor. Curr. Biol., 12, 1027–1033. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. and Nurse,P. (1999) Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature, 400, 461–464. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yokobayashi,S., Yamamoto,M. and Nurse,P. (2001) Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature, 409, 359–363. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Okayama,H. and Nurse,P. (2000) Fission yeast Fizzy-related protein srw1p is a G1-specific promoter of mitotic cyclin B degradation. EMBO J., 19, 3968–3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., West,R.R., McIntosh,J.R. and Hiraoka,Y. (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J. Cell Biol., 145, 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano H., Gannon,J. and Hunt,T. (1996) The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J., 15, 5268–5279. [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Muszynski,M.G. and Kelly Dawe,R. (1999) The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol., 145, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]