Abstract
Kid is a kinesin-like DNA-binding protein known to be involved in chromosome movement during mitosis, although its actual motor function has not been demonstrated. Here, we describe the initial characterization of Kid as a microtubule-based motor using optical trapping microscopy. A bacterially expressed fusion protein consisting of a truncated Kid fragment (amino acids 1–388 or 1–439) is indeed an active microtubule motor with an average speed of ∼160 nm/s, and the polarity of movement is plus end directed. We could not detect processive movement of either monomeric Kid or dimerizing chimeric Kid; however, low levels of processivity (a few steps) cannot be detected with our method. These results are consistent with Kid having a role in chromosome congression in vivo, where it would be responsible for the polar ejection forces acting on the chromosome arms.
Keywords: chromosome movement/kinesin-like protein/microtubule motor protein/mitosis/polar ejection force
Introduction
Mitosis is the process by which the genetic material is divided correctly between two daughter cells. Mitosis proceeds via the construction of and subsequent structural changes in the spindle. These processes are believed to be actuated by a growing number of recently discovered microtubule-based motor members of the kinesin superfamily and cytoplasmic dynein.
To separate chromosomes correctly into the opposite spindle poles, at least two microtubule-dependent forces are needed: the poleward force that moves chromosomes towards the spindle poles, and the polar ejection force that moves chromosomes towards the metaphase plate. The balance of two opposite forces must be controlled strictly at each stage in mitosis. It was reported previously that in addition to the poleward microtubule flux (Mitchison, 1989), several motors are most probably involved in the production of poleward-directed force at the kinetochore (Rieder and Salmon, 1998), including MCAK/XKCM1 (Maney et al., 1998; Desai et al., 1999), CENP-E (Lombillo et al., 1995; Schaar et al., 1997) and cytoplasmic dynein (Pfarr et al., 1990; Steuer et al., 1990; Echeverri et al., 1996), whilst chromokinesins (chromosome arm-localizing kinesins), such as human Kid (Tokai et al., 1996; Levesque and Compton, 2001), Xenopus XKid (Antonio et al., 2000; Funabiki and Murray, 2000), Drosophila nod (Zhang et al., 1990; Afshar et al., 1995a), Xenopus Xklp1 (Vernos et al., 1995) and human KIF4 (Lee et al., 2001), appear to be involved in the generation of the anti-polar force along the length of the chromosome arms. Recent reports have shown that MCAK/XKCM1 regulates microtubule dynamics in vivo and in vitro via its capacity for microtubule destabilization (Desai et al., 1999; Moores et al., 2002), and that nod (Matthies et al., 2001) and Xklp1 (Walczak et al., 1998) stabilize microtubule–chromosome interactions. Therefore, not all kinesin-like proteins are motors. It is important to investigate the molecular function of each kinesin-like protein in the spindle so as to elucidate the mechanism of the mitotic process.
Kid was identified originally as a kinesin-like DNA-binding protein in humans (Tokai et al., 1996). Kid is a member of the chromokinesin family (Lawrence et al., 2002). Subsequently Xkid, a Xenopus homolog of Kid, was identified (Antonio et al., 2000; Funabiki and Murray, 2000). This protein, Kid, is predicted to be composed of three domains, based on amino acid sequence: an N-terminal kinesin-like motor domain that contains the nucleotide- and microtubule-binding sites; a ‘stalk’ domain containing an α-helical region that may form a coiled-coil; and a C-terminal non-helical ‘tail’ domain containing a DNA-binding region. This amino acid sequence of the Kid motor domain shows ∼35% identity to the motor domains of other members of the kinesin superfamily, whilst the sequence of the remainder of the molecule shows little homology with those of other members, except that of Drosophila nod (Zhang et al., 1990). Nod shows significant homology in its DNA-binding helix–hairpin–helix 1 (HhH1) domain to Kid. This non-motor functional C-terminal sequence homology has been taken to suggest a correlation in function in vivo between Kid and nod. So far, Kid protein has been reported to possess a microtubule-activated ATPase and an ATP-dependent microtubule-binding affinity (Tokai et al., 1996); however, its characteristics as a motor protein are unknown.
The localization of Kid changes drastically during different cell cycle stages (Tokai et al., 1996; Levesque and Compton, 2001). In interphase, Kid is found exclusively in the nucleus, and the localization pattern is diffuse. In mitotic prophase, a fraction of Kid gradually accumulates at the microtubule-organizing center (MTOC), and then, in metaphase, Kid localizes around chromosomes and spindle poles. At this stage, Kid has been shown to distribute all along the length of mitotic chromosomes. On subsequent entry into anaphase, Kid is enriched at the spindle pole-proximal side of the chromosomes. These previous reports suggest that Kid plays a specific role in separating chromosomes into daughter cells.
Recently, it has been shown that Kid was required for chromosomal movement during mitosis. In vertebrate cultured cells, microinjection of Kid-specific antibodies results in loss of function in chromosome orientation and oscillation (Levesque and Compton, 2001). In Xenopus, chromosome congression is inhibited in the absence of Xkid (Antonio et al., 2000; Funabiki and Murray, 2000). These observations imply that Kid is essential for producing the polar ejection force that pushes chromosome arms away from the spindle poles. Additionally, Funabiki and Murray (2000) have shown that Xkid is degraded at anaphase based on the detailed examination of its molecular composition at metaphase and anaphase, and that its destruction by ubiquitin-mediated proteolysis is coupled to anaphase chromosome segregation. In vertebrate cells, Kid has been shown to interact with SIAH-1 protein [human homolog of the Drosophila seven in absentia (Sina)], which is involved in ubiquitin-mediated proteolysis of different target proteins, and ubiquitylation of Kid has been shown to be mediated by SIAH-1 protein (Germani et al., 2000), implying a possible role for Kid in controlling the progression of mitosis.
Interestingly, it was reported very recently that nod, which was thought to be involved in the polar ejection force, lacks motile properties and instead functions as a brake which attaches chromosomes to microtubules (Matthies et al., 2001). Although apparently not a motor, nod retains its ability to bind to microtubules and its microtubule-activated ATPase. Since the function of nod (Rasooly et al., 1991; Afshar et al., 1995b; Karpen et al., 1996) is thought to be similar to that of Kid, it has been unclear how the polar ejection force might be produced by Kid.
To understand more precisely the function of Kid in the process of spindle formation and the origin of the polar ejection force, it is important to determine whether Kid can actually act as a microtubule motor protein because no chromokinesin have been reported to be able to drive microtubule gliding in vitro. We expressed proteins containing the Kid motor domain, and then investigated the motor activity in an in vitro bead assay using optical tweezers. We found that Kid was indeed a microtubule motor, and that the polarity of movement was plus end-directed. A statistical analysis showed that monomeric and dimeric chimeric Kid was only slightly if at all mechanically processive under load. These results limit possible models for the in vivo function of Kid and are consistent with a proposed role for Kid protein in generating polar ejection forces that act on chromosome arms, as well as in chromosome orientation, oscillation and possibly congression.
Results
Expression and purification of recombinant Kid proteins
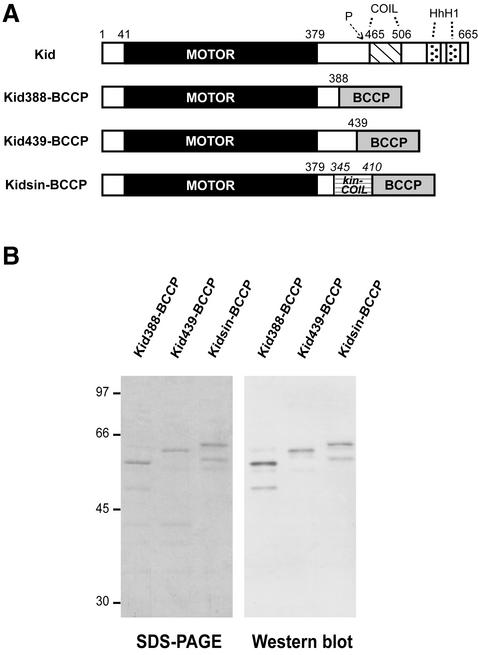
We prepared Kid388–BCCP and Kid439–BCCP which contain the N-terminal 388 and 439 residues, respectively, of human Kid fused to the Escherichia coli biotin carboxyl carrier protein (BCCP) (Figure 1A and B). BCCP was introduced to tether motor proteins to a bead or glass surface via streptavidin. Two kinds of bacterially expressed proteins were prepared by microtubule affinity as shown in Figure 1B. Cross-reaction on a corresponding immunoblot with an antibody against biotin confirmed the expression and purification of the BCCP-fused Kid protein (Figure 1B). These proteins are assumed to be single headed because of a lack of a coiled-coil structure (amino acids 465–506) as predicted by SMART (http://smart.embl-heidelberg.de). Since Kid has a cdc2 kinase phosphorylation consensus sequence (amino acids 463–466), Kid protein including the predicted coiled-coil amino acid sequence (amino acids 465–506) may be subject to regulation by phosphorylation. To avoid some effects of the possible regulation and to construct a dimeric Kid protein, we prepared a chimera (Kidsin–BCCP) that was composed of the Kid head domain (amino acids 1–379) and the Drosophila kinesin stalk domain (amino acids 345–410) (Figure 1A and B). Additionally, Drosophila truncated kinesin (kin410–BCCP and kin351–BCCP) and ncd (BCCP–MC1) were prepared (data not shown).
Fig. 1. Kid constructs used in this study. (A) Schematic representation of the Kid and chimera constructs compared with full-length Kid protein. The numbers show amino acid positions. MOTOR, kinesin-like motor domain including ATP hydrolysis site and microtubule-binding sites; P, cdc2 phosphorylation consensus site; COIL, predicted coiled-coil domain; HhH1, predicted helix–hairpin–helix domain; kin-COIL, kinesin’s coiled-coil domain; BCCP, biotin carboxyl carrier protein. (B) The expressed, purified proteins. Left: Coomassie Blue staining. Molecular weight markers (×103) are indicated on the left. Right: western blot probed with the anti-biotin antibody.
Oligomeric states of Kid proteins
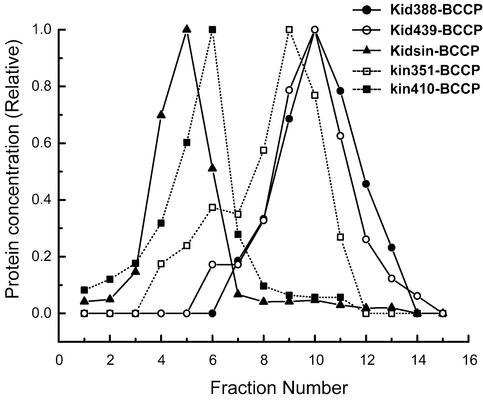
To determine whether Kid388–BCCP, Kid439–BCCP and Kidsin–BCCP are monomers or dimers, the physical properties of these proteins were analyzed by sedimentation. Previous reports have indicated that kin351–BCCP (Inoue et al., 2001) was single headed, whilst kin410– BCCP (Inoue et al., 2001) and BCCP–MC1 (Chandra et al., 1993; Endow and Higuchi, 2000) were doubled headed. Kid388–BCCP, Kid439–BCCP and Kidsin– BCCP were sedimented in sucrose density gradients. Kin351–BCCP, kin410–BCCP and bovine serum albumin (BSA) were used as standards. The sedimentation profiles of the proteins are shown in Figure 2. The peaks of Kid388–BCCP and Kid439–BCCP concentration were in similar positions to the peak fractions of kin351–BCCP and located at a lower mass position than that of BSA, whilst the peak fraction of Kidsin–BCCP was the same as that of kin410–BCCP and at a higher mass position than that of BSA. These results indicated that Kid388–BCCP and Kid439–BCCP exist as a monomer while Kidsin– BCCP exists as a dimer in vitro.
Fig. 2. Sedimentation profiles of motor proteins by sucrose density gradient centrifugation. Open squares, kin351–BCCP; filled squares, kin410–BCCP; filled circles, Kid388–BCCP; open circles, Kid439– BCCP; filled triangles, Kidsin–BCCP. The relative concentrations of motor proteins in each fraction were estimated from scans of Coomassie Blue-stained SDS–polyacrylamide gels. The peaks of Kid388–BCCP and Kid439–BCCP were in similar positions to the peak of kin351–BCCP used as a monomer control. Both sedimented at a lower mass position than the BSA standard peak at fraction number 8. The peak of Kidsin–BCCP was in the same position as kin410–BCCP, the dimer control, and at a higher mass position than that of BSA.
Bacterially expressed Kid has a microtubule motor activity
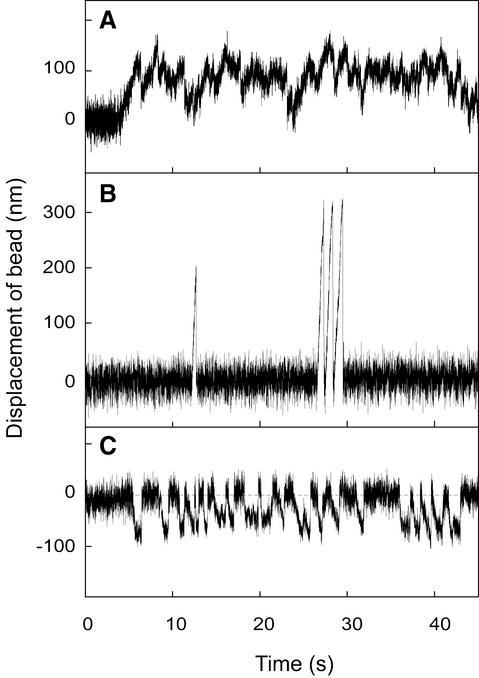
To test whether Kid protein exhibits motile activity on a microtubule, we attempted to demonstrate the ability of Kid to move along a microtubule using optical tweezers. Kid439–BCCP contains an in vivo post-translationally biotinylated domain at its C-terminus. This fusion protein is able to be adsorbed to the streptavidin-coated bead surface (Itakura et al., 1993), thereby avoiding the direct interaction of Kid’s N-terminal motor domain with the bare bead surface causing the inactivation of motor activity. When a bead coated with multiple molecules of Kid439–BCCP was positioned onto the microtubule with the use of optical tweezers, the bead in an optical trap moved along a microtubule adsorbed onto a glass surface and showed repeat ed attachment, force exertion and detachment (Figure 3A). In this way, Kid showed an obvious motile activity on a microtubule. Where Kid439–BCCP molecules were mixed in an ∼5000-fold molar excess over the beads, ∼50% of the beads in the trial were active, and bead velocity declined with increased load on the motors. The Kid388–BCCP- or Kidsin–BCCP-coated beads also showed similar motile behavior (data not shown). To determine the mean velocity of the Kid439–BCCP-coated beads, the traces of 25 bead displacements were averaged as described previously (Inoue et al., 1997), demonstrating that the velocity was ∼160 nm/s at forces of <1 pN (Table I).
Fig. 3. Bead displacement driven by motor proteins within an optical trap. (A) Kid439–BCCP, (B) kin410–BCCP and (C) BCCP–MC1. The traces refer to displacements along the microtubule axis, and plus endward displacements are positive. The trap stiffness was 0.019 pN/nm.
Table I. Summary of motile properties.
| Constructs | Oligomeric statea | Directionality | Velocity on a microtubule (nm/s) | Mechanical processivity |
|---|---|---|---|---|
| Kin410–BCCP | Dimer | Plus endb | 609 ± 16 (5)c | Processive |
| BCCP–MC1 | ND | Minus endb | 298 ± 10 (25) | Non-processive |
| Kid388–BCCP | Monomer | Plus endd | ND | Non-processive |
| Kid439–BCCP | Monomer | Plus endb | 156 ± 13 (25) | Non-processive |
| Kidsin–BCCP | Dimer | Plus endd | 160 ± 14 (20) | Non-processive |
aOligomeric states were estimated by sedimentation (see text and Figure 2).
bPolarity was determined with two methods (see text and Figure 4).
cMean ± SEM; values in parentheses are the numbers of the averaged traces of bead displacements at <1 pN.
dPolarity was determined using asymmetrically labeled fluorescent axoneme–microtubles.
ND, not determined in this study.
The motor polarity of Kid protein on a microtubule
To determine the direction of the Kid movement, we conducted a bead assay using dimly labeled fluorescent microtubules grown from the plus end of a brightly labeled fluorescent axoneme fragment. The axoneme fragment served only to mark the polarity. The Kid protein caused the bead to move towards the plus end of the microtubule (Figure 3A) in the same direction as kinesin (Vale et al., 1985) (Figure 3B) opposite to that of ncd (McDonald et al., 1990; Walker et al., 1990) (Figure 3C), indicating that Kid protein is a plus end-directed motor.
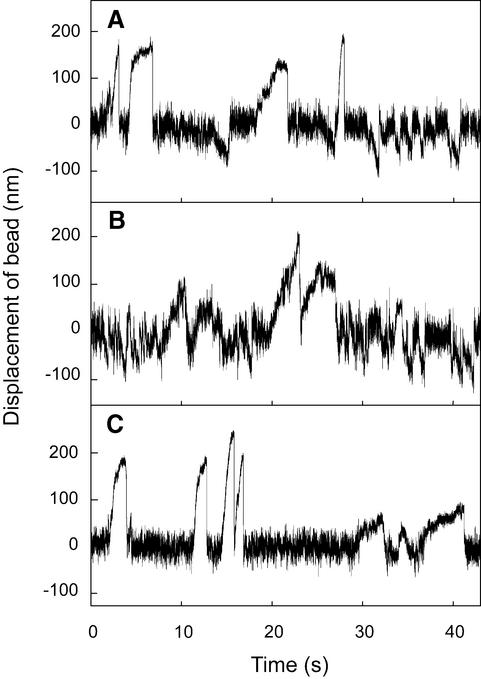
We confirmed the direction of Kid-induced movement using an independent method to observe the displaced direction of a bead attached to two kinds of motor molecules. In this assay, every pairwise combination of Kid439–BCCP, kin410–BCCP and BCCP–MC1 was mixed with streptavidin-coated beads at an appropriate ratio for observation of both motor activities. The bead coated with kinesin and ncd moved in both directions, but the velocity of bead movement was faster towards one end of the microtubule (Figure 4A). Presumably the faster displacements were generated by kinesin, the plus end-directed motor, and slower displacements by ncd, the minus end-directed motor. When coated with Kid and ncd, the beads moved bidirectionally with similar velocities (Figure 4B), indicating that there are oppositely directed motors on the bead. The motility of beads coated with Kid and kinesin was unidirectional, with fast and slow velocity phases (Figure 4C), suggesting that there are two kinds of motors of identical directionality on the bead. Taken together, we conclude that Kid is a plus end-directed motor, as is the case for some other N-terminal kinesin-related motor proteins.
Fig. 4. Displacement of the bead coated with two kinds of motor proteins. (A) Kin410–BCCP and BCCP-MC1 (1:10), (B) Kid439–BCCP and BCCP–MC1 (1:1), (C) Kid439–BCCP and kin410–BCCP (10:1). The molar ratios of the two proteins in solution before binding to the beads are in parentheses. The trap stiffness was 0.022 pN/nm.
The estimation of processivity of Kid
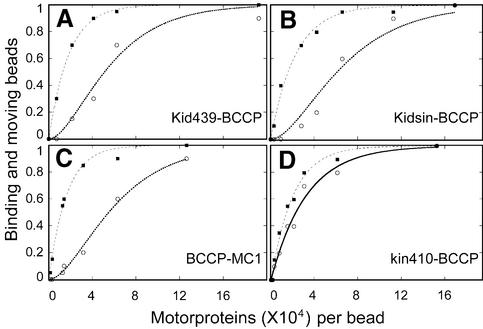
To examine whether single monomeric Kid439–BCCP molecules can move a bead (>30 nm), we prepared motor-coated beads at various molar ratios of motor proteins to beads and tested bead binding and movement in the presence of 1 mM adenylylimido-diphosphate (AMPPNP) or ATP, respectively. In the presence of AMPPNP, single kinesin-related motor proteins are expected to bind strongly to microtubules (Crevel et al., 1996). The fractions of binding and moving beads are shown in Figure 5. For Kid439–BCCP (Figure 5A), the plots of moving beads (open circles in Figure 5A) fit better to the curve inferred from the behavior of a unit of two or more molecules (χ2 = 0.025) than to the curve for the behavior of a single molecule unit (χ2 = 0.050, fitting curve not shown), while the plots of binding beads (filled squares in Figure 5A) fit better to the curve inferred from the behavior of a single molecule unit (χ2 = 0.001) than to that inferred from the behavior of a unit of two or more molecules (χ2 = 0.014, fitting curve not shown) (Svoboda and Block, 1994). Similar results were also obtained with Kidsin–BCCP and BCCP–MC1 (Figure 5B and C; and see legend). For kin410–BCCP, the minimum functional unit for both moving and binding beads was one (Figure 5D; and see legend), as found in previous reports (Inoue et al., 1997). These results indicated that the bead binding is due to a single molecular event of monomeric or dimeric chimeric Kid protein, and that the bead movement is due to multiple molecular events.
Fig. 5. Probability of moving and binding to beads at various molar ratios of four kinds of motor proteins at mixing. Filled squares and open circles denote the fraction of beads binding in 1 mM AMPPNP and moving in 1 mM ATP, respectively. (A) Kid439–BCCP, (B) Kidsin–BCCP and (C) BCCP-MC1. The plots are fitted to 1 – exp(–x/b) for binding beads (broken line) and 1 – (–x/m)exp(–x/m) for moving beads (dotted line), where x is the molar ratio of motor protein to beads following mixing, and b and m are the respective fitting parameters; b and m for Kid439–BCCP are 1849 and 3054, for Kidsin–BCCP are 2302 and 3622, and for BCCP–MC1 are 1643 and 3280. (D) Kin410–BCCP; the plots are fitted to 1 – exp(–x/2039) for binding beads (broken line) and 1 – exp(–x/3250) for moving beads (solid line).
Discussion
Kid has been shown previously to be involved in chromosome arm orientation, chromosome oscillation and congression on the metaphase plate of the mitotic spindle (Levesque and Compton, 2001). Also, previous work predicted that Kid would participate in the generation of the polar ejection force, although no motile activity has been detected hitherto (Tokai et al., 1996; Levesque and Compton, 2001). These studies indicated that if Kid is a plus end-directed motor, its DNA-binding properties suggest that it may directly push chromosomes away from the spindle poles. Our findings show that Kid is indeed a plus end-directed microtubule motor and strongly support previous proposals that the polar ejection force acting on chromosome arms is generated by the plus end-directed motor, Kid. Xkid in Xenopus is thought to be the homolog of human Kid, based on high sequence similarity, similar localization and identical microtubule- and DNA-binding properties (Antonio et al., 2000; Funabiki and Murray, 2000). Since the motor activity of human Kid in vitro is now shown, and since our construct Kid388, which excluded the DNA-binding domain probably unrelated to motility, has ∼60% identity with a corresponding Xkid sequence, we predict that Xkid too is a plus end-directed motor.
To examine the motile activity and polarity of Kid on microtubules, we initially tried a microtubule gliding assay and failed to demonstrate the motile behavior of Kid proteins under the conditions examined, even though kin410–BCCP, kin351–BCCP and BCCP–MC1 showed normal microtubule gliding speed, as previously reported (Berliner et al., 1994; Mazumdar and Cross, 1998; deCastro et al., 1999). When a bead assay with optical tweezers was carried out, the Kid-coated bead moved on microtubules. The reason for the different results from the two assays is unclear. Possibilities include a small number of contaminating inactive molecules, some difference between the surfaces, or particular aspects of Kid’s motor kinetics. The fact that Kid proteins showed the motor activity in a bead assay but not in a microtubule gliding assay indicates the usefulness of the bead assay equipped with optical tweezers when searching for motor activity of new (uncharacterized) kinesin-like proteins. A further advantage is that the bead assay provides a method to determine the direction of the movement without using a polarity-marked microtubule. By mixing the plus (kinesin) or minus (ncd) end-directed motor with the target motor protein and beads at the same time, the behavior of the bead indicates the motor polarity as described above. We do not know the mechanism by which one motor predominates over another for a while and then switches; but we note that these features are reminiscent of the directional instability of microtubule gliding driven by two motors of opposite polarity (Vale et al., 1992).
Kid’s localization over all the chromosome arms and on the spindle microtubules (Tokai et al., 1996; Levesque and Compton, 2001) suggested that multiple molecules of Kid work in teams as described previously (Howard, 1997; Rogers et al., 2001). Our findings that single monomeric and dimeric chimeric Kid molecules show no (or a very low level) mechanical processivity suggest that, in vivo, multiple Kid molecules are involved in chromosome movement. If Kid is a non-processive motor, working in teams may be important for the ‘processive’ chromosome movement, since members must act cooperatively in order to translocate chromosomes along mitotic microtubules. However, we do not know whether native Kid shows low mechanical processivity, because our truncated Kid (Kid388 and Kid439) and dimeric chimera (Kidsin) do not cover the entire sequence of the Kid molecule. Although our Kid constructs including BCCP appeared, by western blot, to be partially degraded (Figure 1B), we know that all the Kid protein in the preparation interacts with microtubules in an ATP-dependent manner, because the protein was purified through microtubule affinity. If the degraded material showed ATP-dependent binding but no motility, plots of binding and of moving beads would show a different form (Figure 5A and B). The fact that the plots of moving beads (open circles in Figure 5A and B) fit well to the curve (dotted line in Figure 5A and B) indicates that single intact Kid molecules cannot move processively. Additionally, our data cannot rule out the possibility of a low level of processivity: i.e. Kid molecules might take two or three discrete steps of 8 nm, because our microscopy cannot register the first ∼25 nm of travel after the initial binding of Kid to the microtubule. Another important point is that motor proteins were subjected to an external load in the optical trap (Schnitzer et al., 2000). If single Kid molecules are very sensitive to load, then Kid could not produce the relatively high force necessary to show processive movement in the optical trap. Our data indicate that the monomeric Kid and the dimeric chimera (Kidsin) have no, or very low, mechanical processivity under moderate external loads. Further study will be necessary to investigate the loaded (deCastro et al., 2000) and zero load (Yajima et al., 2002) processivity of native Kid.
There are other chromokinesins (Lawrence et al., 2002) that are able to bind DNA, including Drosophila nod protein. So far, Kid and nod protein are believed to play similar roles in the cell, except that Kid works in mitosis and nod in Drosophila meiosis. It was reported that nod bound to and was released from microtubules in a nucleotide-dependent way, but did not show motility in gliding assays (Matthies et al., 2001). Nod lacks the RXRP motif thought to be essential for motility, and Matthies et al. showed that inserting the nod motif into kinesin slowed down motility. Nonetheless, it remains in our view possible that nod is a motor, which for technical reasons does not show motility in sliding assays. The point can only be resolved by trying bead assays with nod. Kid protein, which has this conserved adenine-binding motif, actually produces vectorial transport as reported here. This result suggests that the roles of Kid and nod in regulation of the chromosome and spindle assembly/disassembly are different from each other, and that different chromokinesins play distinct roles, acting as brakes that cross-link chromosomes to microtubules, or to generate the anti-polar force necessary to move chromosomes away from spindle poles along microtubules. The motile activity, directionality and processivity of all chromosome arm chromokinesins except for nod and Kid are still unknown. The examination of motility of all chromokinesins in vitro will help us to understand the molecular mechanism of the chromosome movement in mitosis in vivo.
Materials and methods
Construction of plasmids for fusion proteins
To construct plasmids for expression of GST–Kid388–BCCP and GST–Kid439–BCCP (encoding residues 1–388 and 1–439, respectively), the corresponding regions of the Kid gene were amplified from pGEX-Kid (Tokai et al., 1996) using 5′ primers containing the BamHI sites and 3′ primers containing the EcoRI sites. Each PCR fragment was inserted into the BamHI–EcoRI sites of a modified pGEX-2T vector (termed pGEX–BCCP) to be tagged at its C-terminus with BCCP.
Plasmids expressing GST–kin351–BCCP and GST–kin410–BCCP were constructed from pGEX-KIN (Yang et al., 1990) (kind gift of L.S.B.Goldstein, University of California) by the same strategy as described above.
MC1 (encoding residues 195–685) was amplified from pBS-NCD (McDonald et al., 1990) (kind gift of L.S.B.Goldstein, University of California) with the 5′ primer containing a SmaI site and the 3′ primer containing a KpnI site. The resulting PCR product was cloned into the NruI–KpnI sites of the PinPoint Xa-1 vector (Promega). MC1 was thereby tagged at its N-terminus with BCCP.
The chimeric protein Kidsin, encoding residues 1–379 of Kid and 345–410 of kinesin, was constructed as follows. First, the neck region of kinesin (residues 345–410) was amplified from pGEX-KIN with the 5′ primer containing a SmaI site and the 3′ primer containing an EcoRI site. The resulting PCR product was cloned into the EcoRI–SmaI sites of pUC119 [termed pUC-(KN345–410)]. Next, the motor domain of Kid (residues 1–379) was amplified from pGEX-Kid with the 5′ primer containing a BamHI site and the 3′ primer containing a SmaI site. This PCR product was cloned into the BamHI–SmaI sites of pUC-(KN345–410) to create pUC-(Kid1-379)-(KN345–410). The SmaI site between Kid and kinesin was then eliminated by site-directed mutagenesis using the Mutant-K kit (Takara). Finally, the chimeric gene was inserted into the BamHI–EcoRI sites of pGEX–BCCP vector. The final chimeric gene product was composed of GST, a complete catalytic domain of human Kid, a coiled-coil neck domain of Drosophila kinesin and BCCP.
Expression and preparation of the recombinant proteins
To express GST–Kid388–BCCP, GST–Kid439–BCCP, GST–kin351– BCCP, GST–kin410–BCCP and GST–Kidsin–BCCP, the corresponding plasmids were transformed into Nova Blue cells (Novagen). Twelve hour cultures were diluted 1:25 into LB medium containing 50 µg/ml ampicillin and grown at 37°C until mid-log phase. After induction for 12 h in the presence of 0.1 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 23°C, the cells were harvested, and resuspended in lysis buffer (20 mM sodium phosphate pH 7.0, 4 mM MgCl2, 40 mM NaCl, 1 mM EGTA, 20 µM ATP and protease inhibitors), and then disrupted by sonication. High-speed supernatants containing the GST-fused proteins were incubated with glutathione–agarose beads (Sigma) for 30 min on ice. The fusion proteins were eluted from the glutathione beads with elution buffer [50 mM Tris–HCl pH 7.2, 150 mM NaCl, 1 mM MgCl2, 20 µM ATP, 5 mM dithiothreitol (DTT) and 10 mM glutathione). After binding to microtubules in the presence of 0.5 mM AMPPNP and 10 µM paclitaxel, the fusion proteins were digested by thrombin (6.25 U/ml). The motor domains (without GST) were then precipitated with microtubules and finally recovered in the supernatants following centrifugation in dissociation buffer (200 mM K-acetate, 10 mM Tris– acetate pH 7.5, 8 mM ATP, 8 mM MgCl2 and 10 µM paclitaxel). The concentration of proteins was determined by the method of Read and Northcote (1981) with the use of BSA as a standard.
BCCP–MC1 was expressed by a similar method, except that JM109 was used as the host and growth was at 25°C. BCCP–MC1 was purified by ion-exchange column chromatography on SP-Sepharose (Amersham Pharmacia Biotech). The 0.3 M NaCl fraction was used for microtubule affinity (Shimizu et al., 1995).
Western blots
The proteins were separated by SDS–PAGE and then transferred onto Immobilon transfer membrane (Millipore). After blocking with 2% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T), the membrane was first incubated with rabbit anti-biotin antibody (Enzo Diagnostics) and then phosphatase-conjugated anti-rabbit IgG antibody (Kirkegaard and Perry Laboratories) in TBS-T. The immunocomplexes were detected using the BCIP/NBT phosphatase substrate system (Kirkegaard and Perry Laboratories).
Sedimentation analysis
To examine the sedimentation profiles of Kid388–BCCP, Kid439–BCCP and Kidsin–BCCP, the purified proteins (1–10 µM) were analyzed by centrifugation (21 000 g, 24 h, 2°C) in 5–20% (w/v) sucrose density gradients in 50 mM Tris–HCl pH 7.6, 400 mM NaCl, 1 mM MgSO4 and 1 mM ATP using a swinging bucket rotor (P50S2, Hitachi). BSA, kin351–BCCP and kin410–BCCP were used as markers for standard protein, single- and double-headed kinesin. After centrifugation, the gradients were divided into 17 equal volume fractions. The gradient fractions were examined by SDS–PAGE (Laemmli, 1970). To quantitate the relative concentration of motor protein in each fraction, the Coomassie Blue-stained gels were scanned using a CCD camera within a linear response, and the integrated intensity quantitated using Densitograph (Atto).
Preparation of rhodamine-labeled and polarity-marked microtubules
Tubulin was prepared from porcine brain (Weingarten et al., 1974) and labeled with X-rhodamine succinimidyl ester (Molecular Probes) (Hyman et al., 1991). Fluorescent rhodamine-labeled microtubules were prepared by co-polymerizing fluorescent and non-fluorescent tubulin in a molar ratio of 1:20 and were stabilized by 20 µM paclitaxel (Hirakawa et al., 2000). Asymmetrically labeled fluorescent axoneme–microtubules were prepared as described (Vale and Toyoshima, 1988; Endow and Higuchi, 2000).
Microtubule gliding assay
Motility assays were performed as outlined (Itakura et al., 1993), with some modifications. Flow chambers of volume <15 µl were constructed by using two thin lines of silicone vacuum grease to form a channel between the slide glass and the coverslip (Matsunami). The following were added in sequence: 0.3 mg/ml biotinylated BSA was applied to a flow chamber, incubated, rinsed twice with K buffer (80 mM PIPES, 1 mM EGTA, 2 mM MgCl2 pH 6.8) containing 0.5 mg/ml BSA, rinsed twice with 0.5 mg/ml streptavidin, incubated, rinsed twice with K buffer, rinsed twice with 0.1 mg/ml BCCP-fused motor protein, incubated for 5 min, rinsed twice with 20 µM ATP in K buffer, and rinsed once with ∼0.015 mg/ml microtubules in K buffer containing 1 mM ATP and 20 µM paclitaxel. Microtubules were observed at 25–27°C under a dark field microscope (Olympus). Images were taken with an intensified CCD camera (ICD-6100, Ikegami) and recorded onto S-VHS videotape.
Bead motility assay
Bead motility assays were performed as described previously (Higuchi et al., 1997; Inoue et al., 1997). Data for displacements of the beads were passed through a 50 Hz low pass filter. The sampling time of each point was 2.5 ms. Experiments were performed at 25–27°C in K buffer for all the proteins, but for BCCP–MC1 in 50 mM K-acetate, 10 mM Tris-acetate pH 7.3, 1 mM MgCl2 and 1 mM EGTA. Both buffers contained 1 mM ATP or AMPPNP, 1.0 mg/ml casein, 20 µM paclitaxel and an oxygen-scavenging system (Higuchi et al., 1997). Motor protein-coated beads were prepared as described (Inoue et al., 1997). Streptavidin-coated beads were mixed with a solution of BCCP fusion proteins for 20 min at 25°C in the presence of ATP and then kept on ice. To determine the motor directionality on a microtubule, two kinds of motor proteins were incubated simultaneously with streptavidin-coated beads.
Statistical analysis of binding and moving motor protein-coated beads
We defined two kinds of behaviors of motor protein-coated beads in the optical trap on a microtubule: one behavior in which the beads remained bound to a microtubule in the presence of 1 mM AMPPNP (binding beads) and another in which it moved along a microtubule in the presence of 1 mM ATP (moving beads). For the statistical analysis of binding and moving beads, streptavidin-coated beads (9–40 pM) mixed with each motor protein (1–1000 nM) at a different molar ratio of protein to beads were tested in assays by placing a trapped bead in contact with a microtubule on the glass surface. Binding and movement by the bead were judged after placing the bead at three different positions of three microtubules for 20 s each. Data for binding and moving fractions were fitted with both the curve predicted for the case where binding and motility is due to single molecules and that predicted for the case where binding and motility requires two or more molecules. In both cases, it was assumed that the probability of binding and moving conformed to Poisson statistics (Svoboda and Block, 1994). Based on this kind of analysis, the number of minimum functional units participating in the attachment to a microtubule and in the movement along a microtubule could be estimated.
Acknowledgments
Acknowledgements
We thank H.Higuchi for valuable advice on optical trapping microscopy, L.S.B.Goldstein for kinesin and ncd clones, K.Shiroguchi and L.Yamaguchi for fluorescently labeled axoneme–microtubules, and R.Cross, N.Carter and D.Drummond for valuable discussion. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sport and Culture of Japan (to Y.Y.T.).
References
- Afshar K., Scholey,J. and Hawley,R.S. (1995a) Identification of the chromosome localization domain of the Drosophila nod kinesin-like protein. J. Cell Biol., 131, 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar K., Barton,N.R., Hawley,R.S. and Goldstein,L.S. (1995b) DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell, 81, 129–138. [DOI] [PubMed] [Google Scholar]
- Antonio C., Ferby,I., Wilhelm,H., Jones,M., Karsenti,E., Nebreda,A.R. and Vernos,I. (2000) Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell, 102, 425–435. [DOI] [PubMed] [Google Scholar]
- Berliner E., Mahtani,H.K., Karki,S., Chu,L.F., Cronan,J.E. and Gelles,J.,Jr (1994) Microtubule movement by a biotinated kinesin bound to streptavidin-coated surface. J. Biol. Chem., 269, 8610–8615. [PubMed] [Google Scholar]
- Chandra R., Salmon,E.D., Erickson,H.P., Lockhart,A. and Endow,S.A. (1993) Structural and functional domains of the Drosophila ncd microtubule motor protein. J. Biol. Chem., 268, 9005–9013. [PubMed] [Google Scholar]
- Crevel I.M.-T.C., Lockhart,A. and Cross,R.A. (1996) Weak and strong states of kinesin and ncd. J. Mol. Biol., 257, 66–76. [DOI] [PubMed] [Google Scholar]
- deCastro M.J., Ho,C.H. and Stewart,R.J. (1999) Motility of dimeric ncd on a metal-chelating surfactant: evidence that ncd is not processive. Biochemistry, 38, 5076–5081. [DOI] [PubMed] [Google Scholar]
- deCastro M.J., Fondecave,R.M., Clarke,L.A., Schmidt,C.F. and Stewart,R.J. (2000) Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat. Cell Biol., 10, 724–729. [DOI] [PubMed] [Google Scholar]
- Desai A., Verma,S., Mitchison,T.J. and Walczak,C.E. (1999) Kin I kinesins are microtubule-destabilizing enzymes. Cell, 96, 69–78. [DOI] [PubMed] [Google Scholar]
- Echeverri C.J., Paschal,B.M., Vaughan,K.T. and Vallee,R.B. (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol., 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A. and Higuchi,H. (2000) A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature, 406, 913–916. [DOI] [PubMed] [Google Scholar]
- Funabiki H. and Murray,A.W. (2000) The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell, 102, 399–402. [DOI] [PubMed] [Google Scholar]
- Germani A., Bruzzoni-Giovanelli,H., Fellous,A., Gisselbrecht,S., Varin-Blank,N. and Calvo,F. (2000) SIAH-1 interacts with α-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene, 19, 5997–6006. [DOI] [PubMed] [Google Scholar]
- Higuchi H., Muto,E., Inoue,Y. and Yanagida,T. (1997) Kinetics of force generation by single kinesin molecules activated by laser photolysis of caged ATP. Proc. Natl Acad. Sci. USA, 94, 4395–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa E., Higuchi,H. and Toyoshima,Y.Y. (2000) Processive movement of single 22S dynein molecules occurs only at low ATP concentrations. Proc. Natl Acad. Sci. USA, 97, 2533–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. (1997) Molecular motors: structural adaptations to cellular functions. Nature, 389, 561–567. [DOI] [PubMed] [Google Scholar]
- Hyman A., Drechsel,D., Kellogg,D., Salser,S., Sawin,K., Steffen,P., Wordeman,L. and Mitchison,T. (1991) Preparation of modified tubulins. Methods Enzymol., 196, 478–485. [DOI] [PubMed] [Google Scholar]
- Inoue Y., Toyoshima,Y.Y., Iwane,A.H, Morimoto,S., Higuchi,H. and Yanagida,T. (1997) Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc. Natl Acad. Sci. USA, 94, 7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Iwane,A.H., Miyai,T., Muto,E. and Yanagida,T. (2001) Motility of single one-headed kinesin molecules along microtubules. Biophys. J., 81, 2838–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura S., Yamakawa,H., Toyoshima,Y.Y, Ishijima,A., Kojima,T., Harada,Y., Yanagida,T., Wakabayashi,T. and Sutoh,K. (1993) Force-generating domain of myosin motor. Biochem. Biophys. Res. Commun., 196, 1504–1510. [DOI] [PubMed] [Google Scholar]
- Karpen G.H., Le,M.H. and Le,H. (1996) Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science, 273, 118–122. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lawrence C.J., Malmberg,R.L., Muszynski,M.G. and Dawe,R.K. (2002) Maximum likelihood methods reveal conservation of function among closely related Kinesin families. J. Mol. Evol., 54, 42–53. [DOI] [PubMed] [Google Scholar]
- Lee Y.M., Lee,S., Lee,E., Shin,H., Hahn,H., Choi,W. and Kim,W. (2001) Human kinesin superfamily member 4 is dominantly localized in the nuclear matrix and is associated with chromosomes during mitosis. Biochem. J., 360, 549–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque A.A. and Compton,D.A. (2001) The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol., 154, 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo V.A., Nislow,C., Yen,T.J., Gelfand,V.I. and McIntosh,J.R. (1995) Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J. Cell Biol., 128, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T., Hunter,A.W., Wagenbach,M. and Wordeman,L. (1998) Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J. Cell Biol., 142, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H.J., Baskin,R.J. and Hawley,R.S. (2001) Orphan kinesin NOD lacks motile properties but does possess a microtubule-stimulated ATPase activity. Mol. Biol. Cell, 12, 4000–4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M. and Cross,R.A. (1998) Engineering a lever into the kinesin neck. J. Biol. Chem., 273, 29352–29359. [DOI] [PubMed] [Google Scholar]
- McDonald H.B., Stewart,R.J. and Goldstein,L.S. (1990) The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell, 63, 1159–1165. [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. (1989) Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol., 109, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores C.A., Yu,M., Guo,J., Beraud,C., Sakowicz,R. and Milligan,R.A. (2002) A mechanism for microtubule depolymerization by KinI kinesin. Mol. Cell, 9, 903–909. [DOI] [PubMed] [Google Scholar]
- Pfarr C.M., Coue,M., Grissom,P.M., Hays,T.S., Porter,M.E and McIntosh,J.R. (1990) Cytoplasmic dynein is localized to kinetochores during mitosis. Nature, 345, 263–265. [DOI] [PubMed] [Google Scholar]
- Rasooly R.S., New,C.M., Zhang,P., Hawley,R.S. and Baker,B.S. (1991) The lethal(1)TW-6cs mutation of Drosophila melanogaster is a dominant antimorphic allele of nod and is associated with a single base change in the putative ATP-binding domain. Genetics, 129, 409–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read S.M. and Northcote,D.H. (1981) Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal. Biochem., 116, 53–64. [DOI] [PubMed] [Google Scholar]
- Rieder C.L. and Salmon,E.D. (1998) The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol., 8, 310–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K.R., Weiss,S., Crevel,I., Brophy,P.J., Geeves,M. and Cross,R. (2001) KIF1D is a fast non-processive kinesin that demonstrates novel K-loop-dependent mechanochemistry. EMBO J., 20, 5101–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar B.T., Chan,G.K., Maddox,P., Salmon,E.D. and Yen,T.J. (1997) CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol., 139, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer M.J., Visscher,K. and Block,S.M. (2000) Force production by single kinesin motors. Nat. Cell Biol., 2, 718–723. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Toyoshima,Y.Y., Edamatsu,M. and Vale,R.D. (1995) Comparison of the motile and enzymatic properties of two microtubule minus-end-directed motors, ncd and cytoplasmic dynein. Biochemistry, 34, 1575–1582. [DOI] [PubMed] [Google Scholar]
- Steuer E.R., Wordeman,L., Schroer,T.A. and Sheetz,M.P. (1990) Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature, 345, 266–268. [DOI] [PubMed] [Google Scholar]
- Svoboda K. and Block,S.M. (1994) Force and velocity measured for single kinesin molecules. Cell, 77, 773–784. [DOI] [PubMed] [Google Scholar]
- Tokai N., Fujimoto-Nishiyama,A., Toyoshima,Y., Yonemura,S., Tsukita,S., Inoue,J. and Yamamoto,T. (1996) Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J., 15, 457–467. [PMC free article] [PubMed] [Google Scholar]
- Vale R.D. and Toyoshima,Y.Y. (1988) Rotation and translocation of microtubules in vitro induced by dyneins from Tetrahymena cilia. Cell, 52, 459–469. [DOI] [PubMed] [Google Scholar]
- Vale R.D., Reese,T.S. and Sheetz,M.P. (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell, 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R.D., Malik,F. and Brown,D. (1992) Directional instability of microtubule transport in the presence of kinesin and dynein, two opposite polarity motor proteins. J. Cell Biol., 119, 1589–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernos I., Raats,J., Hirano,T., Heasman,J., Karsenti,E. and Wylie,C. (1995) Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell, 81, 117–127. [DOI] [PubMed] [Google Scholar]
- Walker R.A., Salmon,E.D. and Endow,S.A. (1990) The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature, 347, 780–782. [DOI] [PubMed] [Google Scholar]
- Walczak C.E., Vernos,I., Mitchison,T.J., Karsenti,E. and Heald,R. (1998) A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol., 8, 903–913. [DOI] [PubMed] [Google Scholar]
- Weingarten M.D., Suter,M., Littman,D.R. and Kirschner,M.W. (1974) Properties of the depolymerization products of microtubules from mammalian brain. Biochemistry, 13, 5529–5537. [DOI] [PubMed] [Google Scholar]
- Yajima J., Alonso,M.C., Cross,R.A. and Toyoshima,Y.Y. (2002) Direct long-term observation of Kinesin processivity at low load. Curr. Biol., 12, 301–306. [DOI] [PubMed] [Google Scholar]
- Yang J.T., Saxton,W.M., Stewart,R.J., Raff,E.C. and Goldstein,L.S. (1990) Evidence that the head of kinesin is sufficient for force generation and motility in vitro. Science, 249, 42–47. [DOI] [PubMed] [Google Scholar]
- Zhang P., Knowles,B.A., Goldstein,L.S. and Hawley,R.S. (1990) A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell, 62, 1053–1062. [DOI] [PubMed] [Google Scholar]