Abstract
The fungal phytotoxin fusicoccin stabilizes the interaction between the C-terminus of the plant plasma membrane H+-ATPase and 14-3-3 proteins, thus leading to permanent activation of the proton pump. This results in an irreversible opening of the stomatal pore, followed by wilting of plants. Here, we report the crystal structure of the ternary complex between a plant 14-3-3 protein, fusicoccin and a phosphopeptide derived from the C-terminus of the H+-ATPase. Comparison with the corresponding binary 14-3-3 complexes indicates no major conformational change induced by fusicoccin. The compound rather fills a cavity in the protein–phosphopeptide interaction surface. Isothermal titration calorimetry indicates that the toxin alone binds only weakly to 14-3-3 and that peptide and toxin mutually increase each others’ binding affinity ∼90-fold. These results are important for herbicide development but might have general implications for drug development, since rather than inhibiting protein–protein interactions, which is difficult to accomplish, it might be easier to reverse the strategy and stabilize protein–protein complexes. As the fusicoccin interaction shows, only low-affinity interactions would be required for this strategy.
Keywords: 14-3-3/crystal structure/drug design/fusicoccin/plant plasma membrane H+-ATPase
Introduction
Fusicoccin (FC) (Figure 1), a diterpene glycoside, is a wilt-inducing phytotoxin produced by the fungus Fusicoccum amygdali (Ballio et al., 1964). Despite the fact that the fungus is host specific, isolated FC exerts its effects in virtually any higher plant (Marre, 1979). The plant plasma membrane H+-ATPase (PMA) has been identified as the molecular target of FC action (Aducci et al., 1995). This P-type ATPase is responsible for building up an electrochemical proton gradient across the plasma membrane that provides the driving force for nutrient uptake and maintenance of cell turgor (Morsomme and Boutry, 2000). Changes of the latter are known to affect the osmotic swelling of the guard cells and consequently the opening of the stomatal pore, that regulates transpiration in plants. The proton pump is composed of 10 transmembrane helices locating both the N- and C-termini at the cytoplasmic face of the plasma membrane (Auer et al., 1998; Kühlbrandt et al., 2002). The enzyme’s C-terminus acts as an intrasteric inhibitor, the autoinhibitory activity of which is relieved by phosphorylation of the penultimate threonine residue and subsequent association with 14-3-3 proteins (Fuglsang et al., 1999; Svennelid et al., 1999; Maudoux et al., 2000).
Fig 1. The structure of the fungal toxin fusicoccin.
Members of the eukaryotic 14-3-3 family are highly conserved proteins that have been implicated in the regulation of diverse physiological processes by protein– protein interactions. 14-3-3 proteins bind to their numerous target proteins in a sequence-specific and phosphorylation-dependent manner (Sehnke et al., 2002; Tzivion and Avruch, 2002; Yaffe, 2002).
The proposed mechanisms of activity of 14-3-3 proteins on their ligands range from altering their substrate affinity, as observed for serotonin-N-acetyltransferase (Obsil et al., 2001), activating and/or inactivating the catalytic activity of a protein kinase such as c-Raf (Daum et al., 1994; Fantl et al., 1994; Morrison and Cutler, 1997), to regulating the intracellular localization as shown for the protein phosphatase Cdc25c (Peng et al., 1997; Dalal et al., 1999; Piwnica-Worms, 1999). Since 14-3-3 proteins are dimeric, they can also serve as scaffolds to bring together interaction partners such as the protein kinases Raf and Bcr (Braselmann and McCormick, 1995), to stabilize a dimeric binding partner or supply two binding sites for a monomeric partner (Yaffe, 2002).
In higher plants, 14-3-3 proteins are known to be involved in a series of cellular processes, such as the import of proteins into chloroplasts (May and Soll, 2000), the regulation of important metabolic enzymes and the regulation of defence mechanisms in response to stress (Finnie et al., 1999). They additionally have been identified as members of transcription complexes (Lu et al., 1992).
Binding of FC to the phosphorylated PMA–14-3-3 complex is thought to stabilize the PMA–14-3-3 interaction, thus leading to permanent activation of the proton pump (Jahn et al., 1997; Oecking et al., 1997). In order to analyse the molecular mode of FC action, we have determined the structure of plant 14-3-3c with and without a phosphorylated PMA peptide in the presence or absence of FC, and have examined the thermodynamic properties of the interaction between the three components.
Results and discussion
The peptide-binding site on tobacco 14-3-3c
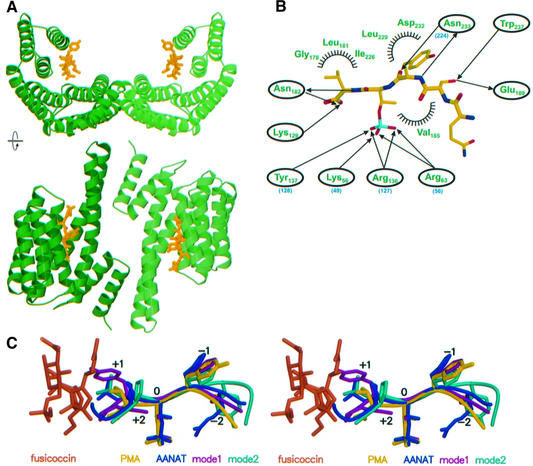
The structure of tobacco 14-3-3c was solved by molecular replacement using phases from human 14-3-3ζ (Liu et al., 1995), and shows 235 from a total of 260 residues. 14-3-3c forms the canonical dimer found in mammalian 14-3-3 proteins, with a root mean square deviation of the backbone Cα-atoms of 0.9 Å as compared with the human 14-3-3ζ. Each monomer consists of nine antiparallel helices that arrange in the form of a ‘U’ to build a large central binding cavity (Figure 2A). Structures of binary and ternary complexes of 14-3-3c with the phosphopeptide Gln-Ser-Tyr-pThr-Val (QSYpTV), conserved in plant H+-ATPases, and with FC were determined using the unliganded 14-3-3c as a starting model. Data for the crystal structure analysis of the different complexes are summarized in Table I.
Fig. 2. Phosphopeptide binding to plant 14-3-3. (A) Ribbon plot of two different orientations of the dimeric tobacco 14-3-3c protein (green) bound to the peptide Gln-Ser-Tyr-pThr-Val (yellow), which constitutes the C-terminal end of PMA2, a H+-ATPase isoform from Nicotiana plumbaginifolia. (B) Scheme of the interaction between peptide and protein, where half circles indicate residues forming van der Waals interactions, and arrows denote hydrophilic interactions between the indicated residues and the corresponding atoms of the peptide. The numbers of some conserved amino acids in human 14-3-3ζ are indicated in parentheses (blue). (C) Stereo representation of the superimposition of phosphopeptides from various 14-3-3 complex structures. The present structure is in yellow, with FC shown in orange, that of 14-3-3ζ with either serotonin N-acetyl transferase (AANAT; Obsil et al., 2001) in blue or model peptides (Yaffe et al., 1997; Rittinger et al., 1999) in turquoise and magenta.
Table I. Crystallographic data collection and refinement statisticsa.
| Native 14-3-3c | Peptide complex | Toxin complex | Ternary complex | |
|---|---|---|---|---|
| Measured reflections | 205 883 | 131 256 | 74 020 | 56 531 |
| Unique reflections | 15 163 | 21 328 | 14 444 | 13 170 |
| Resolution (Å) | 10–2.6 (2.7–2.6) | 10–2.3 (2.4–2.3) | 10–2.6 (2.7–2.6) | 10–2.7 (2.8–2.7) |
| Completeness (%) | 99.1 (99.4) | 95.5 (98.1) | 96.5 (98.3) | 96.8 (97.2) |
| I/σ | 33.1 (8.2) | 18.1 (4.0) | 14.1 (5.0) | 14.0 (4.2) |
| Rsym(%)b | 5.6 (35.4) | 5.8 (35.1) | 7.3 (30.1) | 6.6 (30.0) |
| Rcryst(%)c | 22.2 | 21.0 | 22.2 | 22.4 |
| Rfree(%)d | 25.7 | 24.4 | 25.8 | 26.3 |
| Protein atoms | 1838 | 1875 | 1846 | 1892 |
| Solvent molecules | 68 | 176 | 71 | 62 |
| R.m.s.d.e of bond lengths (Å) | 0.007 | 0.02 | 0.007 | 0.008 |
| R.m.s.d.e of bond angles (°) | 1.2 | 2.0 | 1.2 | 1.2 |
aData for the outermost shell are shown in parentheses.
bRsym = Σ|Ihi – <Ihi>|/ΣIhi, where Ihi is the scaled observed intensity of the ith symmetry-related observation of the reflection h and <Ihi> the mean value.
cRcryst = Σh|Foh – Fch|/ΣhFoh, where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h.
dRfree was calculated as Rcryst with 5% of the data omitted from structure refinement.
eR.m.s.d., root mean square deviations from ideal geometry.
The phosphopeptide occupies the central binding groove of 14-3-3c in an extended conformation. The phosphate moiety of the phosphothreonine forms electrostatic interactions with a positively charged patch formed by residues Lys56, Arg63 and Arg136, and a hydrogen bond to Tyr137 (Figure 2B). This indicates that high-affinity binding of 14-3-3 to PMA is dependent on phosphorylation. Indeed, binding could not be detected by applying a non-phosphorylated 16mer PMA peptide (Fuglsang et al., 1999) as well as a non-phosphorylated version of the peptide used in this study (data not shown). Additionally, there are a number of hydrogen bonds (cut-off limit 3.4 Å) between the peptide, mostly from the main chain, and conserved protein side chains. The peptide’s C-terminal carboxylate is contacted mostly by basic residues, and the side chains of the valine and tyrosine residues flanking the phosphothreonine form van der Waals contacts to the protein (Figure 2B). The structure confirms the notion that the C-terminal YTV motif is highly conserved in plant P-type H+-ATPases. From the 11 isoforms of Arabidopsis thaliana, eight have a YTV C-terminus, the others YTI, YTL and HTV.
A superimposition (Figure 2C) shows that the central portion of the PMA2 phosphopeptide is in a similar conformation and orientation as the 14-3-3-binding motif from a polyoma middle-T peptide (termed ‘mode 1’ peptide) and a library-derived peptide (‘mode 2’ peptide; Yaffe et al., 1997; Rittinger et al., 1999) or the phosphorylated serotonin N-acetyltransferase bound to 14-3-3ζ (Obsil et al., 2001). In these complexes, the interactions of the phosphate with two arginines, a lysine and one tyrosine are well conserved, with the corresponding residues Lys49, Arg56, Arg127 and Tyr128 in 14-3-3ζ. Some of the interactions of the main chain are similarly conserved, such as the double hydrogen bond between Asn233 in 14-3-3c (Asn224 from 14-3-3ζ) and the main chain CO and NH of the –1 peptide residue. The overlay shows that the orientation of the main chain and interactions of the side chains are deviating from the canonical structures in the –2 and beyond the +1 positions. The PMA2-binding peptide QSYpTV-COOH is different from the optimal consensus binding motifs, RSXpS/TXP (mode 1) and RXXXpS/TXP (mode 2; Yaffe et al., 1997), which are recognized by all mammalian 14-3-3 isoforms and are not located at the very C-terminal end of the interaction partner (Petosa et al., 1998; Obsil et al., 2001; Yaffe, 2002). The most significant difference from the canonical 14-3-3 complexes is thus the absence of residues beyond the +1 position, which would interfere sterically with FC binding (Figure 2C, see also below). In the case of the PMA peptide, the C-terminal valine is involved in a number of interactions with 14-3-3c which include the side chain and the carboxylate. The importance of the C-terminal residue for the affinity of FC binding is supported by mutagenesis studies (see below).
The ternary complex
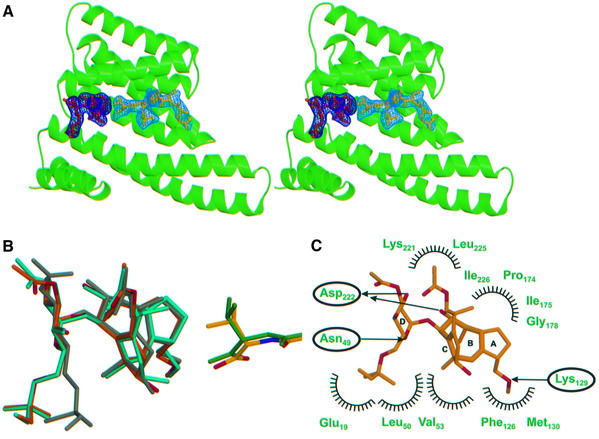
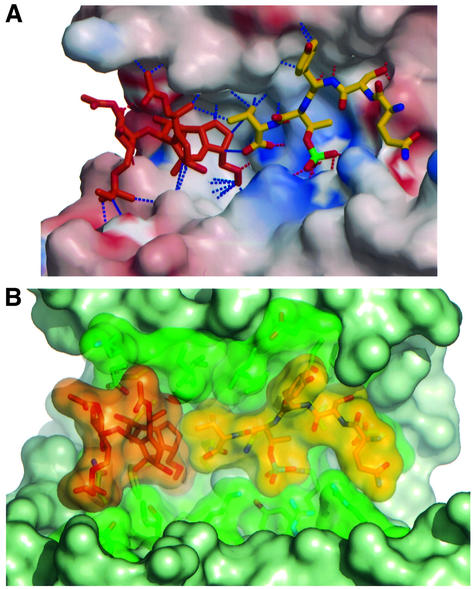
The structure of the ternary 14-3-3–FC–phosphopeptide complex shows that the toxin is accommodated in the large binding groove of 14-3-3 right next to the C-terminus of the peptide (Figure 3A). Comparison of the peptide conformation in the binary and ternary complexes indicates, as far as we can deduce from the present resolution, that the C-terminal valine adopts a different rotameric conformation to accommodate the toxin (Figure 3B). Whereas the glycosidic part of the phytotoxin (ring D in Figure 3C) is solvent exposed and forms two hydrogen bonds to Asn49 and Asp222 as well as some hydrophobic interactions (Figure 3C), the diterpene part is buried and makes extensive hydrophobic contacts to 14-3-3c, with two additional hydrogen bonds to Asp222 and Lys129 (Figure 3C). The more hydrophobic mode of FC binding to 14-3-3 is in contrast to the mostly hydrophilic nature of interaction of the PMA peptide. This is reflected in the electrostatic surface potential of the binding groove of 14-3-3, which is hydrophilic and positively charged in the peptide-binding area and mostly hydrophobic and uncharged in the FC-binding site (Figure 4A).
Fig. 3. The ternary 14-3-3–FC–peptide complex. (A) Ribbon diagram in stereo of a 14-3-3 monomer (green) with both peptide (yellow) and FC (orange) in the active site; the corresponding Fo – Fc OMIT maps (calculated with FC and the peptide omitted from the model) were contoured at the 2.0 σ level and are drawn in dark (FC) and light blue (peptide). (B) Superimposition of peptide (atom colours for ternary, green for binary) and FC (orange and turquoise) from the ternary and binary complexes, respectively. Also shown is the structure of unbound FC determined by X-ray crystallography (grey; Ballio et al., 1968), which has a similar conformation to the unbound FC NMR structure (Ballio et al., 1991). (C) Contacts between the toxin and the 14-3-3, with symbols as in Figure 22B; carbon and oxygen atoms are in orange and red, respectively.
Fig. 4. The fusicoccin effect. (A) Electrostatic potential surface of 14-3-3, showing the strong charge complementarity of the peptide- binding mode and the more hydrophobic nature of FC binding. Hydrogen bonds are indicated by dashed red lines, and major van der Waals contacts by blue dashed lines. (B) Van der Waals surface representation of the active site, showing the close interaction between the two ligands and how they fill the cavity of 14-3-3.
In a search for possible herbicides, a large number of FC derivatives have been either chemically synthesized or isolated from culture filtrates of F.amygdali (summarized by Ballio, 1979). Structure–activity relationships of these compounds generally support our structural model. Minor modifications of the carbocyclic framework are prohibitive for biological activity and their ability to compete with FC for binding. Thus, epimerization of the ligand in position C3 in ring A, or position C9 in ring B leads to a complete loss of binding activity as measured by the inhibition of binding of radioactively labelled FC to purified plasma membranes. The same is true for the introduction of a double bond between positions 2 and 6, which alters the spatial orientation of ring A and B and wipes out the binding affinity. In contrast, more extensive changes can be introduced into the glycosidic part of the molecule. Thus, the hydrogenation of the t-pentenyl moiety, which is used for the radioactive labelling of the molecule, or its complete removal or its derivatization have no influence on binding (Ballio, 1979). The removal of the 3′ substituent has a minor effect, and even the complete deletion of the glycosidic part results in an, albeit reduced, biological activity. Our structure corroborates the findings that the glycosidic part has less importance for 14-3-3 binding since it makes fewer intimate contacts with 14-3-3. It presumably is more important for improving the water solubility and/or bioavailibility of the toxin, since the phytotoxic activity of FC is less tolerant to the changes indicated above (Ballio et al., 1971, 1981; Ballio, 1977).
The peptide and FC contact each other very closely and together fill the central cavity of 14-3-3 (Figure 4B). The interaction involves the C-terminal valine of the peptide and the five- and eight-membered carbocycles of FC. These rings are arranged so as to form an additional hydrophobic pocket on the 14-3-3 interaction surface for the C-terminal valine of the PMA peptide. These contacts bury an extra exposed solvent-accessible surface of ∼50 Å2 when compared with the corresponding binary complexes.
FC has been reported to require PMA for binding to 14-3-3. However, the structural model of the ternary complex and thermodynamic considerations (see below) argue for an, albeit weak, binding site for FC on 14-3-3 in the absence of the H+-ATPase. By soaking FC into the crystal, we indeed were able to determine the structure of the binary 14-3-3–FC complex. The toxin occupies the same site as found in the ternary complex, and there are only minor rearrangements of its conformation between the binary and ternary complex (Figure 3B). Notably, comparison with the structure of FC in solution determined by NMR (Ballio et al., 1991) shows a similar conformation also for unbound FC.
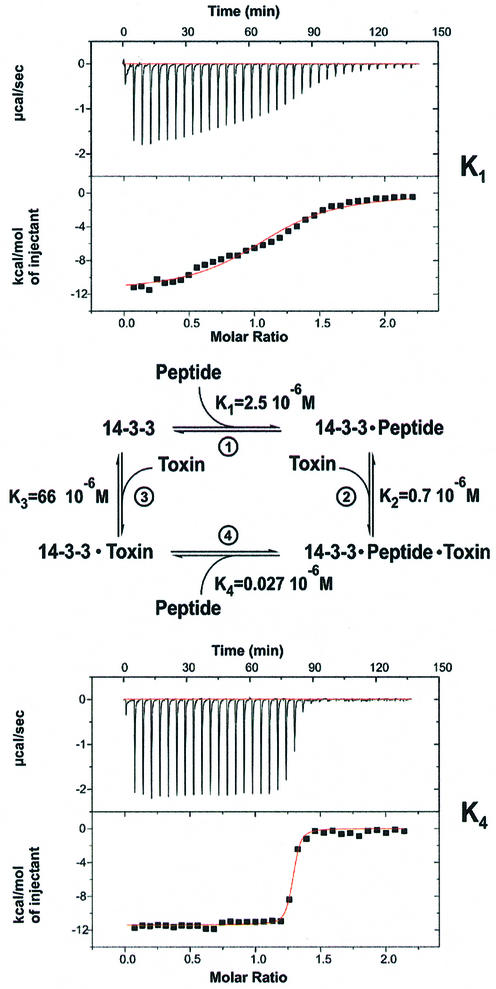
The thermodynamic binding cycle
Previous studies have failed to show the binding of FC to 14-3-3 in the absence of PMA. However, since the consecutive binding of various C-terminal PMA peptides and FC has been well documented, it is mandatory from thermodynamic considerations that the reverse order of binding reactions should produce the same complex, and that FC alone should thus bind, albeit weakly, to 14-3-3. This is supported by the structure of the 14-3-3–FC complex presented above. We thus defined four reactions (1–4) for the binding of the phosphorylated pentapeptide and FC to 14-3-3 defined by the equilibrium constants K1–K4, as shown in Figure 5, and set out to determine the equilibrium dissociation constants and other thermodynamic parameters by means of isothermal titration calorimetry (ITC; Table II). For the interaction between peptide and protein, a dissociation constant KD (K1 in Figure 5) of 2.5 µM was obtained (Figure 5, upper panel). This binding affinity is weaker than values previously reported for larger fragments of the H+-ATPase. The affinity of a phosphorylated 16mer was found to be 88 nM measured under different conditions using surface plasmon resonance (Fuglsang et al., 1999). Micromolar affinities have also been measured for the interaction of phosphorylated c-Raf model peptides from either the 259 or 621 regulatory region of the kinase, whereas peptides with the mode 2 consensus sequence bound with ∼10-fold higher affinity (Yaffe et al., 1997).
Fig. 5. Thermodynamic measurements. Thermodynamic cycle of the coupled equilibria between 14-3-3, the PMA peptide and fusicoccin, and the corresponding dissociation constants for each equilibrium, centre (the value for K3 was calculated). Two examples for the ITC measurements, which show that the binding between 14-3-3 and the peptide (top panel) is much tighter, as seen by the sharp increase in signal (lower panel) after saturation of the binding site with FC.
Table II. Thermodynamic constants obtained by isothermal titration calorimetry for the coupled equilibria as shown in Figure 5.
| KD (µM) | ΔG (kcal/mol) | ΔH (kcal/mol) | –TΔS (kcal/mol) | |
|---|---|---|---|---|
| K1 | 2.5 | –8.3 ± 0.2 | –10.0 ± 1.1 | 1.7 ± 1.3 |
| K2 | 0.7 | –9.2 ± 0.4 | –5.5 ± 1.6 | –3.7 ± 2.0 |
| K3a | 66 | –6.1 ± 1.2 | –3.7 ± 3.5 | –2.4 ± 4.4 |
| K4 | 0.027 | –11.4 ± 0.6 | –11.8 ± 0.8 | 0.4 ± 1.1 |
aCalculated data.
Titration of FC to a saturated binary 14-3-3–peptide complex results in a dissociation constant K2 of 0.7 µM. As predicted, we could show direct binding of the toxin, and its affinity for 14-3-3 (K3) was determined to be of the order of 50 µM. The low affinity and the insolubility of FC in aqueous solutions made direct determination somewhat unreliable. However, from the affinity of the peptide for the 14-3-3–FC complex (K4 = 0.027 µM, Figure 5, lower panel) and since K1 × K2 = K3 × K4, (Figure 5), we can calculate a more reliable value of 66 µM for K3.
Considering the complete binding cycle, we can conclude that FC increases the binding affinity of the peptide 93-fold (K1/K4), and that its own affinity is increased correspondingly (K1/K4 = K3/K2) by the peptide. Since it has been shown that the binding site of PMA on 14-3-3 involves features other than just the C-terminal end (Jelich-Ottmann et al., 2001), the phosphopeptide affinities measured by ITC may not quantitatively reflect the FC effect. However, since increases in affinity for FC have been described for any N-terminally extended fragment of the PMA C-terminus (Fuglsang et al., 1999; Borch et al., 2002), we are confident that the affinities measured here faithfully reproduce and give a molecular explanation for the physiological effect of FC on the proton ATPase.
In order to probe for the nature of the stabilizing effect of FC, ITC data were analysed for other thermodynamic parameters of the binding cycle (Table II). The negative enthalpy (ΔH) of peptide binding to the unliganded 14-3-3 is increased from –10 to –11.8 kcal/mol for the 14-3-3–FC complex. The full increase in binding affinity is due additionally to relieving the unfavourable entropy change of peptide binding to 14-3-3, as –TΔS decreases from 1.7 to 0.4 kcal/mol. Similarly, the tighter binding of FC to 14-3-3 is due to both a more favourable enthalpy and entropy change in the presence of the peptide. We can only speculate as to the nature of the observed effect that leads to the stabilization of PMA binding to 14-3-3. However, since the tight juxtaposition of the two ligands in the binding site buries ∼50 Å2 surface, and does not involve major conformational changes, we would propose that the observed effects are due to the additional interactions between FC and the peptide’s C-terminal valine, and the release of ordered water molecules from the 14-3-3-binding cavity.
PMA binding to 14-3-3 is unique compared with other 14-3-3 binding interactions (Figure 2C) in that the penultimate C-terminal threonine is the phosphorylated residue (YpTV-COOH). The structure as well as ITC measurements show that the C-terminal valine including its carboxylate is important for binding to 14-3-3 and that FC binding would clash with binding of the 14-3-3 consensus motifs that involve residues C-terminal to the +1 position. Binding of peptides to 14-3-3c in the absence of FC shows that addition of a proline to the C-terminus of the peptide has no significant effect, whereas removal of valine considerably weakens the binding (KD = 15 µM), as compared with the standard peptide QSYpTV (KD = 2.5 µM, Figure 6A). This shows that addition of a proline, which presumably also binds in the 14-3-3 cavity, cannot mimic the effect of the toxin. FC binding to 14-3-3c pre-loaded with the C-terminally extended phosphopeptide Gln-Ser-Tyr-pThr-Val-Pro, which resembles more a consensus 14-3-3-binding peptide, was severely impaired, such that the affinity could not be measured reliably (Figure 6B). Furthermore, binding of FC actually requires the interaction with the C-terminal valine, as deletion of the latter again results in a significantly lower binding affinity (Figure 6B). Taken together, this explains why there is no effect of FC on any other known 14-3-3–ligand interaction. To date, all plant proteins characterized as interacting directly with 14-3-3 homologues contain the consensus binding motifs, with the notable exception of the plant PMA. In addition to steric interference, the weak binding of FC to unliganded 14-3-3 is likely to prevent interference with 14-3-3–protein interactions other than the proton pump.
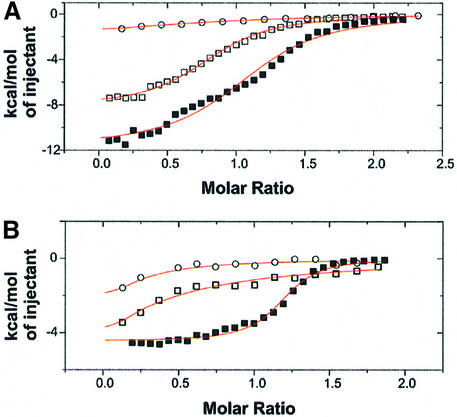
Fig. 6. Effect of mutated PMA peptides, as measured by ITC. (A) Binding of peptides to 14-3-3c in the absence of FC shows that whereas addition of a proline to the C-terminus of the peptide (open squares; KD = 2.7 µM) has no significant effect, removal of valine (open circles; KD = 15 µM) considerably weakens binding to 14-3-3c, as compared with the standard peptide QSYpTV (closed squares; KD = 2.5 µM). Notice the smaller ΔH effect of the mutant peptides, which made evaluation especially of the minus valine peptide less accurate. (B) FC binding to 14-3-3c in the presence of peptide is drastically weakened by addition of a proline (open squares) or removal of valine (open circles) compared with the standard peptide (closed squares).
Implications for drug development
Our results show that the binding of a small molecule can induce tighter association of a protein–protein complex, even though by itself it has low affinity for the individual components of the complex. In the present case, the fungal toxin fills a solvent-exposed cavity in the 14-3-3 complex, thereby increasing the affinity of the complex by almost two orders of magnitude. This increase in affinity stabilizes the activated state of the proton ATPase. The resulting hyperpolarization of the membrane potential activates K+ channels and leads to opening of the stomata. The subsequent elevation of transpiration causes the wilting of plants and explains the drastic physiological effects of FC.
In spite of the large number of protein–protein interactions in the cell and their importance for drug development, it is generally considered to be very difficult to develop inhibitors of protein–protein interactions. This is partly due to the size of a typical protein–protein interface and its flatness, and our inability to understand general thermodynamic principles of the interactions. Correspond ingly, only a small number of lead compounds based on this principle are available to date (Cochran, 2000). Based on the biochemical and structural findings of the FC effect and the available literature on the action of many other naturally occurring compounds, we would propose that it might actually be easier to reverse the strategy and start looking for stabilizers of such interactions. If such an interaction happens to be inhibitory, its stabilization would have a beneficial (positive) effect on a pathway or reaction and would constitute an ideal lead compound for drug development.
This is not without precedent, as many natural compounds have in fact been found, sometimes unexpectedly, to stabilize protein–protein interactions. Brefeldin A, a small molecule inhibitor of Golgi function, has been found to stabilize an otherwise very transient ternary complex between Arf-GDP and its guanine nucleotide exchange factor (Peyroche et al., 1999). Although structural or detailed biochemical data are not available for this system, its implications for drug development have been recognized (Chardin and McCormick, 1999). Likewise, taxol and other diterpenoids such as epothilone are very effective anticancer drugs which stabilize microtubules and prevent their dynamic de- and repolymerization (Jennewein and Croteau, 2001). Another diterpene, forskolin, an active ingredient of traditional medicine in India, has been found to stabilize the subunits of adenylyl cyclase (Tesmer et al., 1997). The antibiotics kirromycin and fusidic acid inhibit prokaryotic ribosome function by stabilizing Ef-Tu or Ef-G on the ribosome, respectively (Stark et al. 1997; Agrawal and Frank, 1999; Ramakrishnan, 2002), thus preventing the cycling of the translational machinery. The immunosuppressants FK506 and rapamycin work on a somewhat related principle in that they glue together two different proteins that otherwise have no measurable affinity for each other (Griffith et al., 1995; Choi et al., 1996; Ivery et al., 2000).
These examples, in combination with the structural and thermodynamic analysis of the FC effect on PMA presented here, suggest that it is a promising alternative in drug development to look systematically and devise screening technology for possibilities of stabilizing rather than inhibiting protein–protein interactions. The results presented here suggest that even weakly binding compounds, which would not have been found by conventional drug target screens, can have a powerful physiological effect.
Materials and methods
Crystallization
Tobacco 14-3-3c (DDBJ/EMBL/GeneBank accession No. AAC49892, amino acids 1–260 with an N-terminal His tag) was expressed recombinantly in Escherichia coli and purified via standard procedures. FC was purchased from Sigma. Phosphopeptides were synthesized by Biosyntan (Berlin). Crystals were grown by the hanging drop method in solutions containing 21% polyethylene glycol (PEG) 400, 0.1 mM citrate buffer pH 4.7, 0.2 mM ammonium acetate pH 7.0 and 10 mM DTT, and belong to the hexagonal space group P6522 with unit cell dimensions a = b = 109.0 Å, c = 135.8 Å and one 14-3-3 monomer in the asym metric unit. They were soaked with ligands for 4 h (FC) or 20 min (peptide) in precipitant solution supplemented to 30% PEG 400 and cryoprotectant (precipitant solution supplemented to 35% PEG 400, 8% isopropanol) prior to freezing in liquid N2.
Structure determination and refinement
Native 14-3-3c crystals were measured at the X13 beamline at DESY, Hamburg (wavelength 0.8459 Å). Complex crystals were measured at the ID29 beamline of ESRF, Grenoble (0.979 Å). Data was processed with XDS (Kabsch, 1993). The structure was solved with AmoRe (CCP4, 1994; Navaza, 2001) and refined with CNS (Brünger et al., 1998). Maps were analysed with Xfit (McRee, 1999) and the structures validated with PROCHECK (Laskowski and MacArthur, 1993). Models contain residues 5–239 of 14-3-3c. Some amino acids in the loop between helices 8 and 9 could not be observed in the electron density maps of the structures. Ninety-nine percent of the residues in all structures were located in the most favourable and favourable (φ, ψ) areas of the Ramachandran diagram. Models have main chain and side chain structural parameters consistently equal to or better than those expected from their respective resolution. Figures were drawn by Molscript (Kraulis, 1991), GRASP (Nicholls et al., 1991), DINO (DINO, 2002) and PyMOL (DeLano, 2002), and rendered with Raster3D (Merritt and Bacon, 1997).
Isothermal titration calorimetry
Binding of ligands to 14-3-3c was measured with an MCS isothermal titration calorimeter (MicroCal Inc., Northampton). Ligands (FC 0.4 mM; phosphopeptides 0.5 mM) were titrated in 8–20 µl steps by injection into solutions containing 14-3-3c (0.05 mM) alone or 14-3-3c saturated with one of the ligands in 25 mM HEPES buffer pH 6.5, 10 mM MgCl2, 5 mM CaCl2, 5 mM DTT at 35°C. Binding isotherms were fitted using a single binding site model and used to calculate the binding enthalpy (ΔH) and association constant (Ka) of the binding reaction. Dissociation constants (KD = 1/Ka), Gibbs free energy changes (ΔG = –RTlnKa) and entropy changes (TΔS = ΔH – ΔG) were calculated from ΔH and Ka. For the binding of peptide to an FC–14-3-3c complex, we also used non-saturating conditions of FC, and observed two binding events which were fitted independently to K1 and K4. All measurements were repeated at least three times.
Coordinates
The atomic coordinates and structure factors of the four structures have been deposited in the Protein Data Bank under the ID codes 1O9C, 1O9D, 1O9E and 1O9F.
Acknowledgments
Acknowledgements
We would like to thank I.Schlichting for help during data collection, ESRF and DESY/EMBL (European Community–Access to Research Infrastructure Action of the Improving Human Potential Programme to the EMBL Hamburg Outstation) for allocation of beamline time, C.Herrmann for helpful comments and discussions, E.W.Weiler for long-term support, and the DFG for financial support to C.O.
References
- Aducci P., Marra,M., Fogliano,V. and Fullone,M.R. (1995) Fusicoccin receptors: perception and transduction of the fusicoccin-signal. J. Exp. Bot., 46, 1463–1478. [Google Scholar]
- Agrawal R.W. and Frank,J. (1999) Structural studies of the translational apparatus. Curr. Opin. Struct. Biol., 9, 215–221. [DOI] [PubMed] [Google Scholar]
- Auer M., Scarborough,G.A. and Kühlbrandt,W. (1998) Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature, 392, 840–843. [DOI] [PubMed] [Google Scholar]
- Ballio A. (1977) Fusicoccin: structure–activity relationships. In Marré,E. and Ciferi,O. (eds), Regulation of Cell Membrane Activities in Plants. Elsevier, Amsterdam, The Netherlands, pp. 217–223.
- Ballio A. (1979) Chemistry and plant growth regulating activity of fusicoccin derivatives and analogues. In Geissbühler,H. (ed.), Advances in Pesticide Science. Pergamon, Oxford, UK, pp. 366–372.
- Ballio A. et al. (1964) Fusicoccin: a new wilting toxin produced by Fusicoccum amygdali Del. Nature, 203, 297. [Google Scholar]
- Ballio A. et al. (1968) The structure of fusicoccin A. Experientia, 24, 631–635. [DOI] [PubMed] [Google Scholar]
- Ballio A., Bottalico,A., Framondino,M., Graniti,A. and Randazzo,G. (1971) Fusicoccin: structure–phytotoxicity relationships. Phytopathol. Mediterr., 10, 26–32. [Google Scholar]
- Ballio A., De Michelis,M.I., Lado,P. and Randazzo,G. (1981) Fusicoccin structure–activity relationships: stimulation of growth by cell enlargement and promotion of seed germination. Physiol. Plant., 52, 471–475. [Google Scholar]
- Ballio A. et al. (1991) 1H NMR conformational study of fusicoccin and related compounds: molecular conformation and biological activity. Phytochemistry, 30, 137–146. [Google Scholar]
- Borch J., Bych,K., Roepstorff,P., Palmgren,M. and Fuglsang,A.T. (2002) Phosphorylation-independent interaction between 14-3-3 protein and the plant plasma membrane H+-ATPase. Biochem. Soc. Trans, 30, 411–415. [DOI] [PubMed] [Google Scholar]
- Braselmann S. and McCormick,F. (1995) BCR and RAF form a complex in vivo via 14-3-3 proteins. EMBO J., 14, 4839–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chardin P. and McCormick,F. (1999) Brefeldin A: the advantage of being uncompetitive. Cell, 97, 153–155. [DOI] [PubMed] [Google Scholar]
- Choi J., Chen,J., Schreiber,S.L. and Clardy,J. (1996) Structure of the FKBP12–rapamycin complex interacting with the binding domain of human FRAP. Science, 273, 239–242. [DOI] [PubMed] [Google Scholar]
- Cochran A.G. (2000) Antagonists of protein–protein interactions. Chem. Biol., 7, R85–R94. [DOI] [PubMed] [Google Scholar]
- Dalal S.N., Schweitzer,C.M., Gan,J. and DeCaprio,J.A. (1999) Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol., 19, 4465–4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G., Eisenmann-Tappe,I., Fries,H.W., Troppmaier,J. and Rapp,U.R. (1994) The ins and outs of Raf kinases. Trends Biochem. Sci., 19, 474–480. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA.
- DIN O (2002) Visualizing structural biology. http://www.dino3d.org
- Fantl A. et al. (1994) Activation of Raf-1 by 14-3-3 proteins. Nature, 371, 612–614. [DOI] [PubMed] [Google Scholar]
- Finnie C., Borch,J. and Collinge,D.B. (1999) 14-3-3 proteins: eukaryotic regulatory proteins with many functions. Plant Mol. Biol., 40, 545–554. [DOI] [PubMed] [Google Scholar]
- Fuglsang A. et al. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947). J. Biol. Chem., 274, 36774–36780. [DOI] [PubMed] [Google Scholar]
- Griffith J.P. et al. (1995) X-ray structure of calcineurin inhibited by the immunophilin-immunosuppressant FKBP12–FK506 complex. Cell, 82, 507–522. [DOI] [PubMed] [Google Scholar]
- Ivery M.T.G. (2000) Immunophilins: switched on protein binding domains? Med. Res. Rev., 20, 452–484. [DOI] [PubMed] [Google Scholar]
- Jahn T. et al. (1997) The 14-3-3 protein interacts directly with the C-terminal region of the plant plasma membrane H+-ATPase. Plant Cell, 9, 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelich-Ottmann C., Weiler,E.W. and Oecking,C. (2001) Binding of regulatory 14-3-3 proteins to the C terminus of the plant plasma membrane H+-ATPase involves part of its autoinhibitory region. J. Biol. Chem., 276, 39852–39857. [DOI] [PubMed] [Google Scholar]
- Jennewein S. and Croteau,R. (2001) Taxol: biosynthesis, molecular genetics and biotechnological applications. Appl. Microbiol. Biotechnol., 57, 13–19. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Kühlbrandt W., Zeelen,J. and Dietrich,J. (2002) Structure, mechanism and regulation of the Neurospora plasma membrane H+-ATPase. Science, 297, 1692–1696. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A. and MacArthur,M.W. (1993) A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Liu D. et al. (1995) Crystal structure of the ζ isoform of the 14-3-3 protein. Nature, 376, 191–194. [DOI] [PubMed] [Google Scholar]
- Lu G., DeLisle,A.F., De Vetten,N.C. and Ferl,R.J. (1992) Brain protein in plants: an Arabidopsis homolog to neurotransmitter pathway activators is part of a DNA binding complex. Proc. Natl Acad. Sci. USA, 89, 11490–11494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marre E. (1979) Fusicoccin: a tool in plant physiology. Annu. Rev. Plant Physiol., 30, 273–312. [Google Scholar]
- Maudoux O. et al. (2000) A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem., 275, 17762–17770. [DOI] [PubMed] [Google Scholar]
- May T. and Soll,J. (2000) 14-3-3 proteins form a guidance complex with chloroplast precursor proteins in plants. Plant Cell, 12, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRee D.E. (1999) XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol., 125, 156–165. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Morrison D.K. and Cutler,R.E. (1997) The complexity of Raf-1 regulation. Curr. Opin. Cell Biol., 9, 174–179. [DOI] [PubMed] [Google Scholar]
- Morsomme P. and Boutry M. (2000) The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim. Biophys. Acta, 1465, 1–16. [DOI] [PubMed] [Google Scholar]
- Navaza J. (2001) Implementation of molecular replacement in AMoRe. Acta Crystallogr. D, 57, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Obsil T., Ghirlando,R., Klein,D.C., Ganguly,S. and Dyda,F. (2001) Crystal structure of the 14-3-3ζ:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell, 105, 257–267. [DOI] [PubMed] [Google Scholar]
- Oecking C., Piotrowski,M., Hagemeier,J. and Hagemann,K. (1997) Topology and target interaction of the fusicoccin-binding 14-3-3 homologs of Commelina communis. Plant J., 12, 441–453. [Google Scholar]
- Peng C.Y. et al. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science, 277, 1501–1505. [DOI] [PubMed] [Google Scholar]
- Petosa C. et al. (1998) 14-3-3ζ binds a phosphorylated Raf peptide and an unphosphorylated peptide via its conserved amphipathic groove. J. Biol. Chem., 273, 16305–16310. [DOI] [PubMed] [Google Scholar]
- Peyroche A. et al. (1999) Brefeldin A acts to stabilize an abortive ARF-GDP–Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell, 3, 275–285. [DOI] [PubMed] [Google Scholar]
- Piwnica-Worms P. (1999) Fools rush in. Nature, 401, 535–537. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Rittinger K. et al. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Sehnke P.C., Delille,J.M. and Ferl,R.J. (2002) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell, 14, S339–S354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H. et al. (1997) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature, 389, 403–406. [DOI] [PubMed] [Google Scholar]
- Svennelid F. et al. (1999) Phosphorylation of Thr-948 at the C terminus of the plasma membrane H+-ATPase creates a binding site for the regulatory 14-3-3 protein. Plant Cell, 11, 2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesmer J.J., Sunahara,R.K., Gilman,A.G. and Sprang,S.R. (1997) Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsα·GTPγS. Science, 278, 1907–1916. [DOI] [PubMed] [Google Scholar]
- Tzivion G. and Avruch,J. (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem., 277, 3061–3064. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. (2002) How do 14-3-3 proteins work? Gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett., 513, 53–57. [DOI] [PubMed] [Google Scholar]
- Yaffe M.B. et al. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell, 91, 961–971. [DOI] [PubMed] [Google Scholar]