Abstract
Successful maturation determines the intracellular fate of secretory and membrane proteins in the endoplasmic reticulum (ER). Failure of proteins to fold or assemble properly can lead to their retention in the ER and redirects them to the cytosol for degradation by the proteasome. Proteasome inhibitors can yield deglycosylated cytoplasmic intermediates that are the result of an N-glycanase activity, believed to act prior to destruction of these substrates by the proteasome. A gene encoding a yeast peptide:N-glycanase, PNG1, has been cloned, but this N-glycanase and its mammalian homolog were reported to be incapable of deglycosylating full-length glycoproteins. We show that both the yeast PNG1 enzyme and its mammalian homolog display N-glycanase activity towards intact glycoproteins. As substrates, cytosolic PNGase activity prefers proteins containing high-mannose over those bearing complex type oligosaccharides. Importantly, PNG1 discriminates between non-native and folded glycoproteins, consistent with a role for N-glycanase in cytoplasmic turnover of glycoproteins.
Keywords: ER quality control/N-glycanase/glycoprotein/proteasome/siRNA
Introduction
N-linked glycosylation of polypeptides that enter the secretory pathway contributes to their proper folding, assembly and trafficking (Helenius and Aebi, 2001). At the cell surface, glycans mediate cell–cell interactions, stabilize proteins and protect them against proteases. While the function of the glycosylation machinery may therefore seem obvious, the necessity for an enzyme that releases N-linked glycans, peptide-N4-(N-acetyl-β-d- glucosaminyl)asparagine amidase (PNGase), is less clear. PNGase was first observed in almond seeds and in the bacterium Flavobacterium meningosepticum (Takahashi, 1977; Plummer et al., 1984). Later, PNGase activity was reported in medaka (fish) embryos and mammalian cells (Seko et al., 1991; Suzuki et al., 1993). A possible role for this enzyme in the cytosol of mammalian cells emerged from a strategy used by the human cytomegalovirus (HCMV) to evade detection by the immune system of its host (Wiertz et al., 1996a,b).
Recognition of intracellular pathogens requires presentation of pathogen-derived peptides at the cell surface by products of the major histocompatibility complex (MHC) (Heemels and Ploegh, 1995). Human class I MHC heavy chains are type I membrane proteins that carry a single N-linked glycan. In the endoplasmic reticulum (ER), the class I MHC heavy chain forms a heterotrimeric complex with β2-microglobulin (β2m) and an 8–10 residue antigenic peptide, which is displayed at the cell surface. Cells that present foreign peptides may then be killed by cytotoxic T cells. The HCMV gene products US2 and US11 interfere with this antigen presentation pathway by dislocating the class I MHC heavy chains from the ER to the cytosol (Wiertz et al., 1996a,b), where they are degraded by the proteasome (Baumeister et al., 1998). For US2- and US11-mediated degradation of class I MHC heavy chains, inhibition of proteasomal proteolysis results in the accumulation of deglycosylated heavy chains in the cytosol. Several other substrates that also undergo this mode of degradation, such as the α-chain of the T-cell receptor (TCRα) (Huppa and Ploegh, 1997), tyrosinase (Halaban et al., 1997) and the cystic fibrosis conductance regulator (Johnston et al., 1998), also yield deglycosylated intermediates when proteasomal proteolysis is blocked, consistent with the action of an N-glycanase prior to their destruction by the proteasome.
Lennarz and co-workers have pursued the identification of an N-glycanase activity in Saccharomyces cerevisiae (Suzuki et al., 2000). They characterized the PNG1 gene product as a polypeptide capable of deglycosylating the fetuin-derived asialoglycopeptide (CH3)2Leu-Asn(Glc NAc5Man3Gal3)-Asp-Ser-Arg. Neither the yeast Png1p nor its murine counterpart were found capable of attacking full-length glycoprotein substrates (Suzuki et al., 2000). These experiments, therefore, did not establish the identity of the mammalian N-glycanase implicated in the type of glycoprotein turnover exemplified by US2- and US11-mediated degradation of class I molecules, or that of TCRα.
To resolve the molecular identity of the PNGase involved in the cytosolic degradation of proteins from the ER, we used a model system that has served as a paradigm for this pathway: in COS-1 cells, TCRα is degraded in a proteasome-dependent manner when expressed in the absence of the subunits required for the formation of a functional TCR (Huppa and Ploegh, 1997; Yu et al., 1997). We used TCRα as a substrate to monitor mammalian N-glycanase activity and its partial purification from COS cells. Comparison of the N-glycanase activity observed in COS cells with the product encoded by the murine PNG1 homolog shows that the murine PNG1 in fact possesses biochemical properties similar to the activity seen in COS cells. By introducing a small interfering (si) RNA complementary to mammalian PNG1 into COS cells, we could silence the observed N-glycanase activity, demonstrating that these enzymatic activities are one and the same. It is noteworthy that both the yeast Png1p and the activity detected in COS cells distinguish folded from unfolded proteins and high-mannose from complex type oligosaccharides, properties consistent with the proposed role for PNG1 in glycoprotein turnover.
Results
Mammalian cell extracts contain an activity that deglycosylates TCRα
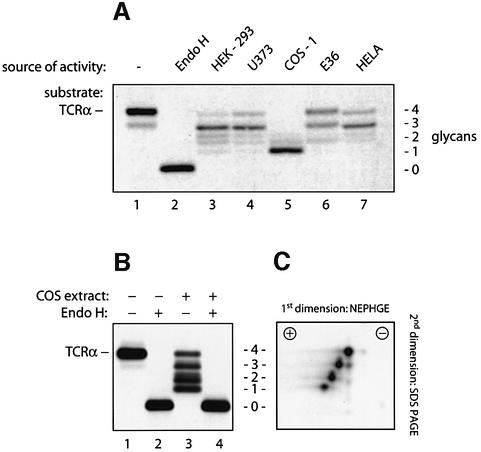
To detect N-glycanase activity present in mammalian tissue culture cells, we incubated detergent (NP-40) lysates from different cell lines with [35S]methionine-labeled TCRα as a substrate. TCRα was obtained from cells expressing the α-subunit of the TCR only. Under these conditions, TCRα fails to progress to the Golgi and retains its high-mannose N-glycans. By SDS–PAGE, the input TCRα appeared as a single band with a mol. wt of 38 kDa (Figure 1A, lane 1). Complete removal of its four N-linked glycans by digestion with endoglycosidase-H (Endo H) reduced the mol. wt of TCRα to 28 kDa (lane 2). Incubation of TCRα with different cell extracts converted the 38 kDa band to a ladder of faster migrating polypeptides, suggesting partial deglycosylation of TCRα (lanes 3–7). Not all cell lines produce the exact same set of intermediates, an observation we attribute to different levels of glucosidase and mannosidase activities in these extracts. The observed loss in molecular weight could be explained either by proteolysis of the polypeptide backbone of TCRα or by its partial deglycosylation. When TCRα was first exposed to COS-1 cell extracts and then treated with Endo H, the ladder of polypeptides was converted to a single band with a molecular weight indistinguishable from that of TCRα treated with Endo H alone (Figure 1B). The observed reduction in molecular weight is thus attributable to a partial deglycosylation of the protein and not to proteolytic digestion of the peptide backbone. NP-40 extracts from COS-1 cells therefore contain an activity that cleaves N-linked oligosaccharides from TCRα. Since lysates from COS cells yielded the largest shift in molecular weight for TCRα, COS cells express higher levels of the activity or a form of the enzyme that is more active on TCRα.
Fig. 1. Mammalian cell extracts contain an N-glycanase activity that deglycosylates TCRα. (A) [35S]methionine-labeled TCRα was immuno precipitated after incubation with buffer alone (lane 1), with Endo H (lane 2) or with detergent extracts containing 750 µg of protein obtained from the indicated cell lines (lanes 3–7). Immunoprecipitates were resolved by SDS–PAGE, and proteins were visualized by fluorography. The number of N-linked glycans attached to TCRα is indicated. The doublet observed occasionally for the TCRα carrying two residual glycans is most probably due to heterogeneous deglycosylation of the polypeptide since the four N-linked attachment sites of TCRα allow six different glycosylation patterns. (B) [35S]methionine-labeled TCRα was obtained by immunoprecipitation after incubation with buffer alone (lanes 1 and 2) or after incubation with extracts from COS-1 cells (lanes 3 and 4). Removal of all N-linked glycans in lanes 2 and 4 by digestion with Endo H resulted in polypeptides with identicalelectrophoretic mobility. The number of N-linked glycans is indicated in (C). To obtain the complete ladder of partially deglycosylated TCRα molecules, only 300 µg of COS cell extracts were used for this deglycosylation. (C) [35S]methionine-labeled TCRα was incubated with COS-1 cell extracts and analyzed by two-dimensional gel electrophoresis (as described in Materials and methods). For each glycan lost, a single negative net charge is acquired.
The deglycosylation of TCRα leads to conversion of the glycosylated asparagine to aspartate
Two types of enzymatic activity could account for the observed deglycosylation. To release oligosaccharides, an endoglycosidase might cleave the diacetylchitobiose core of high-mannose oligosaccharides, as does Endo H. Alternatively, an N-glycanase could cleave the bond be tween the innermost GlcNAc and the asparagine residue of the N-linked glycoprotein, as does the bacterial PNGase F.
Diagnostic of the reaction catalyzed by an N-glycanase is the conversion of the asparagine residue that carries the oligosaccharide to an aspartate upon hydrolysis of the glycoamide bond (Tarentino and Plummer, 1994). The removal of an N-linked glycan by N-glycanase therefore introduces an additional negative charge into the protein, demonstrable by isoelectric focusing (IEF) followed by a separation according to mass by SDS–PAGE. When TCRα, partially deglycosylated by COS extracts, was analyzed by two-dimensional gel electrophoresis, we saw a stepwise decrease in isoelectric point with decreasing number of glycans (Figure 1C). For each glycan lost, a single negative charge was gained. We conclude that the enzymatic activity observed in NP-40 extracts from COS cells is an N-glycanase.
N-glycanase is located predominantly in the cytosol
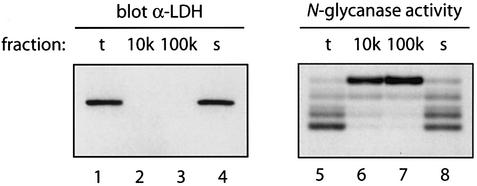
There are conflicting reports on the intracellular location of N-glycanase. In fish oocytes and in mouse L929 cells, the activity has been described as cytosolic (Seko et al., 1991; Suzuki et al., 1993), but others have reported N-glycanase activity in the lumen of rat liver microsomes (Weng and Spiro, 1997). To establish the intracellular location of the observed N-glycanase activity, COS cells were homogenized and fractionated into three subcellular fractions by differential centrifugation: a 10 000 g pellet, a 100 000 g pellet and cytosol (100 000 g supernatant). The cytosolic marker lactate dehydrogenase (LDH) was found only in the unfractionated homogenate and in the cytosolic fraction, but not in the 10 000 g and 100 000 g pellets, as assessed by immunoblotting (Figure 2, lanes 1–4), indicating that no cytosol was trapped in the particulate fractions. Almost all of the deglycosylating activity present in the total fraction was seen in the cytosol. The 100 000 g pellet contained no N-glycanase activity. Trace amounts of partially deglycosylated TCRα were seen in the 10 000 g pellet, indicating that some of the activity might be associated with membrane fragments present in the 10 000 g pellet.
Fig. 2. N-Glycanase is located predominantly in the cytosol. COS-1 cells were subjected to subcellular fractionation (as described in Huppa and Ploegh, 1997). Separation of fractions by SDS–PAGE was followed by immunoblotting with an α-LDH antibody as cytosolic marker (lanes 1–4). In parallel, fractions were tested for N-glycanase activity by incubation with TCRα as a substrate in the described assay (lanes 5–8).
N-glycanase is a single enzymatic activity
To characterize N-glycanase further, a purification scheme was established (Figure 3). On five successive chromatographical separations, the enzyme behaved as a monodisperse peak, consistent with a single enzymatic activity. Since the method of detection is a qualitative assay, we were unable to determine the extent of purification. The activity had a mol. wt of ∼200 kDa as judged from size estimates by gel filtration. We were unable to obtain amounts of a sufficiently purified preparation suitable for identification of the active species by mass spectrometry.
Fig. 3. The N-glycanase activity in COS cells is a single enzymatic entity. The purification scheme for N-glycanase from COS cells consisted of seven steps. First, COS-1 cytosol was obtained by differential centrifugation. Then N-glycanase was enriched by ammonium sulfate precipitation. The precipitate was resuspended and separated on a hydroxylapatite column. The flowthrough, which contained the N-glycanase activity, was separated on four chromatographic colums. Individual fractions obtained after each chromatographic separation were assayed for N-glycanase activity using TCRα as the substrate. Fractions containing activity were pooled as indicated and used in the next purification step.
PNG1 mRNA encodes the deglycosylation activity observed in COS-1 and 3T3 cells
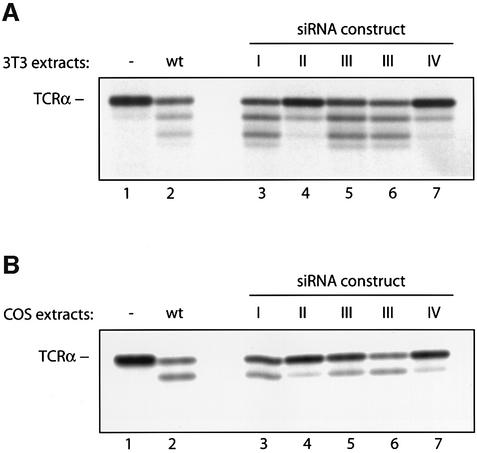
Is the N-glycanase activity observed in COS cells attributable to the mammalian homolog of the yeast PNG1 gene? While we have produced antibodies against recombinant murine PNG1, these antibodies do not cross-react with primate PNG1. The levels of endogenous PNG1 in mouse cells are below the levels of detection (2.5 pg/1000 cells), based on a comparison with recombinant mouse PNG1 as a standard. To verify the relationship between the PNG1 gene and the activity we observe in COS cells, we resorted to an alternative approach: we infected COS-1 and mouse 3T3 cells with an siRNA-encoding retrovirus to obtain cell lines with a reduced expression (knockdown) of the PNG1 mRNA (Brummelkamp et al., 2002). The encoded siRNAs were designed to recognize four different regions of the PNG1 mRNA conserved from mouse to human. The PNG1 knockdown cell lines were tested in the deglycosylation assay using TCRα as a substrate. When compared with wild-type cells, 3T3 cells infected with two different siRNA constructs showed significantly reduced levels of deglycosylation activity (see Figure 4A, constructs II and IV). When applied to COS-1 cells, the same two retrovirus-encoded siRNAs also significantly decreased deglycosylation activity (see Figure 4B). Taken together, these RNA knockdown results show that the PNG1 gene encodes the deglycosylation activity detected in COS-1 and 3T3 cells, establishing its activity as that encoded by the mammalian ortholog of the yeast Png1 gene.
Fig. 4. The deglycosylation activity observed in 3T3 and COS-1 cells is encoded by PNG1 mRNA. PNG1 knockdowns were generated in 3T3 (A) and COS-1 (B) cells as described in Materials and methods. Extracts from generated cell lines were tested in the deglycosylation assay, using [35S]methionine-labeled TCRα as a substrate. In (A), 400 µg of 3T3 cell extracts were used, in (B) 600 µg of COS-1 cell extracts. Lane 1: TCRα substrate incubated without cell extracts. Lane 2: TCRα incubated with wild-type cell extracts. Lanes 3–7: TCRα substrate incubated with extracts from PNG1 knockdown cell lines, infected with constructs I–IV (see Materials and methods for details). Note that extracts from cells infected with knockdown construct III were used from two independent DNA isolates, since the complete nucleotide sequence of this construct could not be determined. The N-glycanase activity observed in the wild-type COS extracts is lower than that seen in Figure 1, lane 5, because for the experiment shown here the extracts have been frozen prior to performing the deglycosylation assay.
The yeast and the mammalian PNG1 enzyme can deglycosylate TCRα
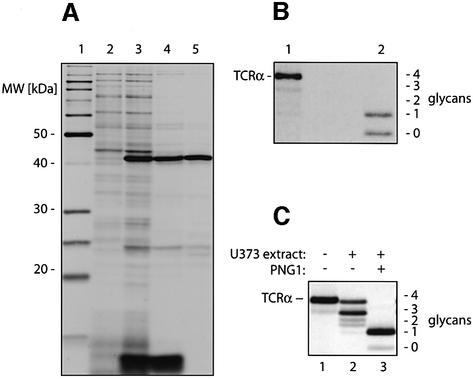
It was reported that the yeast Png1p enzyme could act only on short glycopeptides but not on full-length glycoproteins (Suzuki et al., 2000). To examine how the yeast enzyme behaved in our deglycosylation assay, the 43 kDa yeast Png1p was expressed in Escherichia coli and purified by anion exchange chromatography, followed by a gel filtration step (Figure 5A). Lysates from bacteria containing no Png1p did not deglycoslylate TCRα, whereas lysates from bacteria expressing the PNG1 and the purified enzyme both yielded partially and even the fully deglycosylated TCRα molecule (Figure 5B). We also cloned the 74 kDa mouse PNG1 by RT–PCR; we then tested the encoded protein in the N-glycanase assay, using TCRα as a substrate. Since U373 astrocytoma cells serve as the standard for US2- and US11-mediated degradation of class I molecules (Wiertz et al., 1996a,b), we expressed mammalian PNG1 in these cells. U373 cells stably transfected with mouse PNG1 cDNA had significantly higher N-glycanase activity compared with mock-transfected cells (Figure 5C). Lysates from U373 cells stably expressing PNG1 assayed on TCRα yielded predominantly the form lacking three N-linked glycans and even some fully deglycosylated TCRα, demonstrating that the mammalian PNG1 can also remove all four N-glycans from TCRα.
Fig. 5. The yeast and the mammalian PNG1 can deglycosylate full-length TCRα. (A) PNG1 was expressed in BL21 cells and purified in two steps. An aliquot from each step was separated by SDS–PAGE, and protein bands were visualized by silver staining. Lane 1 shows a molecular weight marker, lane 2 contains lysates from uninduced cells, and lane 3 lysates from cells after induction. The bacterial lysates were purified over a Uno Q-12 anion-exchange column (lane 4) and sub sequently separated by gel filtration on a Superdex 75 column (Pharmacia) (lane 5). (B) [35S]methionine-labeled TCRα was incubated with either lysates from uninduced BL21 cells (lane 1) or 10 µg of the purified Png1p (lane 2). Incubation with the purified yeast Png1p yielded a TCRα molecule that had lost either all or three of its four N-linked glycans. (C) [35S]methionine-labeled TCRα was immunoprecipitated after incubation with buffer alone (lane 1), extracts from mock transfected U373 cells (lane 2) or extracts from U373 cells expressing the murine PNG1 (lane 3).
Cys306 is part of a transglutaminase domain that forms the active site of N-glycanase
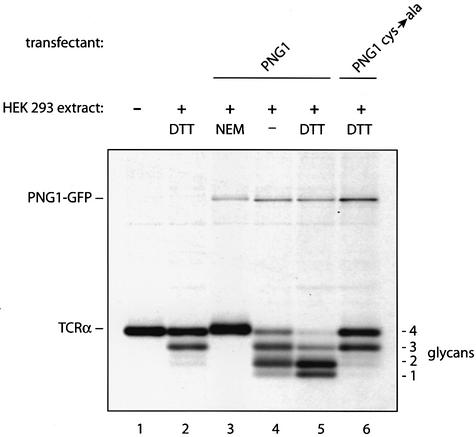
The N-glycanase sequence shows homology to the catalytic domain of the transglutaminase-like superfamily found in animal transglutaminases and bacterial proteases (Katiyar et al., 2002). The transglutaminase-like superfamily is characterized by three motifs that center around three conserved amino acids: a cysteine, a histidine and an aspartate residue (Makarova et al., 1999). In the human blood-clotting factor XIIIa, one of the structurally characterized transglutaminases, these three amino acids form a catalytic triad required for activity (Yee et al., 1994). We conducted an alkylation experiment to verify that a cysteine is indeed part of the catalytic center of N-glycanase activity as detected in our assay. HEK-293 cells were transiently transfected with murine PNG1 C-terminally tagged with green fluorescent protein (GFP) (Figure 6). We used HEK-293 cells for these experiments, because the levels of N-glycanase activity observed in HEK-293 cells are significantly lower than in COS cells (compare Figure 1, lanes 3 and 5). Their low levels of endogenous N-glycanase activity and their easy transfectability make HEK-293 cells the system of choice to study the effect of different PNG1 constructs by transient transfection. Extracts from [35S]methionine-labeled HEK-293 cells were tested in the TCRα deglycosylation assay, and the effects of the alkylating agent N-ethylmaleimide (NEM) or the reducing agent dithiothreitol (DTT) on enzyme activity were measured. After the incubation, TCRα and GFP-tagged N-glycanase were immunoprecipitated at the same time and analyzed by SDS–PAGE. Mock-transfected HEK-293 cells had low levels of endogenous N-glycanase activity even in the presence of DTT, while no GFP-reactive material was recovered (Figure 6, lane 2). The activity of GFP-tagged PNGase and endogenous PNGase was abolished by inclusion of NEM (Figure 6, lanes 3 and 4). Addition of DTT enhanced enzymatic activity (Figure 6, lanes 4 and 5). Together, these results show that a free thiol is required for PNG1 activity. Cys306 of the mouse PNG1 protein corresponds to the cysteine that is part of the catalytic triad of the human blood-clotting factor XIIIa. We changed Cys306 to Ala306 by site-directed mutagenesis, and the mutant GFP-tagged construct was then examined for activity. The Cys306-Ala mutant reduced the N-glycanase activity to background level (Figure 6, lanes 2 and 6). While Cys306 is indeed required for PNG1 function, this mutation does not have a dominant-negative effect on endogenous N-glycanase activity as assayed in vitro.
Fig. 6. Cys306 is part of a transglutaminase domain that forms the active site of N-glycanase. [35S]methionine-labeled TCRα was incubated either with buffer alone (lane 1) or with [35S]methionine-labeled extracts obtained from HEK-293 cells that were mock transfected, (lane 2), from transfectants expressing PNG1–GFP (lanes 3–5) or from transfectants expressing PNGAla306–GFP (lane 6). NEM or DTT was added to the incubations as indicated. After the incubations, immunoprecipitations with anti-TCRα and anti-GFP antibodies were performed simultaneously.
N-glycanase recognizes high-mannose type glycans on misfolded proteins
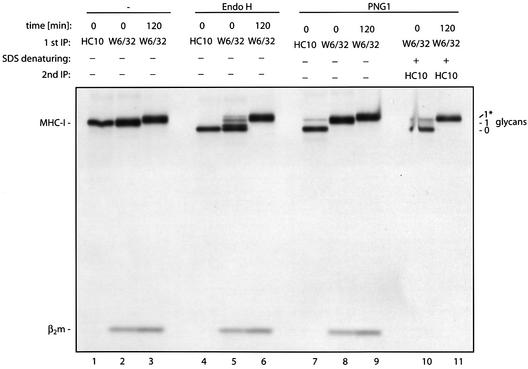
We also examined the class I MHC heavy chain as a suitable N-glycanase substrate. After its release from the ER, the peptide-loaded heavy chain associated with β2m travels to the cell surface via the Golgi apparatus, where its single high-mannose N-linked glycan is converted to a complex type oligosaccharide (see Figure 9). This modification renders the heavy chain resistant to digestion by Endo H. We can therefore define three relevant biosynthetic substrates for N-glycanase: free heavy chains, the assembled class I complex bearing high-mannose oligosaccharides, and assembled class I molecules with complex type glycans. Free heavy chains carry high-mannose oligosaccharides exclusively. To see if these three intermediates differ in their susceptibility to deglycosylation by N-glycanase, lymphoblastoid B cells (FH17) known to express high levels of class I MHC molecules, and thus a convenient source of the relevant substrates, were metabolically labeled for 15 min. Class I MHC heavy chains were immunoprecipitated at the 0 or 120 min chase points. At the 0 min time point, unfolded heavy chains bearing high-mannose N-linked glycans were recovered using the HC10 antibody, specific for free heavy chains (Stam et al., 1990), and the trimeric complex was recovered using the conformation-specific antibody W6/32 (Barnstable et al., 1978) at the 0 and 120 min time points (Figure 7, lanes 1–3). Some of the trimeric heavy chain molecules recovered at the onset of the chase (0 min) have already acquired resistance to Endo H digestion and, after 120 min of chase, all N-linked glycans are complex type oligosaccharides and fully resistant to Endo H (Figure 7, lanes 4–6). When these immunoprecipitates were exposed to the purified yeast Png1p enzyme, only the free class I MHC heavy chain from the 0 min time point was susceptible to digestion by the N-glycanase (Figure 7, lane 7), whereas class I MHC heavy chains that had formed a complex with β2m were completely resistant to digestion with N-glycanase (Figure 7, lanes 8 and 9). To see if free heavy chains bearing a complex type glycan can be deglycosylated by yeast Png1p, the W6/32 immunoprecipitates from the 0 and 120 min chase points were denatured by boiling with SDS, followed by re-immunoprecipitation of free heavy chains with the HC10 antibody. Exposure of these immunoprecipitates to yeast N-glycanase resulted in deglycosylation of high-mannose-containing heavy chains from the 0 min chase point, but no deglycosylation of heavy chains bearing a complex type glycan could be observed (Figure 7, lanes 10 and 11). Removal of sialic acids with neuramidase did not facilitate subsequent deglycosylation by N-glycanase (data not shown). Similar observations were made when class I heavy chain molecules were incubated with N-glycanase prior to immunoprecipitation (data not shown). The differential behavior of N-glycanase towards free class I heavy chains or fully assembled class I trimer is therefore not due to the different antibodies used for retrieval of class I molecules. Partially purified mammalian N-glycanase behaved indistinguishably from the yeast enzyme (data not shown), indicating that both the mammalian and the yeast PNG1 discriminate between folded and unfolded glycoproteins, as well as between high-mannose and complex type oligosaccharides.
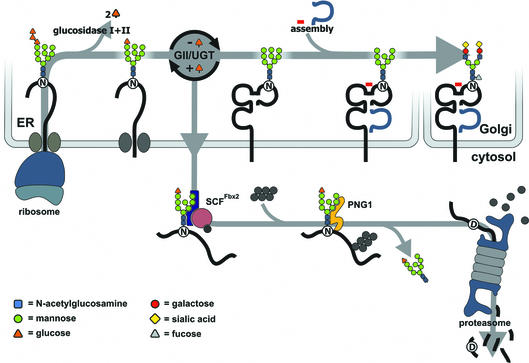
Fig. 9. A proposed model for the degradation of MHC heavy chains from the ER. Class I MHC heavy chain molecules are inserted into the ER where the N-linked oligosaccharide is transferred from a dolicholpyrophosphate carrier onto the Asn-X-Ser/Thr acceptor sequence in the nascent chain (Silberstein and Gilmore, 1996). The N-linked oligosaccharide carries three glucose residues that are removed sequentially by ER glucosidases I and II (GI/GII). The concerted action of glucosidase II and UDP-glucose: UGT constitutes a cycle that deglucosylates and reglucosylates the oligosaccharide on the folding polypeptide chain. Calnexin and calreticulin bind to the monoglucosylated form of the oligosaccharide and assist in folding of the polypeptide. Binding of β2m and a peptide allows the heavy chain to exit to the Golgi where the high-mannose type N-linked glycan is converted to a complex type glycan before the fully assembled complex reaches the cell surface. Proteins that fail to fold properly or do not assemble with their appropriate binding partners are retained in the ER and become a substrate of ER mannosidase I (not shown) after a certain lag time, as do properly folded glycoproteins, yielding a Man8 structure. A Man8 structure on a misfolded protein signals that the polypeptide has resided in the ER for some time without acquiring its native structure. Such proteins are extracted from the ER and become ubiquitylated by ubiquitylating enzymes that may reside at the cytosolic face of the ER membrane. A recently discovered E3 ubiquitin ligase, Fbx2, recognizes N-linked glycans in the cytosol and may trigger selective ubiquitylation of glycoproteins upon their arrival in the cytosol (Yoshida et al., 2002). The 26S proteasome degrades the glycoprotein after its deglycosylation by N-glycanase. The deglycosylation results in the conversion of the asparagine that carried the N-linked glycan to an aspartate. It has not yet been proven whether cytosolic oligosaccharides carry a terminal Glc residue.
Fig. 7. Only free heavy chains bearing high-mannose type oligosaccharides are a target of N-glycanase. Lymphoblastoid B cells (FH17) were pulse labeled for 10 min. Free class I MHC heavy chain molecules were obtained by immunoprecipitation immediately (0 min) after the pulse using the HC10 antibody (lane 1), and heavy chains complexed with β2m were immunoprecipitated by the conformation-specific W6/32 antibody (lane 2). At 120 min after the pulse, heavy chains were immunoprecipitated by the W6/32 antibody (lane 3). The number of glycans is indicated; the asterisk represents glycans of the complex type. Samples were treated with Endo H (lanes 4–6) or digested with 10 µg of the yeast Png1p (lanes 7–9). Fully assembled class I MHC molecules from the 0 and 120 min time points were denatured by boiling with SDS and incubated with yeast Png1p after re-immunoprecipitation with the HC10 antibody (lanes 10 and 11).
Truncated forms of high-mannose type glycans are not recognized by N-glycanase
To examine whether truncated forms of the high-mannose type N-linked glycan are a suitable substrate for N-glycanase, we digested class I MHC heavy chains with Endo H or with jack bean mannosidase. The resulting glycoproteins were then used as substrates in the deglycosylation assay. After the deglycosylation reaction, the products were analyzed by SDS–PAGE and IEF (Figure 8). Treatment of heavy chains with bacterial Endo F released the N-glycan and converted the asparagine side chain that carried the glycan to an aspartate residue as judged by SDS–PAGE and IEF (Figure 8, lanes 2 and 9). Incubation of class I heavy chains with mammalian N-glycanase resulted in partial deglycosylation and a shift in isoelectric point (Figure 8, lanes 3 and 10). Exposure of Endo H-treated heavy chains to mammalian N-glycanase did not alter the isoelectric point of the heavy chains (Figure 8, lanes 5 and 12). Heavy chains that carry a single N-acetylglucosamine are therefore not a substrate for mammalian N-glycanase. Treat ment of class I heavy chains with with jack bean mannosidase yielded a preparation with truncated oligosaccharides, predicted to be a Manβ1–4GlcNAc–GlcNAc core trisaccharide (Figure 8, lanes 6 and 13) (Barber et al., 1996). No shift in isoelectric point was seen after mannosidase treatment. Exposure of the jack bean mannosidase-treated samples to COS cell N-glycanase did not alter the mobility of free heavy chains (Figure 8, lanes 7 and 14), suggesting that a Manβ1–4GlcNAc– GlcNAc core trisaccharide does not serve as a substrate for COS cell N-glycanase.
Fig. 8. Truncated forms of high-mannose N-glycans are not recognized by N-glycanase. Class I MHC heavy chains were obtained from U373 cells by immunoprecipitation. After digestion with the indicated glycosidases, samples were analyzed either directly by SDS–PAGE (top panel) or IEF (bottom panel), or a subsequent digestion with mammalian PNG1 was performed prior to gel electrophoresis where indicated. Incubation with mannosidase yielded a doublet band, which is presumably due to incomplete digestion of the substrate as indicated by the two structures shown on the left. The faster migrating protein (closed arrowhead for lane 1) is a glycosidase-sensitive proteolytic degradation product (open arrowhead for lanes 2–5) of the heavy chain molecule.
Discussion
Peptide:N-glycanase is thought to be involved in the cytosolic turnover of glycoproteins, since a number of misfolded glycoproteins that are degraded from the ER appear as deglycosylated but otherwise intact intermediates in the cytosol (Wiertz et al., 1996a; Halaban et al., 1997; Huppa and Ploegh, 1997; de Virgilio et al., 1998; Johnston et al., 1998). These intermediates are observed only when proteasomal proteolysis is blocked, suggesting that N-deglycosylation precedes destruction by the proteasome. Alternatively, inhibition of the proteasome could redirect glycoproteins to an altogether different pathway, where the N-linked glycans are then targeted by N-glycanase. While inhibition of the proteasome yields deglycosylated protein intermediates in the cytosol, it also stabilizes to some extent their precursors in the ER (Halaban et al., 1997; de Virgilio et al., 1998; Meerovitch et al., 1998; Yang et al., 1998). Proteasome inhibitors should therefore decrease the amount of free oligosaccharides released into the cytosol if N-glycanase is an integral component of the ER quality control system. Karaivanova and Spiro (2000) explored the relationship between ER-associated degradation and the release of free polymannose oligosaccharides in the cytosol. Indeed, they found a reduction of free oligosaccharides in the cytosol when the proteasome was blocked, and degradation of ER resident glycoproteins was induced with castanospermine. The appearance of deglycosylated glycoprotein breakdown intermediates in the cytosol is therefore not an artifact induced by the addition of proteasome inhibitors.
Of immediate immunological relevance to the involvement of N-glycanase in glycoprotein turnover is the observation that several TAP (transporter associated with antigen processing)-dependent, class I MHC-restricted epitopes derived from glycoproteins require the post-translational conversion of the asparagine that carried the oligosaccharide moiety to aspartate (Skipper et al., 1996; Bacik et al., 1997; Mosse et al., 1998), a modification that occurs during the deamidation reaction catalyzed by N-glycanase. This observation, together with the finding that proteasome inhibitors decrease the amount of free oligosaccharides in the cytosol, demonstrates that removal of N-linked glycans and glycoprotein degradation are coupled events.
The properties of PNG1 as reported here support a role for this enzyme in the degradation of proteins from the ER. The activity is located in the cytosol where it may assist the proteasome in the task of eliminating glycoproteins derived from the ER. N-glycanase is capable of deglycosylating TCRα, CD3δ (data not shown) and free class I MHC heavy chains. All of these proteins are well characterized substrates of this degradation pathway. We do not know why deglycosylation of full-length proteins by PNG1 has not been observed before. It is possible that commercially available substrates represent mostly properly folded glycoproteins. In this context, it is of particular interest that both mammalian and yeast PNG1 distinguish free class I MHC heavy chain molecules from heavy chains in a complex with β2m. The N-linked glycan of a heavy chain in a complex with β2m extends outward from the protein, and the glycan is therefore expected to be fully accessible to N-glycanase (see Figure 10A). Indeed, both Endo H and Endo F readily deglycosylate heavy chains in a complex with β2m, without the need for inclusion of denaturants such as SDS. This suggests that PNG1 possesses an additional trait that allows it to distinguish properly folded glycoproteins from substrates that must be attacked. This feature is reminiscent of the properties of one of the key enzymes in ER quality control: UDP-glucose:glycoprotein glucosyltransferase (UGT), which serves as the folding sensor in the calnexin/calreticulin cycle (Helenius and Aebi, 2001). Calnexin and calreticulin are two ER resident lectins that interact specifically with monoglucosylated glycoproteins. By binding and release of proteins bearing monoglucosylated glycans, calnexin and calreticulin promote folding and assembly of unfolded proteins in the lumen of the ER. Interaction of a folding substrate with calnexin or calreticulin and glucosidase II results in removal of the last glucose from the N-linked glycan. This event releases the protein from the calnexin/calreticulin cycle (Figure 9). Proteins that have failed to reach their native conformation are reglucosylated by UGT and re-enter the calnexin/calreticulin cycle. Our data suggest that N-glycanase not only recognizes high-mannose oligosaccharides and distinguishes them from complex type glycans, but may do so preferentially on non-native proteins.
Fig. 10. Structure of class I MHC heavy chain with an N-linked glycan and the proteasome degrading a glycopeptide. (A) Modeling of the HLA-A2–Tax peptide complex with β2m and a high-mannose N-glycan attached to Asn86. The HLA-A2 chain is rendered in red, β2m in green, Tax peptide in white and the oligosaccharide in yellow. Asn86 is blue. (B) Cross-section of the 20S proteasome modeled with the nine-residue N-terminal tail of the α3 subunit deleted to open the outer pore (Groll et al., 2000). Seven α-subunits (green) form the outer pore that leads to the antechamber; the inner cavity is formed by seven β-subunits (blue). The glycopeptide consists of residues 70–95 from the HLA-A2 sequence (red) and an N-glycan (yellow) attached to Asn86 (blue).
Analysis of the substrate specificity of mammalian PNG1 demonstrated that the enzyme can attack the triglucosylated N-glycan that is transferred co-translationally onto the nascent polypeptide chain and all subsequent processing intermediates that occur naturally in the ER (data not shown). Truncated forms of the high-mannose oligosaccharide such as those generated by digestion with jack bean α-mannosidase were not recognized by this N-glycanase. Class I MHC heavy chains bearing glycans of the complex type were not deglycosylated at all by N-glycanase, consistent with a role for this enzyme in ER quality control. Proteins that are degraded via this pathway enter the cytoplasm from a pre-Golgi compartment and never acquire complex type N-glycans. Efficient degradation of certain glycoproteins may require their transport from the ER to the Golgi and back. These substrates can receive an α1,6 mannose addition in the cis-Golgi (Haynes et al., 2002), but they do not acquire Endo H resistance (Vashist et al., 2001). Cytosolic PNG1 is therefore unlikely to encounter glycoproteins bearing complex type N-glycans. The complex type oligosaccharide of properly assembled human MHC class I chain is α1,6 fucosylated in the Golgi (Barber et al., 1996), which may sterically hinder N-glycanase from attacking complex oligosaccharides of this type. While some of the experiments shown were carried out using partially purified mammalian PNG1, we also observed deglycosylation of class I MHC molecules and TCRα using the purified yeast PNG1 enzyme. Deglycosylation by PNG1 therefore does not require any additional factors. Our gel filtration experiments suggest that PNG1 is multimeric: mammalian PNG1 has a mol. wt of ∼200 kDa, whereas the cloned murine PNG1 has a predicted mol. wt of only 74 kDa. The active enzyme may form a homomultimeric complex or may associate with other subunits not required for enzymatic activity. While we have not been able to co-immunoprecipitate additional polypeptides with the GFP-tagged PNG1, Suzuki and co-workers have reported an association of both the mammalian and the yeast PNG1 with Rad23. Rad23 contains an N-terminal ubiquitin-like domain that can bind to the proteasome (Suzuki et al., 2001), which suggests a physical interaction between PNG1 and the proteasome. A yeast two-hybrid screen likewise showed that mouse PNG1 interacts with mHR23B and a subunit of the 19S cap of the proteasome (Park et al., 2001).
The appearance of deglycosylated intermediates in the cytosol and the ability of PNG1 to digest full-length glycoproteins imply that deglycosylation precedes degradation. Consistent with this hypothesis, the Rad23 protein mediates the association of PNG1 with the proteasome. To enter the central cavity that contains the active sites of the proteasome, a substrate must pass through an axial channel constricted by two narrow pores. The outer pore is gated and opens only upon binding of a regulatory particle such as the 19S or the 11S particle. The complex of the yeast proteasome with a heterologous 11S particle shows an opened outer pore (Whitby et al., 2000). Further, a deletion of the N-terminal tail of the α3-subunit opens the pore occluded in wild-type proteasomes (Groll et al., 2000). In both cases, the dimensions of the outer pore are comparable with those of the ungated pore of the Thermoplasma acidophilum proteasome, which has a diameter of 13 Å (Lowe et al., 1995). The inner pore that leads to the central cavity has a diameter of 20 Å (Rechsteiner et al., 2000). We modeled a stretch of the HLA-A2 (amino acids 70–95) peptide carrying the high-mannose type N-glycan-attached Asn86 into the antechamber of a proteasome where the outer pore is in an open state (Figure 10B). The degree of flexibility attributed to oligosaccharides (Rudd et al., 1999) might allow an N-linked glycan attached to a polypeptide to reach the inner chamber of the proteasome. It was proposed that small openings located at the interface between the α- and β-subunits of the 20S proteasome serve as an exit for the breakdown products generated by the proteasome (Groll et al., 1997). These fenestrations have a diameter of 10 Å and are unlikely to allow the exit of an N-glycan. Peptides generated by the proteasome might escape from the inner chamber via the channel that is also used for protein entry (Kohler et al., 2001). Since the outer pore is opened only when a regulatory particle has bound, peptides presumably leave the proteasome via the site from which substrates are fed into the inner chamber. This two-way traffic would reduce even further the space effectively available for an N-linked glycan. Many glycoproteins contain more than one N-linked glycan, and degradation of a protein such as TCRα would result in two or more N-linked glycans traveling in opposite directions if this model were correct. Therefore, N-glycanase digestion should facilitate glycoprotein degradation by the proteasome.
Is N-glycanase essential for glycoprotein turnover? A yeast strain deficient in the PNG1 gene is viable and capable of degrading a mutant form of carboxypeptidase Y (Suzuki et al., 2000), consistent with the possibility of complete glycoprotein breakdown by the proteasome. While both models need not be mutually exclusive, we favor the idea that N-glycanase acts upstream of the proteasome. The failure rate of protein synthesis is considerable (Schubert et al., 2000) and, especially under stressful conditions, N-glycanase may be more important to prevent overload of the cytoplasm with incompletely degraded protein fragments. The availability of siRNA capable of reducing N-glycanase activity will be a useful tool to explore the role of this enzyme in cellular physiology.
Materials and methods
Antibodies, chemicals and enzymes
α-LDH antibody, NEM and jack bean mannosidase were purchased from Sigma, and DTT was purchased from Boehringer Ingelheim. Endo F and Endo H were from New England Biolabs and used according to the manufacturer’s instructions. The polyclonal antisera directed against TCRα (Huppa and Ploegh, 1997) and GFP (Fiebiger et al., 2002) were used as described before.
Cell lines and transfection
U373-MG astrocytoma cells, HEK-293 cells, COS-1 cells and CEM-NKR cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS). The lymphoblastoid B-cell line FH17 was grown in RPMI with 10% FCS. CEM-NKR cells expressing TCRα were generated by electroporation with pcDNA 3.1 containing the cDNA of TCRα. Transient transfection of HEK-293 cells was performed using Lipofectamine (Gibco) according to the manufacturer’s instructions. Metabolic labeling, immunoprecipitations and gel electrophoresis were performed as described (Huppa and Ploegh, 1997).
Molecular cloning
Expression plasmids for the mouse PNG1 protein were cloned by RT–PCR. The primers used were PNG1-S (5′-CCGGAATTCCGG ATGGCGTCGGCCACACTGGGCAGC-3′) and PNG1-AS (5′-CC GCTCGAGCGGTCAGAGGTCATTGAACGTTATAATTATTTCC-3′). The PCR product was cloned into the pGEM-T(+) vector (Promega) and, after digestion with EcoRI and XhoI, subcloned into the pcDNA 3.1(+) vector (Invitrogen). Mouse PNG1 cDNA was cloned between the XhoI and EcoRI site of pEGFP-C1 (Clontech) to generate the GFP-tagged constructs. Site-directed mutagenesis of the active site cysteine to alanine of PNG1 cDNA was achieved by PCR. The primers used were PNG1-S306-A (5′-CAAGATGTGGACGCGCTGGTGAATGGGCC-3′) and the reverse complement of PNG1-S306-A.
Deglycosylation assay
The source for the [35S]methionine-labeled TCRα was CEM-NKR cells that were stably transfected with TCRα. CEM-NKR cells were metabolically labeled for 45 min and, for the assay, TCRα either was purified by immunoprecipitation or whole lysates from CEM-NKR were used directly after the cells were treated with NEM to inhibit endogenous N-glycanase activity. MHC class I molecules were obtained from U373 astrocytoma cells or the lymphoblastoid B-cell line FH17. The substrates were incubated with the fractions to be tested for activity or with NP-40 lysates from COS-1 cells containing 750 µg of protein in a final volume of 1 ml of 25 mM Tris pH 7.4, 0.5% NP-40 (v/v), 5 mM DTT and 5 mM MgCl2 overnight at 4°C. Substrates were then immunoprecipitated and analyzed by SDS–PAGE.
Protein purification
COS-1 cells were detached by trypsinization and washed twice in phosphate-buffered saline supplemented with 1 mM phenylmethylsulfonyl fluoride. Cells (108) were resuspended in 50 ml of 250 mM sucrose, 25 mM imidazole pH 7.0, 5 mM DTT, 0.1 mM EDTA, and homogenized by passing three times through a French press at 0.5 MPa. Cytosol was prepared from the homogenate by centrifugation for 30 min at 10 000 g in an SW34 rotor (Sorvall). The supernatant was spun at 100 000 g for 1 h in a Ti50.1 rotor (Beckmann). Adjusting the (NH4)2SO4 concentration to 50% precipitated proteins in the supernatant. Precipitated proteins were collected by centrifugation in an SW34 rotor (Sorvall) at 10 000 g for 15 min. The protein pellet was solubilized in 50 mM sodium phosphate buffer (pH 6.8, 10% glycerol, 5 mM DTT, 0.1 mM EDTA) and applied to a hydroxyapatite column (Bio-Rad Hydroxyapatite Bio-Gel HT Gel, 5 × 3 cm). The flowthrough fraction was dialyzed overnight against buffer A (25 mM imidazole pH 7.0, 10% glycerol, 5 mM DTT, 0.1 mM EDTA) and 400 mM (NH4)2SO4. The dialyzed material was applied on a Toyopearl MD P Butyl Hydrophobic Interaction column (Toso Haas). Bound proteins were eluted by decreasing the (NH4)2SO4 concentration to 0 mM over 15 column volumes. Fractions (1 ml) were collected and analyzed for N-glycanase activity. Active fractions were pooled, and proteins present in the active fractions were precipitated by adjusting the (NH4)2SO4 concentration to 50%. The precipitate was solubilized in buffer A + 0.25% Triton X-100 (w/v) and loaded on a Bio-Rad Uno-Q1 Anion Exchange column. N-glycanase activity was eluted by an NaCl gradient from 0 to 400 mM NaCl over 10 column volumes. Fractions of 0.5 ml were collected and assayed for N-glycanase activity. Active fractions were dialyzed overnight against buffer A + 0.25% Triton X-100 (w/v) and applied on a Mono P HR 5/5 Chromatofocusing Column (Pharmacia). Bound protein was eluted at pH 5.5. The pH of each fraction was adjusted to pH 7.0 by adding imidazole pH 7.0. Fractions containing N-glycanase activity were concentrated and loaded on a Superdex 200A gel filtration column (Pharmacia). Fractions of 0.5 ml were collected.
Generation of mouse 3T3 and COS-1 knockdown cell lines
We chose four regions of the PNG1 mRNA that are evolutionarily conserved from mouse to human, and selected 19 nucleotides of each region. These 19-nucleotide sequences were used to clone pRETRO (Brummelkamp et al., 2002). Transcription of the retroviral construct gives rise to the selected 19-nucleotide sequence separated by a nine-nucleotide spacer from the reverse complement of the same selected 19-nucleotide sequence (for details see Brummelkamp et al., 2002). Selected 19-nucleotide sequences were as follows (numbers in parentheses indicate PNG1 nucleotide coordinates based on the mouse cDNA sequence NM021504, GenBank): construct I, GATGAGGTAGTTGA TGTCA (1171–1193); construct II, TTGTGGAGCTTGTTGAATT (1340–1362); construct III, TGGGCTTTGAAGAGGGAGA (269–291); and construct IV, GTTCTTCAGTCCAACATTC (547–569).
After puromycin selection, infected cells were expanded, and 400–600 µg of NP-40 cell extracts were tested in the deglycosylation assay described above.
Bacterial expression and purification of Png1p
BL21 cells were transformed with the pET28b vector containing the PNG1 sequence (provided by Tadashi Suzuki, Stony Brook, NY), and expression was induced by adding 1 mM isopropyl-β-d-thiogalacto pyranoside at an OD600 of 0.8. After 3 h induction, cells were harvested by centrifugation and incubated on ice for 30 min in buffer A supplemented with 1 mg/ml lysozyme, followed by sonication. The extract was centrifuged at 10 000 g for 20 min and the supernatant was loaded on a Bio-Rad Unopeek Q-12 column. Bound protein was eluted by a gradient from 0 to 500 mM NaCl over 10 column volumes. Fractions containing Png1p were pooled, concentrated and subsequently separated on a Superdex 75 column.
Molecular modeling
The high-mannose type N-linked glycan from the adhesion domain of human CD2 (Wyss et al., 1995) was attached in silico to Asn86 of the HLA-A2–Tax peptide complex (Gewurz et al., 2001). Residues 71–95 of the HLA-A2 sequence with the CD2 N-linked glycan were modeled into the structure of the yeast 20S proteasome (Groll et al., 2000). Molecular modeling and rendering of the images were performed using Pymol software (DeLano, 2002).
Acknowledgments
Acknowledgements
We thank Dr R.Agami for donating pSUPER and pRETRO required to generate the siRNA knockdown constructs. C.H. is a fellow of the Boehringer Ingelheim Fonds, Germany. This work was supported in part by grants from the NIH and the Mizutani Foundation for Glycoscience, Tokyo, Japan.
References
- Bacik I. et al. (1997) Introduction of a glycosylation site into a secreted protein provides evidence for an alternative antigen processing pathway: transport of precursors of major histocompatibility complex class I-restricted peptides from the endoplasmic reticulum to the cytosol. J. Exp. Med., 186, 479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber L.D. et al. (1996) Unusual uniformity of the N-linked oligosaccharides of HLA-A, -B and -C glycoproteins. J. Immunol., 156, 3275–3284. [PubMed] [Google Scholar]
- Barnstable C.J., Bodmer,W.F., Brown,G., Galfre,G., Milstein,C., Williams,A.F. and Ziegler,A. (1978) Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens—new tools for genetic analysis. Cell, 14, 9–20. [DOI] [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- de Virgilio M., Weninger,H. and Ivessa,N.E. (1998) Ubiquitination is required for the retro-translocation of a short-lived luminal endoplasmic reticulum glycoprotein to the cytosol for degradation by the proteasome. J. Biol. Chem., 273, 9734–9743. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2002) The PyMOL Molecular Graphics System. http://www.pymol.org.
- Fiebiger E., Story,C., Ploegh,H.L. and Tortorella,D. (2002) Visualization of the ER-to-cytosol dislocation reaction of a type I membrane protein. EMBO J., 21, 1041–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gewurz B.E., Gaudet,R., Tortorella,D., Wang,E.W., Ploegh,H.L. and Wiley,D.C. (2001) Antigen presentation subverted: structure of the human cytomegalovirus protein US2 bound to the class I molecule HLA-A2. Proc. Natl Acad. Sci. USA, 98, 6794–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Ditzel,L., Lowe,J., Stock,D., Bochtler,M., Bartunik,H.D. and Huber,R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Groll M., Bajorek,M., Kohler,A., Moroder,L., Rubin,D.M., Huber,R., Glickman,M.H. and Finley,D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol., 7, 1062–1067. [DOI] [PubMed] [Google Scholar]
- Halaban R., Cheng,E., Zhang,Y., Moellmann,G., Hanlon,D., Michalak,M., Setaluri,V. and Hebert,D.N. (1997) Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc. Natl Acad. Sci. USA, 94, 6210–6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes C.M., Caldwell,S. and Cooper,A.A. (2002) An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER–Golgi transport. J. Cell Biol., 158, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemels M.T. and Ploegh,H. (1995) Generation, translocation and presentation of MHC class I-restricted peptides. Annu. Rev. Biochem., 64, 463–491. [DOI] [PubMed] [Google Scholar]
- Helenius A. and Aebi,M. (2001) Intracellular functions of N-linked glycans. Science, 291, 2364–2369. [DOI] [PubMed] [Google Scholar]
- Huppa J.B. and Ploegh,H.L. (1997) The α chain of the T cell antigen receptor is degraded in the cytosol. Immunity, 7, 113–122. [DOI] [PubMed] [Google Scholar]
- Johnston J.A., Ward,C.L. and Kopito,R.R. (1998) Aggresomes: a cellular response to misfolded proteins. J. Cell Biol., 143, 1883–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaivanova V.K. and Spiro,R.G. (2000) Effect of proteasome inhibitors on the release into the cytosol of free polymannose oligosaccharides from glycoproteins. Glycobiology, 10, 727–735. [DOI] [PubMed] [Google Scholar]
- Katiyar S., Suzuki,T., Balgobin,B.J. and Lennarz,W.J. (2002) Site-directed mutagenesis study of yeast peptide:N-glycanase. Insight into the reaction mechanism of deglycosylation. J. Biol. Chem., 277, 12953–12959. [DOI] [PubMed] [Google Scholar]
- Kohler A., Cascio,P., Leggett,D.S., Woo,K.M., Goldberg,A.L. and Finley,D. (2001) The axial channel of the proteasome core particle is gated by the Rpt2 ATPase and controls both substrate entry and product release. Mol. Cell, 7, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Lowe J., Stock,D., Jap,B., Zwickl,P., Baumeister,W. and Huber,R. (1995) Crystal structure of the 20S proteasome from the archaeon T.acidophilum at 3.4 Å resolution. Science, 268, 533–539. [DOI] [PubMed] [Google Scholar]
- Makarova K.S., Aravind,L. and Koonin,E.V. (1999) A superfamily of archaeal, bacterial and eukaryotic proteins homologous to animal transglutaminases. Protein Sci., 8, 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K., Wing,S. and Goltzman,D. (1998) Proparathyroid hormone-related protein is associated with the chaperone protein BiP and undergoes proteasome-mediated degradation. J. Biol. Chem., 273, 21025–21030. [DOI] [PubMed] [Google Scholar]
- Mosse C.A., Meadows,L., Luckey,C.J., Kittlesen,D.J., Huczko,E.L., Slingluff,C.L., Shabanowitz,J., Hunt,D.F. and Engelhard,V.H. (1998) The class I antigen-processing pathway for the membrane protein tyrosinase involves translation in the endoplasmic reticulum and processing in the cytosol. J. Exp. Med., 187, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Suzuki,T. and Lennarz,W.J. (2001) Identification of proteins that interact with mammalian peptide:N-glycanase and implicate this hydrolase in the proteasome-dependent pathway for protein degradation. Proc. Natl Acad. Sci. USA, 98, 11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer T.H. Jr, Elder,J.H., Alexander,S., Phelan,A.W. and Tarentino,A.L. (1984) Demonstration of peptide:N-glycosidase F activity in endo-β-N-acetylglucosaminidase F preparations. J. Biol. Chem., 259, 10700–10704. [PubMed] [Google Scholar]
- Rechsteiner M., Realini,C. and Ustrell,V. (2000) The proteasome activator 11S REG (PA28) and class I antigen presentation. Biochem. J., 345, 1–15. [PMC free article] [PubMed] [Google Scholar]
- Rudd P.M. et al. (1999) Roles for glycosylation of cell surface receptors involved in cellular immune recognition. J. Mol. Biol., 293, 351–366. [DOI] [PubMed] [Google Scholar]
- Schubert U., Anton,L.C., Gibbs,J., Norbury,C.C., Yewdell,J.W. and Bennink,J.R. (2000) Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature, 404, 770–774. [DOI] [PubMed] [Google Scholar]
- Seko A., Kitajima,K., Inoue,Y. and Inoue,S. (1991) Peptide: N-glycosidase activity found in the early embryos of Oryzias latipes (Medaka fish). The first demonstration of the occurrence of peptide:N-glycosidase in animal cells and its implication for the presence of a de-N-glycosylation system in living organisms. J. Biol. Chem., 266, 22110–22114. [PubMed] [Google Scholar]
- Silberstein S. and Gilmore,R. (1996) Biochemistry, molecular biology and genetics of the oligosaccharyltransferase. FASEB J., 10, 849–858. [PubMed] [Google Scholar]
- Skipper J.C. et al. (1996) An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med., 183, 527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam N.J., Vroom,T.M., Peters,P.J., Pastoors,E.B. and Ploegh,H.L. (1990) HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int. Immunol., 2, 113–125. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Seko,A., Kitajima,K., Inoue,Y. and Inoue,S. (1993) Identification of peptide:N-glycanase activity in mammalian-derived cultured cells. Biochem. Biophys. Res. Commun., 194, 1124–1130. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Park,H., Hollingsworth,N.M., Sternglanz,R. and Lennarz,W.J. (2000) PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J. Cell Biol., 149, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park,H., Kwofie,M.A. and Lennarz,W.J. (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J. Biol. Chem., 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- Takahashi N. (1977) Demonstration of a new amidase acting on glycopeptides. Biochem. Biophys. Res. Commun., 76, 1194–1201. [DOI] [PubMed] [Google Scholar]
- Tarentino A.L. and Plummer,T.H.,Jr (1994) Enzymatic deglycosylation of asparagine-linked glycans: purification, properties and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol., 230, 44–57. [DOI] [PubMed] [Google Scholar]
- Vashist S., Kim,W., Belden,W.J., Spear,E.D., Barlowe,C. and Ng,D.T. (2001) Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol., 155, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng S. and Spiro,R.G. (1997) Demonstration of a peptide: N-glycosidase in the endoplasmic reticulum of rat liver. Biochem. J., 322, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby F.G., Masters,E.I., Kramer,L., Knowlton,J.R., Yao,Y., Wang,C.C. and Hill,C.P. (2000) Structural basis for the activation of 20S proteasomes by 11S regulators. Nature, 408, 115–120. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Jones,T.R., Sun,L., Bogyo,M., Geuze,H.J. and Ploegh,H.L. (1996a) The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell, 84, 769–779. [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Tortorella,D., Bogyo,M., Yu,J., Mothes,W., Jones,T.R., Rapoport,T.A. and Ploegh,H.L. (1996b) Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature, 384, 432–438. [DOI] [PubMed] [Google Scholar]
- Wyss D.F., Choi,J.S., Li,J., Knoppers,M.H., Willis,K.J., Arulanandam, A.R., Smolyar,A., Reinherz,E.L. and Wagner,G. (1995) Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science, 269, 1273–1278. [DOI] [PubMed] [Google Scholar]
- Yang M., Omura,S., Bonifacino,J.S. and Weissman,A.M. (1998) Novel aspects of degradation of T cell receptor subunits from the endoplasmic reticulum (ER) in T cells: importance of oligosaccharide processing, ubiquitination and proteasome-dependent removal from ER membranes. J. Exp. Med., 187, 835–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee V.C., Pedersen,L.C., Le Trong,I., Bishop,P.D., Stenkamp,R.E. and Teller,D.C. (1994) Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII. Proc. Natl Acad. Sci. USA, 91, 7296–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y. et al. (2002) E3 ubiquitin ligase that recognizes sugar chains. Nature, 418, 438–442. [DOI] [PubMed] [Google Scholar]
- Yu H., Kaung,G., Kobayashi,S. and Kopito,R.R. (1997) Cytosolic degradation of T-cell receptor α chains by the proteasome. J. Biol. Chem., 272, 20800–20804. [DOI] [PubMed] [Google Scholar]