Abstract
In viral cap-snatching, the endonuclease intrinsic to the viral polymerase cleaves cellular capped RNAs to generate capped fragments that are primers for viral mRNA synthesis. Here we demonstrate that the influenza viral polymerase, which is assembled in human cells using recombinant proteins, effectively uses only CA-terminated capped fragments as primers for viral mRNA synthesis in vitro. Thus we provide the first in vitro system that mirrors the cap-snatching process occurring in vivo during virus infection. Further, we demonstrate that when a capped RNA substrate contains a CA cleavage site, the functions of virion RNA (vRNA) differ from those previously described: the 5′ terminal sequence of vRNA alone is sufficient for endonuclease activation, and the 3′ terminal sequence of vRNA functions solely as a template for mRNA synthesis. Consequently, we are able to identify the vRNA sequences that are required for each of these two separable functions. We present a new model for the influenza virus cap-snatching mechanism, which we postulate is a paradigm for the cap-snatching mechanisms of other segmented, negative-strand and ambisense RNA viruses.
Keywords: cap-snatching/endonuclease cleavage/influenza virus polymerase/viral mRNA synthesis
Introduction
Cap-snatching is the mechanism by which the synthesis of the mRNAs of influenza virus is initiated (Bouloy et al., 1978; Plotch et al., 1979, 1981; Krug et al., 1989). Specifically, the endonuclease intrinsic to the influenza viral polymerase cleaves cellular capped RNAs in the nucleus to generate capped fragments 10–13 nucleotides long that are primers for viral mRNA synthesis. Other segmented, negative-strand and ambisense RNA viruses, both animal and plant, share such a transcription initiation mechanism but use cytoplasmic, rather than nuclear, cellular capped RNA fragments as primers (Vialat and Bouloy, 1992; Huiet et al., 1993; Jin and Elliott, 1993a,b; Garcin et al., 1995; Ramirez et al., 1995; Estabrook et al., 1998; Duijsings et al., 2001). We anticipate that key features of the cap-snatching mechanism are shared among these various RNA viruses.
Important insights into the influenza virus cap-snatching mechanism have been provided using vaccinia virus vectors to express the three viral polymerase proteins PB1, PB2 and PA in human cells (Hagen et al., 1994; Tiley et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001; Leahy et al., 2001a,b). The resulting complex of three polymerase proteins does not exhibit cap-dependent endonuclease activity unless it is first activated by virion RNA (vRNA) sequences. Hence vRNA molecules function not only as templates for mRNA synthesis, but also as essential cofactors that activate the catalytic functions of the polymerase that produce capped RNA primers. All eight vRNA segments have 13 and 12 conserved bases at their 5′ and 3′ termini, respectively, which show partial inverted complementarity (Robertson, 1979; Desselberger et al., 1980). Previous studies have shown that the common 5′ terminal vRNA sequence binds to a sequence in the PB1 protein centered around two arginine residues at positions 571 and 572 (Li et al., 1998). This binding activated two other polymerase sites: (i) a cap-binding site on the PB2 protein, and (ii) an RNP1-like binding site on the PB1 protein for the 3′ terminal vRNA sequence (Li et al., 1998, 2001). These studies showed that subsequent binding of the 3′ terminal vRNA sequence to its cognate PB1 sequence activated the endonuclease that cleaves the bound capped RNAs 10–13 nucleotides from their 5′ ends (Hagen et al., 1994; Li et al., 2001). Activation of endonuclease activity was reported to require (i) base-pairing between bases 11–13 in the 5′ terminus of vRNA and bases 10–12 in the 3′ terminus, and (ii) small hairpin loops in both the 5′ and 3′ termini (Leahy et al., 2001a,b). Endonuclease cleavage resides in a PB1 sequence that contains three essential acidic amino acids (Li et al., 2001), similar to the active sites of other enzymes that cut polynucleotides to produce 3′-OH ends, including transposases and retroviral integrases (Baker and Luo, 1994; Dyda et al., 1994; Rice and Mizuuchi, 1995).
A major caveat about the studies cited above is that the capped RNA fragments generated by the endonuclease in these studies are poorly used as primers to initiate viral mRNA synthesis (Hagen et al., 1994; Leahy et al., 2001a,b; Li et al., 2001). Hence, either these capped fragments are intrinsically poor primers, or the vaccinia virus expressed polymerase is deficient in the transcription initiation reaction. Here we demonstrate that the former explanation is the case. We confirm that the capped RNA fragments generated in previous studies, which contain either AG or UA at their 3′ termini (Hagen et al., 1994; Leahy et al., 2001a,b; Li et al., 2001), are poorly used as primers to initiate viral mRNA synthesis. In contrast, we demonstrate that only those capped RNA fragments that contain a CA 3′ terminus are effectively used as primers. It was established in the early 1980s that the cellular capped primers which are used during virus infection contain predominately CA at their 3′ termini (Beaton and Krug, 1981; Shaw and Lamb, 1984). Thus we provide the first in vitro system mirroring the cap-snatching mechanism that occurs during virus infection in vivo.
Further, we demonstrate that when a capped RNA substrate contains a CA cleavage site, the functions of vRNA sequences in the initiation of viral mRNA synthesis differ from those previously described using other capped RNA substrates. Specifically, with a CA-containing capped RNA substrate, 5′ terminal vRNA alone is sufficient for the activation of the endonuclease, and 3′ terminal vRNA functions solely as a template for the initiation of mRNA synthesis. Consequently, we are able to identify the vRNA sequences that are required for each of these two separable functions. Based on our results, we present a new model for the influenza virus cap-snatching mechanism. We postulate that this model is a paradigm for the cap-snatching mechanisms catalyzed by the RNA polymerases of other segmented, negative strand and ambisense RNA viruses.
Results
CA-terminated capped RNA fragments are preferentially used as primers to initiate influenza viral mRNA synthesis
The influenza virus polymerase can be formed in HeLa cells using vaccinia virus recombinants expressing the influenza virus PB1, PB2 and PA proteins (Hagen et al., 1994; Tiley et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001; Leahy et al., 2001a,b). The cap-dependent endonuclease that is intrinsic to this polymerase was shown to be activated by the addition of both the 5′ and 3′ terminal sequences of influenza vRNA. For our experiments, we use RNA oligonucleotides 16 and 15 nucleotides long containing the 5′ and 3′ terminal sequences of influenza vRNA, respectively, which will be referred to as 5′ vRNA and 3′ vRNA (Li et al., 1998, 2001).
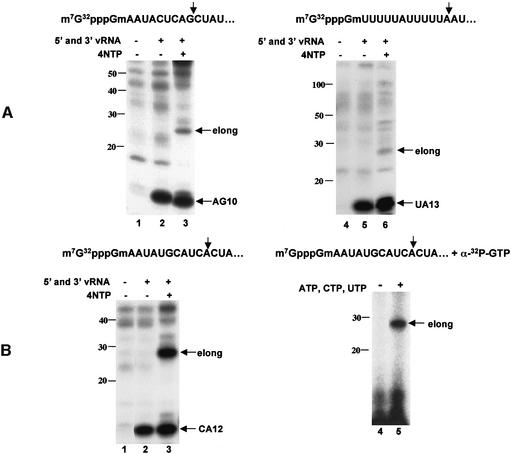
The capped RNA fragments that were generated by the endonuclease in these previous studies were poorly used as primers to initiate viral mRNA synthesis (Hagen et al., 1994; Li et al., 2001). Figure 1A shows representative examples of the poor priming exhibited by two capped RNA fragments used in previous studies, one containing an AG 3′ terminus (Hagen et al., 1994; Cianci et al., 1995; Leahy et al., 2001a,b) and the other containing a UA 3′ terminus (Li et al., 1998, 2001). To assay for both endonucleolytic cleavage and subsequent priming, we used capped RNA substrates that contain 32P only in their 5′ terminal cap structure and have the initial 5′ terminal sequences indicated in Figure 1A. In the absence of the four nucleoside triphosphates (NTPs), these two capped RNAs are efficiently cleaved at the indicated G or A residue to produce the capped RNA fragments, 10 and 13 nucleotides long, respectively, which are designated as AG10 and UA13 (lanes 2 and 5). However, in the presence of the four NTPs, only a very small amount of the capped RNA cleavage product is transcriptionally elongated (lanes 3 and 6), indicating that these two capped RNA fragments are poorly used as primers for the transcription of the 3′ vRNA template.
Fig. 1. CA-terminated capped RNA fragments are preferentially used as primers to initiate influenza viral mRNA synthesis. (A) The 32P-labeled capped RNA substrates containing the indicated initial 5′ terminal sequence were each incubated with a nuclear extract expressing the three P proteins in the absence of 5′ and 3′ vRNA (lanes 1 and 4), or in the presence of 5′ and 3′ vRNA in either the absence (lanes 2 and 5) or the presence (lanes 3 and 6) of the four NTPs. The RNA products were separated on a denaturing 20% polyacrylamide gel. Positions of the RNA markers 10–100 nucleotides long are denoted; elong denotes elongation product. (B) Left panel: the 32P-labeled capped RNA substrate containing a CA site was incubated with the nuclear extract in the absence of 5′ and 3′ vRNA (lane 1), or in the presence of 5′ and 3′ vRNA in either the absence (lane 2) or presence (lane 3) of the four NTPs. Right panel: the unlabeled capped RNA substrate containing a CA site was incubated with the nuclear extract in the presence of both 5′ and 3′ vRNA in the presence of either [α-32P]GTP alone (lane 4) or [α-32P]GTP and unlabeled ATP, CTP and GTP (lane 5).
The cellular capped primers that are used during influenza virus infection contain predominately CA at their 3′ termini (Beaton and Krug, 1981; Shaw and Lamb, 1984). To determine whether this in vitro system exhibits the same specificity, we used a 32P cap-labeled RNA substrate containing a CA sequence at a potential cleavage site (Figure 1B, left panel). This capped RNA is cleaved to produce a capped RNA fragment containing a CA 3′ terminus, designated as CA12 (lane 2). In the presence of the four NTPs, a large amount of an RNA product ∼14 nucleotides longer than the capped RNA fragment is produced (lane 3), demonstrating that the polymerase has efficiently transcribed the 15-nucleotide-long 3′ vRNA oligonucleotide. Thus this capped RNA fragment is an effective primer for the initiation of mRNA synthesis. It is expected that 14, rather than 15, nucleotides are added because transcription initiates at the penultimate, rather than the ultimate, 3′ vRNA base (Plotch et al., 1981; Krug et al., 1989). To confirm that transcription has occurred, we carried out a reaction in which the capped RNA was unlabeled and RNA synthesis was measured using a labeled NTP (α-[32P]GTP) (Figure 1B, right panel); an RNA product of the same size is observed (lane 2). These results demonstrate that our in vitro system mirrors in vivo events in that significant transcription of the 3′ vRNA template occurs with a CA-terminated capped fragment, but not with capped fragments containing two other termini. The capped RNA substrate that contains a CA cleavage site 10–13 nucleotides from the cap and hence generates a CA-terminated capped fragment will be designated capped CA-containing RNA in subsequent experiments.
In the presence of a capped CA-containing RNA substrate, only 5′ vRNA is required for endonuclease activation and 3′ vRNA functions solely as a template for the initiation of mRNA synthesis
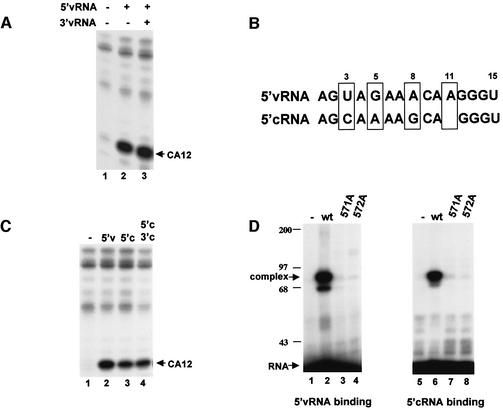
Because of our finding of the unique ability of a capped CA-containing RNA to produce capped RNA primers that initiate viral mRNA synthesis, we revisited the roles of 5′ vRNA and 3′ vRNA in polymerase activation using this capped RNA substrate. Previous studies using other capped RNA substrates found that the addition of both 5′ vRNA and 3′ vRNA is required for the activation of the endonuclease (Hagen et al., 1994; Tiley et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001; Leahy et al., 2001a,b). In contrast, as shown in Figure 2A, with a capped CA-containing RNA substrate, the addition of 5′ vRNA alone efficiently activates the endonuclease (lane 2). The presence of 3′ vRNA as well as 5′ vRNA results in only a small (∼10%) increase in endonuclease activity (lane 3). The same result was also obtained at 10-fold lower concentrations of 5′ vRNA (data not shown). Thus 5′ vRNA binding to the polymerase is sufficient to activate the endonuclease activity for CA-containing RNA substrates. Neither the interaction of 3′ vRNA with the polymerase nor base-pair interactions between 5′ and 3′ vRNA is required.
Fig. 2. (A) With a capped CA-containing RNA, only 5′ vRNA is required for endonuclease activation. 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the absence of 5′ and 3′ vRNA (lane 1), in the presence of 5′ vRNA (lane 2) or in the presence of both 5′ and 3′ vRNA (lane 3). (B) Comparison of the sequences of 5′ vRNA and 5′ cRNA. The four bases that differ between these two sequences are boxed. (C) With a capped CA-containing RNA, 5′ cRNA activates the endonuclease. 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the absence of vRNA and cRNA (lane 1), in the presence of 5′ vRNA (lane 2), in the presence of 5′ cRNA (lane 3) or in the presence of both 5′ and 3′ cRNA (lane 4). (D) 5′ vRNA and 5′ cRNA bind to the same PB1 sequence in the polymerase. A nuclear extract, either lacking viral P proteins (–) (lanes 1 and 5), or containing either wild-type (wt) P proteins (lanes 2 and 6) or the indicated mutant PB1 protein plus wild-type PB2 and PA proteins (lanes 3, 4, 7 and 8), was incubated with 32P-labeled 5′ vRNA (lanes 1–4) or 32P-labeled 5′ cRNA (lanes 5–8) containing a thioU. After UV cross-linking, protein–oligoribonucleotide complexes were separated from free oligoribonucleotide by gel electrophoresis. Positions of molecular weight markers (kDa) are denoted.
The sequence at the 5′ terminus of the full-length copies of vRNA, termed complementary RNA or cRNA, differs from 5′ vRNA at only four positions (Figure 2B). Previous studies using capped RNAs lacking a CA cleavage site showed that, in contrast with 5′ vRNA, 5′ cRNA did not activate endonuclease cleavage, although it activated cap-binding (Cianci et al., 1995; Leahy et al., 2002). However, when we use a capped CA-containing RNA substrate, 5′ cRNA alone also efficiently activates the endonuclease (Figure 2C), although the amount of endonuclease activity is slightly (∼20%) lower than that in the presence of 5′ vRNA. The further addition of 3′ cRNA results in little or no increase in endonuclease activity. RNA-binding assays using oligonucleotides containing thioU show that 5′ cRNA, like 5′ vRNA, forms a complex with the polymerase (Figure 2D, lanes 2 and 6). To determine whether 5′ cRNA and 5′ vRNA bind to the same site in the viral polymerase, we employed mutant polymerases in the RNA-binding assays. Mutagenesis experiments have established the identity of the 5′ vRNA binding site in the PB1 subunit: replacement of a single arginine at either position 571 or position 572 of PB1 with alanine inactivates the 5′ vRNA binding site in the polymerase (Li et al., 1998). As shown in Figure 2D, polymerases containing a PB1 protein subunit with such single mutations do not bind either 5′ vRNA or 5′ cRNA, indicating that 5′ cRNA and 5′ vRNA bind to the same site in the PB1 protein.
Our results establish that with a capped CA-containing RNA substrate, 3′ vRNA functions solely as a template for the initiation of mRNA synthesis. In contrast, the 3′ terminal sequence of cRNA (3′ cRNA) cannot replace 3′ vRNA as a template, whether 5′ vRNA or 5′ cRNA is used to activate the endonuclease (Figure 3B, lanes 4 and 6). Consequently, the activation of the CA-specific endonuclease by 5′ cRNA does not lead to capped RNA-primed transcription of cRNA because 3′ cRNA does not function as a template for such synthesis.
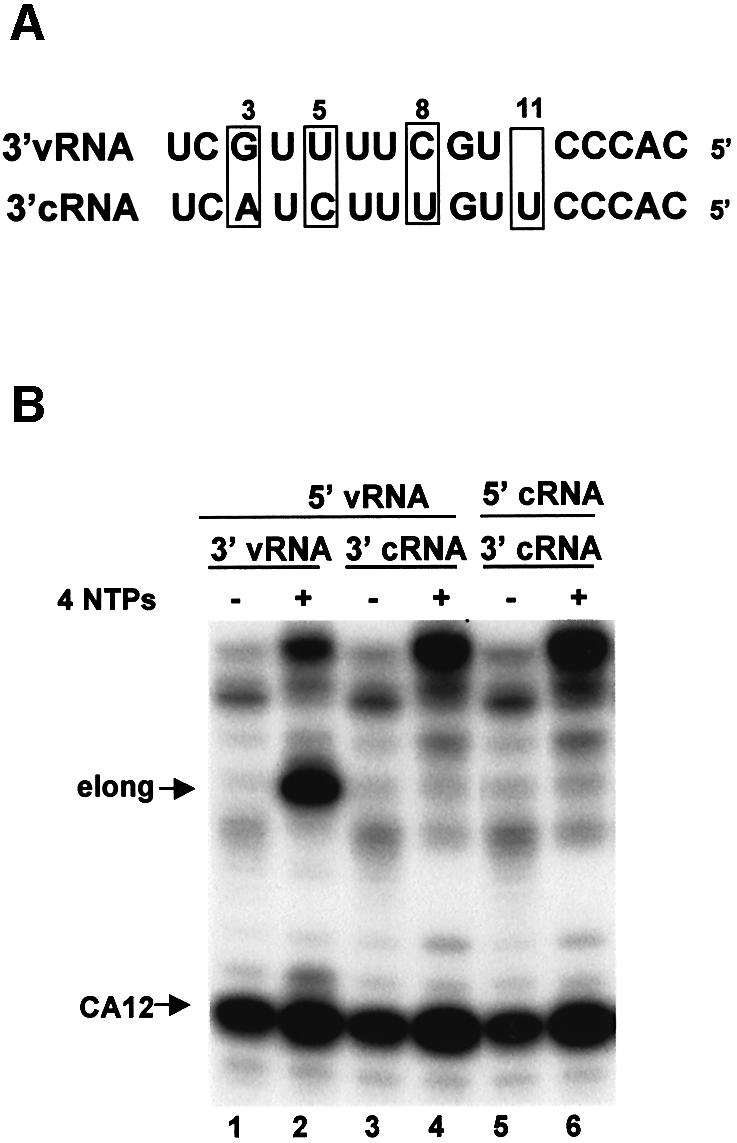
Fig. 3. 3′ vRNA, but not 3′ cRNA, functions as a template for the initiation of mRNA synthesis. (A) Comparison of the sequences of 3′ vRNA and 3′ cRNA. The four bases that differ between these two sequences are boxed. (B) 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the presence of the indicated combinations of 5′ vRNA, 5′ cRNA, 3′ vRNA and 3′ cRNA in the absence (–) or presence (+) of the four NTPs.
Identification of the sequences in 5′ vRNA that are required for activating the CA-specific endonuclease in the absence of 3′ vRNA
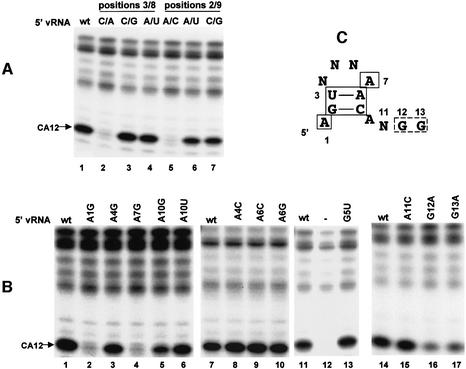
Previous studies using capped RNA substrates lacking a CA cleavage site identified the 5′ vRNA sequences that were required for endonuclease activation in the presence of 3′ vRNA (Leahy et al., 2001a). Here we identify the sequences in 5′ vRNA that are required for activation of the CA-specific endonuclease that is independent of 3′ vRNA. Because either 5′ cRNA or 5′ vRNA activates endonuclease cleavage, the four differences in the sequence (bases 3, 5, 8 and 11) between 5′ vRNA and 5′ cRNA (Figure 2B) have minimal effect on the activation of the CA-specific endonuclease. In fact, the identity of the bases at positions 3 and 8 has no effect on endonuclease activation as long as the base at position 3 can form hydrogen bonds with the base at position 8 (Figure 4A, lanes 1–4). Replacement of the wild-type 3U/8A sequence with either a 3C/8G sequence or a 3A/8U sequence (as found in 5′ cRNA), both of which allow hydrogen-bonding between positions 3 and 8, has no effect on endonuclease activitation, whereas the 3C/8A replacement, which does not allow such hydrogen-bonding, eliminates endonuclease activation. Similarly, the identity of the bases at positions 2 and 9 has no effect on endonuclease activity as long as the base at position 2 can form hydrogen bonds with the base at position 9 (lanes 1 and 5–7). Conse quently, a stem–tetraloop structure involving bases 2–9 is required for endonuclease activation.
Fig. 4. Identification of the bases/structures in 5′ vRNA that are required for endonuclease activation using a capped CA-containing RNA. (A) Role of 5′ vRNA bases 2, 3, 8 and 9. 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the presence of wt 5′ vRNA (lane 1) or in the presence of 5′ vRNA containing the indicated base changes (lanes 2–7). (B) Role of 5′ vRNA bases 1, 4, 5, 6, 7 and 10–13 assayed as described in (A). (C) Summary of the sequence/structure requirements for endonuclease activation by 5′ vRNA with a capped CA-containing RNA. N denotes any base. The bases/structure enclosed in solid boxes are required for endonuclease activity, and the bases enclosed in the broken box strongly affect endonuclease activity.
Of the bases in the tetraloop, the only one whose change leads to the loss of endonuclease activation is the A at position 7 (Figure 4B, lanes 3, 4, 8–10 and 13), indicating that this A is the only base in the 5′ vRNA tetraloop that is required for activation of the CA-specific endonuclease. In addition, the A at position 1, which is not in the stem–tetraloop, is also required, because changing this A to a G eliminates essentially all endonuclease activation (lane 2). Our results concerning the A at position 10 are less clear cut: a change to G reduces endonuclease activity by ∼60%, whereas a change to U has no significant effect (lanes 5 and 6). Finally, we determined whether bases 11–13 in 5′ vRNA play a role in endonuclease activation that is independent of hydrogen-bonding with bases in 3′ vRNA. Changing the A at position 11 to C has no significant effect on endonuclease activation (lane 15). In contrast, changing the G at position 12 or 13 to A reduces endonuclease activity by ∼80–90% (lanes 16 and 17), indicating that these two Gs play a significant role in endonuclease activation. The sequence/structure requirements for endonuclease activation by 5′ vRNA alone using a capped CA-containing RNA substrate are summarized in Figure 4C.
Identification of the sequences in 3′ vRNA that are required for template activity
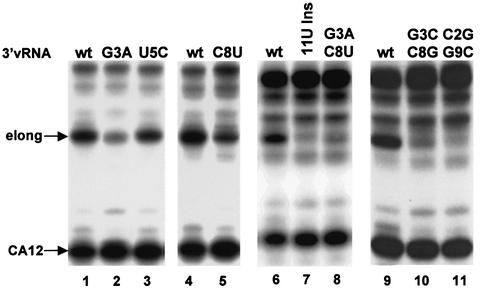
With a capped CA-containing RNA substrate, 3′ vRNA functions solely as a template for the initiation of mRNA synthesis, thereby enabling us to identify the 3′ vRNA sequences that are required specifically for this function. Because 3′ vRNA, but not 3′ cRNA, functions as a template, some or all of the four base differences between these two RNAs (bases 3, 5, 8 and 11) result in complete loss of template activity (Figure 3). To determine which of these bases is required, bases 3, 5 and 8 in 3′ vRNA were individually replaced with the corresponding base in 3′ cRNA (Figure 5). Replacing the U at position 5 with C has little or no effect on template activity (lane 3). In contrast, replacing the G at position 3 with A substantially reduces template activity (∼90%), and replacing the C at position 8 with U reduces template activity by ∼75% (lanes 2 and 5). When these changes were made at both positions 3 and 8, thereby retaining potential hydrogen-bonding, template activity is almost totally lost (lane 8). To confirm that hydrogen-bonding between these two bases does not play a role in template activity, the identities of the bases at positions 3 and 8 were switched, thereby preserving the potential for a G–C base-pair; again, template activity is almost completely lost (lane 10). A similar switching experiment involving positions 2 and 9 (lane 11) demonstrates that hydrogen-bonding between these two bases does not play a role in template activity either. Consequently, the identity of the bases at positions 2, 3, 8 and 9 of 3′ vRNA is essential for template activity. In addition, the extra U at position 11 in the 3′ cRNA sequence is not tolerated, because inserting a U between bases 10 and 11 of 3′ vRNA eliminates all detectable template activity (lane 7).
Fig. 5. Identification of the bases in 3′ vRNA that are required for template activity using capped CA-containing RNA. 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the presence of the four NTPs and 5′ vRNA, and either wt 3′ vRNA (lanes 1, 4, 6 and 9) or 3′ vRNA containing the indicated base changes.
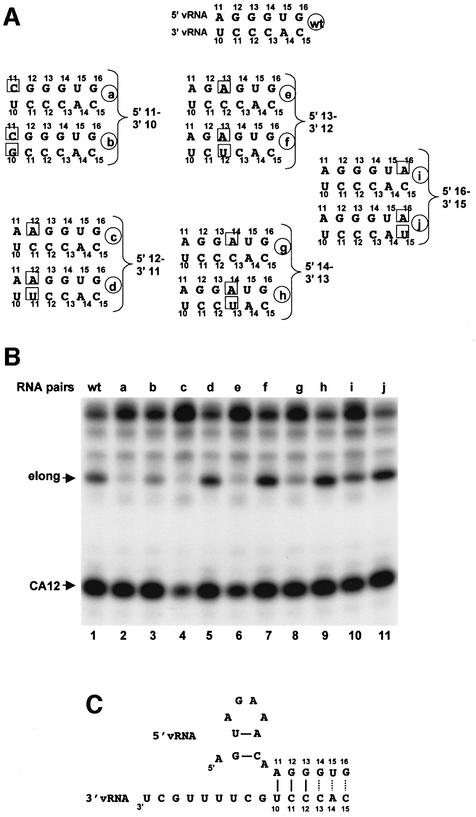
To determine whether hydrogen-bonding of bases 10–12 in 3′ vRNA with bases 11–13 in 5′ vRNA is important for the template activity of 3′ vRNA, we used the series of mutant 5′ and 3′ vRNAs indicated in Figure 6A. The template activity using the various pairs of 5′ and 3′ vRNAs is shown in Figure 6B. Changing the A at position 11 in 5′ vRNA to a C, thereby eliminating the potential for the hydrogen-bonding of this base with the U at position 10 in 3′ vRNA, results in complete loss of template activity (RNA a, lane 2). Some template activity (∼30%) is restored by changing the U at position 10 in 3′ vRNA to a G (RNA pairs b, lane 3), thereby establishing a C–G base-pair between these bases in 5′ and 3′ vRNA. Because template activity is not completely restored, the original A-U base-pair at this position is not only required but also preferred to a G–C base-pair. Changing the G at position 12 in 5′ vRNA to A results in complete loss of template activity, and template activity is restored almost completely by changing the C at position 11 in 3′ vRNA to U (RNA c and d, lanes 4 and 5), indicating that base-pairing between base 11 in 3′ vRNA and base 12 in 5′ vRNA is required for template activity. The same base substitutions corresponding to the 3′ vRNA bases at positions 12, 13 and 15 demonstrated the following: (i) base-pairing between base 12 in 3′ vRNA and base 13 in 5′ vRNA is required for template activity; (ii) base-pairing between base 13 or 15 in 3′ vRNA and base 14 or 16 in 5′ vRNA, respectively, enhances but is not required for template activity, because some template activity occurs in the absence of base-pairing at these positions (RNA pairs e–j, lanes 6–11). We conclude that base-pairing between 3′ vRNA bases 10–12 and 5′ vRNA bases 11–13 is required for template activity, as summarized in Figure 6C.
Fig. 6. The function of 3′ vRNA as template for the initiation of viral mRNA synthesis requires the formation of three base-pairs with 5′ vRNA. (A) The sequence of bases 11–16 and bases 10–15 in 5′ and 3′ vRNA oligonucleotides, respectively, used to assess the role of hydrogen bond formation between 5′ and 3′ vRNA. (B) 32P-labeled capped CA-containing RNA was incubated with the nuclear extract in the presence of the four NTPs and the indicated 5′ and 3′ RNA pairs denoted in (A). (C) Base-pairing between 5′ and 3′ vRNA during the initiation of mRNA synthesis. Solid lines denote base-pairs required for initiation; dotted lines indicate base-pairs that enhance initiation.
As we have shown above, base-pairing between wild-type 5′ vRNA and 3′ vRNA is not required for activation of the endonuclease that cleaves at CA. However, when endonuclease activation is decreased by introducing a mutation into 5′ vRNA at either position 12 or 13, this decrease is ameliorated by forming base-pairs at these positions with mutant 3′ vRNA (Figure 6B, lanes 4–7). These results confirm the importance of bases 12 and 13 in 5′ vRNA for endonuclease activation.
The template activity of 3′ vRNA is mediated by its binding to the polymerase
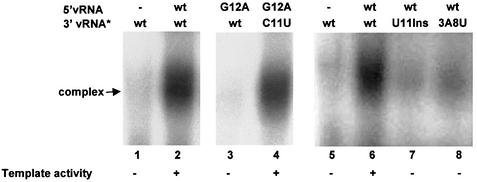
We determined whether the template activity of 3′ vRNA is mediated by its binding to the viral polymerase. Radiolabeled wild-type or mutant 3′ vRNA was incubated with 5′ vRNA-activated polymerase using a capped CA-containing RNA, and polymerase–RNA complexes were separated from free RNA using non-denaturing gels (Figure 7). Binding of 3′ vRNA to the polymerase requires the formation of a base-pair between base 11 in 3′ vRNA and base 12 in 5′ vRNA (lanes 3 and 4). In addition, 3′ vRNA binding to the polymerase is almost completely eliminated by insertion of U between positions 10 and 11 (lane 7), or replacing the bases at both positions 3 and 8 (lane 8). In all cases, the loss of polymerase binding correlates with the loss of template activity, indicating that the binding of 3′ vRNA to the polymerase determines whether it functions as a template for mRNA synthesis. Our results show that this binding is mediated by the interaction of the first nine bases of 3′ vRNA with the polymerase and by the formation of hydrogen bonds between bases 10–12 in 3′ vRNA and bases 11–13 in 5′ vRNA.
Fig. 7. The template activity of 3′ vRNA is mediated by its binding to the viral polymerase in the presence of 5′ vRNA. The nuclear extract was incubated with unlabeled capped CA-containing RNA, 32P-labeled 3′ vRNA (3′ vRNA*, 8 × 104 counts/min) containing either the wild-type (wt) sequence or the indicated mutation, and either no 5′ vRNA (–) or the unlabeled 5′ vRNA containing the indicated sequence. The template activity observed with these pairs of 5′ and 3′ vRNAs is denoted.
Discussion
An in vitro system that mirrors the cap-snatching process in infected cells
Influenza viral mRNA synthesis is initiated by cap-snatching: the endonuclease intrinsic to the viral polymerase cleaves capped RNAs to produce capped fragments 10–13 nucleotides long that are primers for viral mRNA synthesis (Bouloy et al., 1978; Plotch et al., 1979, 1981; Krug et al., 1989). Here we demonstrate that the influenza viral polymerase which is assembled in human cells using recombinant proteins effectively uses CA-terminated capped fragments, but not capped fragments with two other termini, as primers to initiate viral mRNA synthesis in vitro. Primer extension analysis of the cellular sequences on the 5′ ends of viral mRNAs in infected cells, which was carried out in the early 1980s, showed that the vast majority of the cellular sequences terminate in CA (Beaton and Krug, 1981; Shaw and Lamb, 1984), indicating that the viral polymerase in vivo predominately utilizes capped CA-containing RNA fragments as primers for viral mRNA synthesis. Thus we have provided the first in vitro system that mirrors the cap-snatching mechanism that occurs in vivo during virus infection. Consequently, it is likely that this in vitro polymerase system faithfully reproduces in vivo viral mRNA synthesis and should elucidate other aspects of viral mRNA synthesis in infected cells.
Endonuclease activity that cleaves capped RNAs at CA is activated solely by the binding of 5′ vRNA to the polymerase
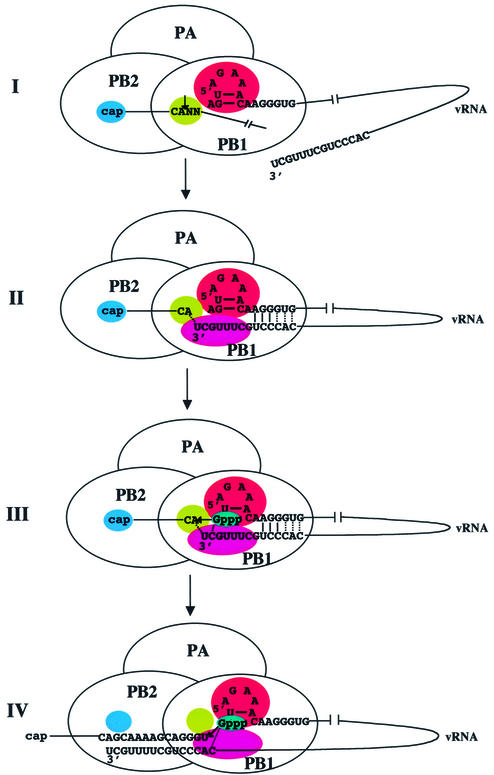
We show that the addition of 5′ vRNA alone to the influenza virus polymerase activates endonuclease cleavage at a CA of a capped RNA substrate. Consequently, this activation does not require interaction of 3′ vRNA with the polymerase, nor base-pair interactions between 5′ and 3′ vRNA. Rather, this activation is mediated solely by the recognition of specific structural elements/bases in 5′ vRNA by the polymerase, specifically by the binding site in the PB1 subunit centered around two arginine residues at positions 571 and 572 (Li et al., 1998). The structural element recognized in 5′ vRNA is a stem– tetraloop resulting from the formation of two base-pairs, between bases 2 and 9, and between bases 3 and 8. The only base in the tetraloop that is required for endonuclease activation is the A at position 7. The other 5′ vRNA bases that are recognized are the A at position 1 and the GG dinucleotide at positions 12 and 13. Recognition of this structure and sequences in 5′ vRNA activates not only the cap-binding site in the PB2 protein but also the endonuclease site in the PB1 protein for cleavage at a CA sequence (Figure 8, panel I). This endonuclease site contains three essential acidic amino acids, similar to the active sites of other enzymes that cut polynucleotides to produce 3′-OH ends (Baker and Luo, 1994; Dyda et al., 1994; Rice and Mizuuchi, 1995). Many of these other enzymes also exhibit specificity for cleavage at CA.
Fig. 8. Model of the influenza virus cap-snatching mechanism, in which the processes of RNA activation of the polymerase, endonuclease cleavage of capped CA-containing RNA substrates and initiation of mRNA synthesis are coupled. Base-pairing between 5′ and 3′ vRNA: solid lines denote base-pairs required for initiation; dotted lines indicate base-pairs that enhance initiation. Dotted lines are also shown for the postulated base-pairing (i) between the 3′ terminal A of the capped RNA fragment and the 3′ terminal U of 3′ vRNA and (ii) between the initiating GTP and the 3′ penultimate C of 3′ vRNA.
Subsequent addition of 3′ vRNA enables the endonuclease to cleave at sites other than CA. Two of the reported requirements for the activation of a viral endonuclease that cleaves at AG, namely pairing of bases 11–13 in 5′ vRNA with bases 10–12 in 3′ vRNA, and a small hairpin loop in 3′ vRNA (Leahy et al., 2001a,b), are clearly not involved in the activation of the CA-specific endonuclease. In fact, we show that such a hairpin loop in 3′ vRNA is not formed during the initiation of viral mRNA synthesis. Because hydrogen-bonding of bases 11–13 in 5′ vRNA with bases 10–12 in 3′ vRNA can enhance endonuclease activity when it is decreased by a mutation in 5′ vRNA, such hydrogen-bonding may also ameliorate the weak cleavage of a capped RNA at sites other than CA. Endonuclease activity that cleaves at either CA or other sites requires a stem–tetraloop in 5′ vRNA (Leahy et al., 2001b, 2002; present study).
The sequences in 3′ vRNA that are required for its function as a template for the initiation of influenza viral mRNA synthesis
We were able to identify bases in 3′ vRNA that are required for its function as a template for viral mRNA synthesis because this is the sole function for 3′ vRNA when the capped RNA substrate contains a CA cleavage site. We show that the identity of the bases at positions 2, 3, 8 and 9, rather than their participation in base-pairing, is essential for template activity. Consequently, the template activity of 3′ vRNA does not require a stem–tetraloop structure resulting from base-pairing between bases 2 and 9, and between bases 3 and 8. The absence of a stem–loop structure in 3′ vRNA should facilitate its transcription during the initiation of viral mRNA synthesis. When two of the bases at these positions are changed, 3′ vRNA does not bind to the polymerase, indicating that these bases mediate the interaction of 3′ vRNA with its binding site on the polymerase. Further, we show that the ability of 3′ vRNA to function as a template also requires that it form three essential base-pairs with 5′ vRNA, specifically between bases 10–12 in 3′ vRNA and bases 11–13 in 5′ vRNA. In the absence of one of these base-pairs, 3′ vRNA does not bind to the polymerase. The formation of these base-pairs between 5′ and 3′ vRNA indicates that the binding sites for these two RNA sequences on the PB1 protein, which are ∼300 amino acids apart (Li et al., 1998), are adjacent to each other in the three-dimensional structure of the active form of the PB1 protein. We conclude that the 3′ vRNA template is bound to the polymerase by both RNA–protein and RNA–RNA interactions; the first nine bases of 3′ vRNA are bound to its specific binding site on the PB1 protein subunit, and bases 10–12 of 3′ vRNA are bound via hydrogen-bonding to bases in 5′ vRNA (Figure 8, panel II). Binding of 3′ vRNA to the polymerase via these two interactions is eliminated by the insertion of a U between bases 10 and 11 in 3′ vRNA. This insertion should not disrupt the potential for the required base-pairing between 5′ and 3′ vRNA, but would change the spacing between the 3′ vRNA bases involved in this base-pairing and the 3′ vRNA bases involved in binding to the PB1 protein. The inhibition of polymerase binding and template activity resulting from the insertion of this U indicates that this spacing is also critical.
Thus we have identified sequences in 5′ and 3′ vRNA that are required for their separable functions in viral mRNA synthesis. Two of these sequence requirements coincide with those previously determined for the composite of viral mRNA synthesis and vRNA replication in vivo, namely (i) a stem–tetraloop in 5′ vRNA formed by base-pairing between bases 2 and 9 and between bases 3 and 8, and (ii) hydrogen-bonding between bases 10–12 in 3′ vRNA and bases 11–13 in 5′ vRNA (Flick et al., 1996). It is not surprising that other results of the in vivo study differ from ours because the in vivo assay, unlike ours, was designed to measure the participation of 5′ and 3′ vRNA in both viral RNA replication and viral mRNA synthesis.
A mechanism for the preferential usage of CA-terminated capped RNA fragments as primers to initiate viral mRNA synthesis
Binding of 5′ vRNA is the required first step in the activation of the polymerase, because 5′ vRNA activates the endonuclease and provides the bases with which 3′ vRNA forms the hydrogen bonds that are required for its template activity (Figure 8, panels I and II). Immediately upon 5′ vRNA binding, capped RNA substrates with CA sites are preferentially cleaved. We postulate that, as a result of this specificity of the PB1 endonuclease site for CA 3′ termini, the CA-terminated capped RNA fragments remain tightly bound to the endonuclease active site, which positions these termini close to both the 3′ end of the vRNA template and the nucleotide addition site of the polymerase [also located on the PB1 subunit (Li et al., 2001)] (Figure 8, panel III). Although other capped RNAs can be cleaved after 3′ vRNA binding, the resulting capped fragments would be weakly bound to the endonuclease active site and hence would not be effectively positioned in the same manner. The A of the CA end of the capped RNA fragment may base-pair with the terminal U of 3′ vRNA (Duijsings et al., 2001). Transcription is initiated by the addition of a G to the CA-terminated capped RNA primer, which is subsequently released from the endonuclease site on the polymerase (Figure 8, panel IV). The 3′ vRNA template is released from both its PB1 binding site and its base-pairing with 5′ vRNA, and is threaded past the nucleotide addition site of the polymerase, resulting in the synthesis of an RNA containing the capped RNA fragment linked to sequences encoded by 3′ vRNA. During this synthesis the cap of the elongating mRNA is released from its PB2 binding site (Braam et al., 1983). Thus the three essential base-pairs between 5′ and 3′ vRNA are formed transiently during the polymerase-catalyzed initiation of mRNA synthesis, consistent with previous experiments, which showed that the juxtaposition of the 5′ and 3′ ends of vRNA requires the polymerase (Klumpp et al., 1997). Figure 8, panels I–IV, presents our new model for the influenza virus cap-snatching mechanism, in which the processes of RNA activation of the polymerase, endonuclease cleavage of capped CA-containing RNA substrates and initiation of mRNA synthesis are coupled.
Specificity for the generation and usage of 3′ CA-terminated RNA or DNA is exhibited by other enzymes, nucleases that produce 3′-OH ends and polynucleotidyl transferases such as transposases and retroviral integrases (Mizuuchi, 1992; Asante-Appiah and Skalka, 1997; Esposito and Craigie, 1998; Lee and Harshey, 2001). Recent evidence indicates that the Mu transposase preferentially uses CA/GT-terminated DNAs for integration into target DNAs because (i) terminal CA–TG base-pairing is flexible, thereby facilitating the unpairing of the DNA ends, and (ii) CA and/or its TG complement specifically interacts with the transposase, thereby stabilizing the DNA–transposase complex and positioning the CA-terminated DNA for integration (Lee and Harshey, 2001). The latter mechanism is similar to our proposed mechanism for the preferential generation and usage of CA-terminated capped RNA primers by the influenza virus polymerase. Interestingly, the integrase active site is similar to the influenza virus endonuclease active site in containing three essential acidic amino acids (Baker and Luo, 1994; Rice and Mizuuchi, 1995; Li et al., 2001).
What is the function of the capped RNA fragments lacking CA termini that are produced by the polymerase in the presence of both 5′ and 3′ vRNA?
Only CA-terminated capped fragments are effectively used as primers to initiate viral mRNA synthesis catalyzed by the influenza viral polymerases that are assembled either in influenza virus infected cells or in human cells using vaccinia virus vectors (Beaton and Krug, 1981; Shaw and Lamb, 1984; present study). Other capped RNA fragments lacking CA termini that are produced by the viral endonuclease in the presence of both 5′ and 3′ vRNA are poorly used as primers by these viral polymerases (Beaton and Krug, 1981; Shaw and Lamb, 1984; Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001; Leahy et al., 2001a,b; present study). However, capped RNA fragments lacking CA termini are effectively used by the viral polymerase that is packaged in virus particles (virions) (Bouloy et al., 1978; Plotch et al., 1979, 1981; Krug et al., 1989). In fact, the cap-snatching mechanism for influenza viral mRNA synthesis was discovered using the virion polymerase and β-globin mRNA, which yields G-terminated capped RNA fragments. It is not known how the virion polymerase is able to utilize capped RNA fragments lacking CA termini as primers. The flexibility resulting from the use by the virion polymerase of a larger assortment of capped RNA fragments as primers facilitates the initial (primary) transcription of vRNA by the virion polymerase. It may also be inferred that results obtained using the influenza virion polymerase are only relevant to primary transcription in infected cells and do not mirror the overall transcriptional events in infected cells, which are catalyzed by newly assembled viral polymerases.
The role of the binding of the 5′ cRNA sequence to the polymerase: selective protection of viral mRNAs from cap-snatching
We show that 5′ cRNA (complementary RNA, the full-length copy of vRNA) activates the CA-specific endonuclease via binding to the same site on the PB1 subunit as does 5′ vRNA. The activation of endonuclease activity by 5′ cRNA does not lead to capped RNA primed transcription of cRNA because 3′ cRNA does not function as a template for such synthesis. Instead, as shown previously (Shih and Krug, 1996), the binding of a 5′ cRNA sequence to the polymerase functions in the selective protection of viral mRNAs from cap-snatching. A 5′ cRNA sequence is found in all viral mRNAs, where it is located immediately 3′ to the 5′ sequence that is snatched from cellular capped RNAs (Krug et al., 1989; Shih and Krug, 1996). This cRNA sequence and the viral mRNA cap bind to the polymerase. As a consequence of this binding, the viral mRNAs cannot be cleaved by other polymerases that have been activated by vRNA sequences (Shih and Krug, 1996). This selective protection of viral mRNAs is essential because if the 5′ ends of viral mRNAs were cleaved and used as primers, net synthesis of viral mRNAs would not occur.
Do the polymerases of other segmented, negative-strand and ambisense RNA viruses share the mechanism of cap-snatching depicted in Figure 8?
The synthesis of the mRNAs of other segmented, negative-strand and ambisense RNA viruses is also initiated by cap-snatching (Vialat and Bouloy, 1992; Huiet et al., 1993; Jin and Elliott, 1993a,b; Garcin et al., 1995; Ramirez et al., 1995; Estabrook et al., 1998; Duijsings et al., 2001). However, the studies so far carried out with these viruses have not included the two experimental approaches which provided the evidence for the cap-snatching mechanism for influenza viral mRNA synthesis depicted in Figure 8. The first experimental approach was the reverse transcriptase catalyzed copying of the total population of two influenza A viral mRNAs synthesized in infected cells, followed by direct sequencing, which demonstrated that CA-terminated capped fragments are preferentially used as primers for influenza viral mRNA synthesis during virus infection in vivo (Beaton and Krug, 1981; Shaw and Lamb, 1984). For other segmented, negative-strand and ambisense RNA viruses, sequence information concerning host capped primers utilized in vivo has only been obtained by sequencing cloned DNAs generated from the viral mRNAs synthesized in vivo (Huiet et al., 1993; Jin and Elliott, 1993a,b; Garcin et al., 1995; Ramirez et al., 1995; Estabrook et al., 1998; Duijsings et al., 2001). The host sequences in these cloned DNAs may not represent the total population of primer sequences snatched from host mRNAs. Consequently, it will be important to determine the total population of these host primer sequences by reverse transcriptase catalyzed copying of viral mRNAs synthesized in infected cells. The second experimental approach utilized influenza virus polymerases that are assembled by expressing the three viral polymerase subunits in human cells in the absence of vRNA (Hagen et al., 1994; Tiley et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001; present study), which most likely mimics the nascent polymerases synthesized in infected cells. These experiments established that the influenza virus endonuclease is activated by 5′ vRNA and that CA-terminated capped fragments are preferentially used as primers for viral mRNA synthesis. Experiments using the already assembled and activated polymerase from influenza virions did not reveal either of these properties (Krug et al., 1989). Only experiments with virion polymerases have been carried out for other segmented, negative-strand or ambisense RNA viruses (Vialat and Bouloy, 1992). Consequently, with these other RNA viruses, it will be important to use recombinant polymerases that are produced in cells in the absence of vRNA to elucidate the mechanism of transcriptional initiation. We postulate that future experiments will establish that key features of the cap-snatching mechanisms catalyzed by the polymerases of these other RNA viruses are similar to the influenza virus cap-snatching mechanism depicted in Figure 8.
Materials and methods
Preparation of recombinant influenza virus polymerase complexes
HeLa cells were infected with three recombinant vaccinia virus vectors encoding the influenza virus PB1, PB2 and PA proteins (Cianci et al., 1995; Li et al., 1998, 2001). Where indicated, the vaccinia virus vector expressing wild-type PB1 protein was replaced by the vector expressing a PB1 protein in which the arginine at either position 571 or 572 was replaced with alanine (Li et al., 1998). At 20–24 h after infection, nuclear extracts were prepared and used as the source of the influenza virus polymerase complex (Hagen et al., 1994; Li et al., 1998, 2001).
Preparation of labeled oligoribonucleotides and RNAs
Alfalfa mosaic virus RNA 4 containing an m7G32pppGm 5′ end was prepared as described previously (Plotch et al., 1981). Two other capped RNAs were prepared by SP6 RNA polymerase catalyzed transcription of pGEM7z (+) and pGEM9z (–) linearized with SmaI and SalI, respectively, resulting in transcripts 67 and 46 nucleotides long, respectively. These transcripts were isolated by gel electrophoresis and incubated with the vaccinia virus capping enzyme and 2′-O-methyl transferase in the presence of [α-32P]GTP to produce m7G32pppGm 5′ ends (Plotch et al., 1981). Where indicated, the cap of the pGEM9z (–) transcript was formed using unlabeled GTP. Oligoribonucleotides containing wild-type and mutant 5′ and 3′ terminal sequences of vRNA and of cRNA were synthesized using an oligonucleotide synthesizer (Dharmacon Inc.) and, where indicated, were labeled at their 5′ ends using T4 polynucleotide kinase and [γ-32P]ATP (Li et al., 1998).
Cap-dependent endonuclease and elongation assay
For cap-dependent endonuclease assay, the polymerase was first activated by the addition of 5′ vRNA alone, both 5′ vRNA and 3′ vRNA, 5′ cRNA alone or both 5′ cRNA and 3′ cRNA. The indicated capped RNA containing an m7G32pppGm 5′ end (0.25 ng, 1 × 105 counts/min) was added, together with 0.1 µg of transfer RNA to inhibit non-specific nucleases in the nuclear extract, and the mixture was incubated for 60 min at 30°C (Hagen et al., 1994; Cianci et al., 1995; Li et al., 1998, 2001). To assay for RNA chain elongation, the four ribonucleoside triphosphates (NTPs), each at 1 mM, were added to the above reaction. In one elongation experiment, the capped RNA was unlabeled and the elongation product was labeled by including [α-32P]GTP in the reaction. The RNA products were separated on a denaturing 20% polyacrylamide–7 M urea gel. Labeled RNAs of 10–100 nucleotides (Ambion Decade Marker System) were run as molecular weight markers.
RNA binding assays
The gel-shift assay for the binding of labeled 3′ vRNA to the polymerase complex was carried out as described previously (Li et al., 1998). Polymerase–RNA complexes were separated from free RNA by electrophoresis on 4% non-denaturing gels. For 5′ vRNA and 5′ cRNA binding, the nuclear extract was incubated for 25 min at room temperature with 32P-labeled 5′ vRNA or 5′ cRNA (2 × 104 counts/min) containing a thioU residue under conditions previously described (Li et al., 1998). The mixtures were exposed to 366 nm ultraviolet light, and cross-linked protein–oligoribonucleotide complexes were separated from free oligoribonucleotide by electrophoresis on 7% denaturing gels.
Acknowledgments
Acknowledgements
We appreciate the advice of Mei-Ling Li. This investigation was supported by grant F-1468 from the Welch Foundation to R.M.K.
References
- Asante-Appiah E. and Skalka,A.M. (1997) Molecular mechanisms in retrovirus DNA integration. Antiviral Res., 36, 139–156. [DOI] [PubMed] [Google Scholar]
- Baker T.A. and Luo,L. (1994) Identification of residues in the Mu transposase essential for catalysis. Proc. Natl Acad. Sci. USA, 91, 6654–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A.R. and Krug,R.M. (1981) Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res., 9, 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouloy M., Plotch,S.J. and Krug,R.M. (1978) Globin mRNA are primers for the transcription of influenza viral RNA in vitro. Proc. Natl Acad. Sci. USA, 75, 4886–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J., Ulmanen,I. and Krug,R.M. (1983) Molecular model of a eukaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell, 34, 609–618. [DOI] [PubMed] [Google Scholar]
- Cianci C., Tiley,L. and Krystal,M. (1995) Differential activation of the influenza virus polymerase via template RNA binding. J. Virol., 69, 3995–3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Racaniello,V.R., Zazra,J.J. and Palese,P. (1980) The 3′ and 5′-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene, 8, 315–328. [DOI] [PubMed] [Google Scholar]
- Duijsings D., Kormelink,R. and Goldbach,R. (2001) In vivo analysis of the TSWV cap-snatching mechanism: single base complementarity and primer length requirements. EMBO J., 20, 2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F., Hickman,A.B., Jenkins,T.M., Engelman,A., Craigie,R. and Davies,D.R. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science, 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- Esposito D. and Craigie,R. (1998) Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein–DNA interaction. EMBO J., 17, 5832–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estabrook E.M., Tsai,J. and Falk,B.W. (1998) In vivo transfer of barley stripe mosaic hordeivirus ribonucleotides to the 5′ terminus of maize stripe tenuivirus RNAs. Proc. Natl Acad. Sci. USA, 95, 8304–8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R., Neumann,G., Hoffmann,E., Neumeier,E. and Hobom,G. (1996) Promoter elements in the influenza vRNA terminal structure. RNA, 2, 1046–1057. [PMC free article] [PubMed] [Google Scholar]
- Garcin D., Lezzi,M., Dobbs,M., Elliott,R.M., Schmaljohn,C., Kang,C.Y. and Kolakofsky,D. (1995) The 5′ ends of Hantaan virus (Bunyaviridae) RNAs suggest a prime-and-realign mechanism for the initiation of RNA synthesis. J. Virol., 69, 5754–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen M., Chung,T.D.Y., Butcher,A. and Krystal.,M. (1994) Recombinant influenza virus polymerase: requirement of both 5′ and 3′ viral ends for endonuclease activity. J. Virol., 68, 1509–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huiet L., Feldstein,P.A., Tsai,J.H. and Falk,B.W. (1993) The maize stripe virus major noncapsid protein messenger RNA transcripts contain heterogeneous leader sequences at their 5′ termini. Virology, 197, 808–812. [DOI] [PubMed] [Google Scholar]
- Jin H. and Elliott,R.M. (1993a) Characterization of Bunyamwera virus S RNA that is transcribed and replicated by the L protein expressed from recombinant vaccinia virus. J. Virol., 67, 1396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. and Elliott,R.M. (1993b) Non-viral sequences at the 5′ ends of Dugbe nairovirus S mRNAs. J. Gen. Virol., 74, 2293–2297. [DOI] [PubMed] [Google Scholar]
- Klumpp K., Ruigrok,R.W. and Baudin,F. (1997) Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J., 16, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M., Alonso-Caplen,F.V., Julkunen,I. and Katze,M. (1989) Expression and replication of the influenza virus genome. In Krug,R.M. (ed.), The Influenza Viruses. Plenum Press, New York, NY, pp. 89–152.
- Leahy M.B., Dobbyn,H.C. and Brownlee,G.G. (2001a) Hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol., 75, 7042–7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy M.B., Pritlove,D.C., Poon,L.L. and Brownlee,G.G. (2001b) Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol., 75, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy M.B., Zecchin,G. and Brownlee,G.G. (2002) Differential activation of influenza A virus endonuclease activity is dependent on multiple sequence differences between the virion RNA and cRNA promoters. J. Virol., 76, 2019–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I. and Harshey,R.M. (2001) Importance of the conserved CA dinucleotide at Mu termini. J. Mol. Biol., 314, 433–444. [DOI] [PubMed] [Google Scholar]
- Li M.-L., Ramirez,C. and Krug,R.M. (1998) RNA-dependent activation of primer RNA production by the influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J., 17, 5844–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.-L., Rao,P. and Krug,R.M. (2001) The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J., 20, 2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. (1992) Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy,M. and Krug,R.M. (1979) Transfer of 5′ terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc. Natl Acad. Sci. USA, 76, 1618–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S.J., Bouloy,M., Ulmanen,I. and Krug,R.M. (1981). A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell, 23, 847–858. [DOI] [PubMed] [Google Scholar]
- Ramirez B.C., Garcin,D., Calvert,L.A., Kolakofsky,D. and Haenni,A.L. (1995) Capped nonviral sequences at the 5′ end of the mRNAs of rice hoja blanca virus RNA4. J. Virol., 69, 1951–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. and Mizuuchi,K. (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- Robertson J.S. (1979) 5′ and 3′ terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Res., 6, 3745–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M.W. and Lamb,R.A. (1984). A specific sub-set of host-cell mRNAs prime influenza virus mRNA synthesis. Virus Res., 1, 455–467. [DOI] [PubMed] [Google Scholar]
- Shih S.R. and Krug,R.M. (1996) Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology, 226, 430–435. [DOI] [PubMed] [Google Scholar]
- Tiley L.S., Hagen,M., Matthews,J.T. and Krystal.,M. (1994) Sequence-specific binding of the influenza virus RNA polymerase to sequences located at 5′ ends of the viral RNAs. J. Virol., 68, 5108–5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialat P. and Bouloy,M. (1992). Germiston virus transcriptase requires active 40S ribosomal subunits and utilizes capped cellular RNAs. J. Virol., 66, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]