Abstract
Diverse functions, including DNA replication, recombination and repair, occur during S phase of the eukaryotic cell cycle. It has been proposed that p53 and BLM help regulate these functions. We show that p53 and BLM accumulated after hydroxyurea (HU) treatment, and physically associated and co-localized with each other and with RAD51 at sites of stalled DNA replication forks. HU-induced relocalization of BLM to RAD51 foci was p53 independent. However, BLM was required for efficient localization of either wild-type or mutated (Ser15Ala) p53 to these foci and for physical association of p53 with RAD51. Loss of BLM and p53 function synergistically enhanced homologous recombination frequency, indicating that they mediated the process by complementary pathways. Loss of p53 further enhanced the rate of spontaneous sister chromatid exchange (SCE) in Bloom syndrome (BS) cells, but not in their BLM-corrected counterpart, indicating that involvement of p53 in regulating spontaneous SCE is BLM dependent. These results indicate that p53 and BLM functionally interact during resolution of stalled DNA replication forks and provide insight into the mechanism of genomic fidelity maintenance by these nuclear proteins.
Keywords: DNA repair/nuclear trafficking/RAD51/sister chromatid exchange
Introduction
Homologous recombination (HR) is one of the primary DNA repair processes that maintain the fidelity of the human genome (Hoeijmakers, 2001). In yeast, the products of the RAD52 epistasis group carry out HR. Functional homologs of these gene products also exist in mammalian cells. RAD51 is perhaps the most essential component because it catalyzes the strand exchange reaction (Haber, 2000). RAD51 is a member of a large complex, termed BASC (BRCA-associated complex), which also contains members of the eukaryotic replication machinery [e.g. proliferating cell nuclear antigen (PCNA)], helicases (e.g. BLM), repair enzymes (e.g. MSH2/6) and other recombination proteins (Y.Wang et al., 2000). In addition, RAD51 physically interacts with BLM, and a subset of BLM nuclear foci that form in response to ionizing radiation co-localize with RAD51 (Wu et al., 2001).
BLM and the Werner syndrome (WS) protein WRN are RecQ helicases that may regulate HR through their ability to suppress inappropriate recombination (van Brabant et al., 2000). Bloom syndrome (BS) patient cells exhibit chromosomal and DNA instability characterized by elevated rates of sister chromatid exchanges (SCEs), insertions, deletions, loss of heterozygosity, telomere associations and quadriradials (Chakraverty and Hickson, 1999). BLM can catalyze branch migration of Holliday junctions (HJs) in vitro, which may prevent the collapse of replication forks (Karow et al., 2000). Because RecQ helicases initiate recombination from internal nicks or small gaps, it has been proposed that they may be involved in ‘replication fork repair’ (Chakraverty and Hickson, 1999). BLM also has been found in a nuclear matrix-bound complex within promyelocytic leukemia nuclear bodies (PML NBs), and it accumulates in the S phase of the cell cycle (Bischof et al., 2001). Recently, it was observed that BLM is required for correct nuclear localization of RAD50–MRE11–NBS1 complexes after replication fork arrest (Franchitto and Pichierri, 2002).
We and others have reported that WRN and BLM bind to p53 (Blander et al., 1999; Spillare et al., 1999; Wang et al., 2001). WS and BS cells have an attenuated DNA damage-activated, p53-mediated apoptotic pathway (Spillare et al., 1999; Wang et al., 2001). Although cultured cells from the majority of BS patients do not appear to have abnormal p53 accumulation after UV- or X-ray-induced DNA damage (Lu and Lane, 1993), BS cells can undergo a delayed and prolonged accumulation, as well as a delayed recovery of the levels of p53 and its transcriptional targets in response to UV radiation (Collister et al., 1998). However, at least some hydroxy urea (HU)-treated BLM-deficient cells exhibit both an intact S phase arrest and accumulation of p53 (Ababou et al., 2002).
Apart from its role in G1/S- and G2/M-dependent cell cycle checkpoints and apoptosis (Oren, 1999), p53 has also been implicated in S phase processes and DNA repair. p53 binds to HJs in DNA and facilitates their cleavage during HR (Lee et al., 1997). In vitro processing of HJs by BLM is regulated by p53 (Yang et al., 2002). During S phase arrest induced by HU treatment, accumulated p53 is transcriptionally inactive (Gottifredi et al., 2001). Functional inactivation of wild-type p53 results in elevated HR rates (Susse et al., 2000; Slebos and Taylor, 2001; Saintigny and Lopez, 2002). Hence, the modulation of HR activity by p53 is independent of its transcriptional activation function (Willers et al., 2000).
Here, we demonstrate that BLM and p53 co-localize at the sites of stalled DNA replication forks and physically interact with RAD51. Using isogenic cell lines, we show that BLM localizes at sites of stalled DNA replication in the absence of p53. However, BLM substantially enhances p53 [either wild-type or mutated (Ser15Ala)] localization at these sites. Once accumulated at these sites, p53 and BLM modulate HR and SCE. These results indicate the dynamic and regulatory interaction between p53 and BLM as a novel mechanism to maintain genomic fidelity by resolving stalled DNA replication forks during S phase of the cell cycle.
Results
p53 and BLM co-localize at sites of stalled DNA replication forks
The human telomerase reverse transcriptase (hTERT)-transformed cell lines investigated were normal human fibroblast GM07532 (NHF), normal human fibroblast GM07532 expressing E6 (NHF E6), BS fibroblast GM03509 (BS) and chromosome 15-corrected BS fibroblast GM03509 (BS A-15). They differ in p53 and BLM status (summarized in Table I). BLM protein levels are low in G1, accumulate in S and persist in G2/M phases of the cell cycle (Bischof et al., 2001). Transcriptionally inactive p53 can also accumulate in S phase (Gottifredi et al., 2001). To induce S phase arrest, we utilized HU, which blocks replication fork progression by depleting deoxyribonucleotide pools and triggering replication fork collapse. HU treatment for 16 h led to a similar accumulation of BLM and p53 in all cells, except those in which BLM or p53 were deficient. N-terminal p53 antibody DO-1 (epitope spanning N-terminal residues 21–25) and 588 (raised against p53 C-terminal residues 320–393; Sengupta and Wasylyk, 2001) were used to visualize p53 induction at the protein level (data not shown). One of the epitopes recognized by the polyclonal antibody 588 spans 371–380 amino acids of p53, also recognized by Pab 421 (see Supplementary figure 1A available at The EMBO Journal Online). The p53 foci detected by 588 were competed out with a wild-type p53 peptide, but not with a peptide phosphorylated at 376 and 378, or by a peptide with a scrambled sequence (Supplementary figure 1B; data not shown). This indicated that dephosphorylated 376 and 378 amino acid residues are necessary for the efficient detection of p53 foci by 588.
Table I. Status of p53 and BLM in different cell lines.
| NHF (GM07532) | NHF E6 (GM07532 E6) | BS (GM03509) | BS A-15 (GM03509 A-15) | BS E6 (GM03509 E6) | BS A-15 E6 (GM03509 A-15 E6) | |
|---|---|---|---|---|---|---|
| p53 | + | – | + | + | – | – |
| BLM | + | + | – | + | – | + |
The status of p53 and BLM as known from the literature and from this study is represented. The presence and absence of the functional gene product is represented by (+) and (–), respectively.
To follow the interplay between BLM and p53 on a single-cell basis, we monitored their localization by immunofluorescence. A subpopulation (8–12%) of asynchronous, untreated NHF cells exhibited nuclear BLM foci (4–15 per cell), which co-localized with RAD51, but not appreciably with p53 (Supplementary figure 2a and b). Focal staining of p53 was observed with 588. Other anti-p53 antibodies (DO-1, Pab 1801 and Pab 421) resulted in diffuse nuclear staining (data not shown), probably due to recognition of different epitopes. Very few p53 foci (2–8 per cell) were visualized in untreated NHF cells. These few foci showed co-localization with RAD51 foci, but not with BLM (Supplementary figure 2b and c). After HU treatment, both p53 and BLM foci increased and were visualized in 45–55% of the cells. The number of BLM and p53 foci increased to 25–60 per cell.
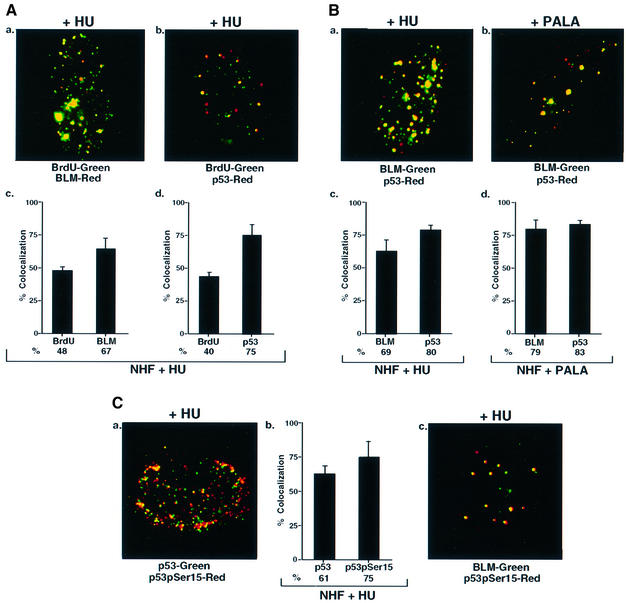
DNase treatment of cells labeled briefly with bromodeoxyuridine (BrdU) leads to focal anti-BrdU staining at sites where reinitiation of stalled DNA replication occurs (Kennedy et al., 2000). After HU treatment, BrdU and p53 co-localized extensively, though not exclusively (Figure 1A, a–d). Further, both p53 and BLM co-localized with PCNA (data not shown). We also found a high degree of co-localization of p53 and BLM in HU-treated NHF cells (Figure 1B, a and c). Treatment with other agents that can stall cells in S phase, including N-phosphonacetyl-l-aspartate (PALA) and aphidicolin, led to similar co-localization of p53 and BLM (Figure 1B, b and d; data not shown).
Fig. 1. BLM and p53 co-localize at stalled DNA replication forks in NHF. (A) BLM and p53 co-localized with BrdU. NHF were incubated with HU for 16 h, washed and incubation continued for 10 min in the presence of BrdU. Immunofluorescence was carried out with antibodies against (a) BrdU/BLM (C-18) and (b) BrdU/p53 (588). Quantitation of (a) and (b) is represented in (c) and (d). (B) BLM and p53 co-localized. NHF was treated with (a) HU or (b) PALA, and immunofluorescence was carried out with antibodies against BLM (C-18)/p53 (588). Quantitation of (a) and (b) is represented in (c) and (d). (C) Co-localization of p53, p53pSer15 and BLM. The same as (A) except that immunofluorescence was carried out with antibodies against (a) p53 (588)/p53pSer15 (16G8) and (c) BLM (C-18)/p53pSer15. Quantitation of (a) is represented in (b).
A high degree of co-localization was observed for p53 foci with the 588 antibody and those observed with a monoclonal anti-phospho-p53 (Ser15) antibody in HU-treated cells (Figure 1C, a and b). This indicated that both modified and unmodified p53 could be present at the sites of stalled DNA replication forks in complex with BLM. As expected, Ser15 phosphorylated p53 also co-localized extensively with BLM (Figure 1C, c).
BLM and p53 physically associate and co-localize with RAD51
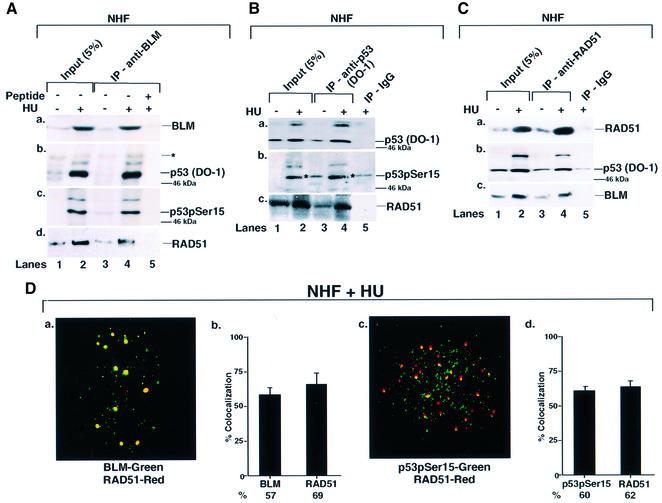
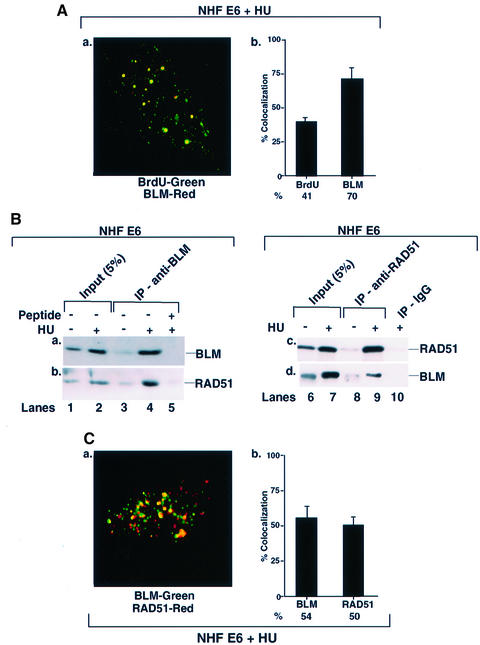
BLM is a member of the BASC supercomplex and physically interacts with proteins involved in DNA repair, recombination and replication (Y.Wang et al., 2000). It has been shown that both WRN and BLM helicase physically associate with p53 (Blander et al., 1999; Wang et al., 2001). To determine whether p53 and BLM physically associate during HU-induced S phase arrest, NHF cells were lysed and immunoprecipitations were carried out with BLM, p53 and RAD51 antibodies (Figure 2). We detected DO-1-recognizable p53 in BLM immunoprecipitates of HU-treated cells (Figure 2A, b), and p53 was at least partially phosphorylated on Ser15 (Figure 2A, c). Unfortunately, the p53 antibodies 588 and anti-phospho-p53 (Ser15) were unsuitable for immunoprecipitation. Moreover, despite the ability to detect p53 in BLM immunoprecipitates, BLM itself is undetectable in p53 immunoprecipitates (Wang et al., 2001). However, RAD51 was present in both BLM and p53 DO-1 immunoprecipitates (Figure 2A, d and B, c), and both BLM and p53 were present in RAD51 immunoprecipitates (Figure 2C, b and c). Consistent with immunoprecipitation data, both BLM and p53 co-localized with RAD51 after HU treatment (Figure 2D).
Fig. 2. BLM, p53 and RAD51 physically interact and co-localize in NHF. (A) Immunoprecipitation (IP) of BLM revealed p53 and RAD51. NHF was either left asynchronous (lane 1) or contact inhibited and treated with HU (lane 2) for 16 h. Lysates (400 µg) were immunoprecipitated with antibodies against BLM (C-18) in the absence (lanes 3 and 4) or presence (lane 5) of the peptide against which the anti-BLM antibody was raised. Input (lanes 1 and 2) indicates 5% of the lysate used for IP. The efficiency of IP was verified with self-antibody (a). Antibodies used for detection of other proteins in BLM IP were (b) anti-p53 (DO-1), (c) anti-phospho-p53 (Ser15) and (d) anti-RAD51 (Ab-1). Asterisks indicates a cross-reactive band. The position of the 46 kDa molecular weight marker is indicated. Lanes 1 and 3, minus HU; lanes 2, 4 and 5, plus HU. (B) IP of p53 revealed RAD51. The same as (A) except that the IP was carried out with antibodies either against p53 (DO-1) (lanes 3 and 4) or against the corresponding IgG (lane 5). Antibodies used for detection of other proteins in p53 IP were (b) anti-phospho-p53 (Ser15) and (c) anti-RAD51 (Ab-1). (C) IP of RAD51 revealed p53 and BLM. The same as (A) except that the IP was carried out with antibodies against RAD51 (Ab-1) (lanes 3 and 4) or against the corresponding IgG (lane 5). Antibodies used for detection of other proteins in RAD51 IP were (b) anti-p53 (DO-1) and (c) anti-BLM (C-18). (D) BLM, p53 and RAD51 co- localized. Contact-inhibited NHF were treated with HU for 16 h. Immunofluorescence was carried out with antibodies against (a) BLM (C-18)/RAD51 (Ab-1) and (b) phospho-p53 (Ser15) 16G8/RAD51 (Ab-1). Quantitation of (a) and (c) is represented in (b) and (d).
BLM is required for efficient p53 localization to sites of BrdU incorporation
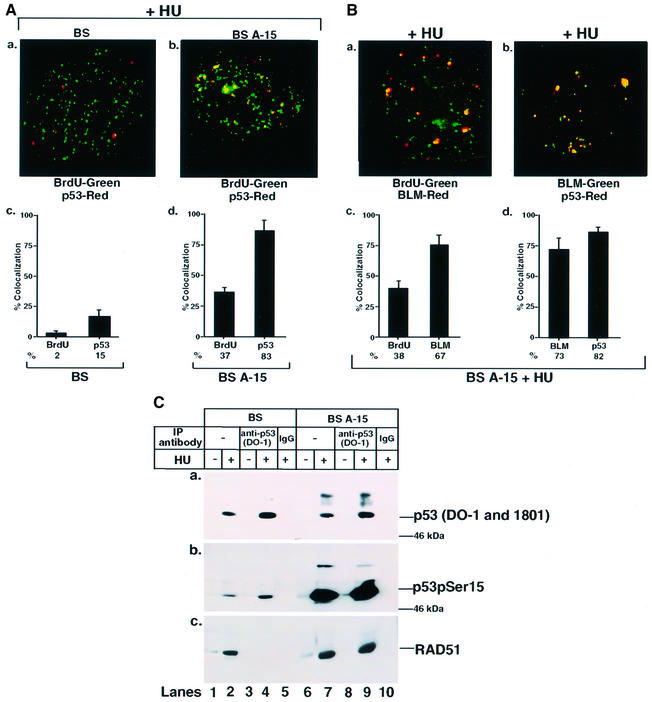
To determine whether p53 localization to sites of stalled DNA replication forks was BLM dependent, related cell lines BS and BS A-15 were used. With HU treatment, RAD51 foci formed to an equal extent in both BS and BS A-15 cells, and co-localized with stalled DNA replication forks (Supplementary figure 2d and e). However, in comparison with cells with wild-type BLM (BS A-15), the number of p53 foci formed in HU-treated BS cells was 3- to 4-fold less. Unlike BS cells, the p53 foci co-localized to a large extent with BrdU in BS A-15 cells (Figure 3A). Similarly, BLM foci co-localized with BrdU at the stalled DNA replication forks, and p53 along with BLM co-localized extensively in BS A-15 cells (Figure 3B). These data indicate that BLM is largely responsible for intranuclear transport of p53 to stalled DNA replication forks.
Fig. 3. Ser15-phosphorylated p53 co-localizes with BLM and physically interacts with RAD51 in BS A-15, but not in BS cells. (A) p53 co-localized with BrdU in BS A-15 cells. Contact-inhibited (a) BS or (b) BS A-15 cells were treated with HU for 16 h, washed and incubation continued for 10 min in the presence of BrdU. Immunofluorescence was carried out with antibodies against BrdU/p53 (588). Quantitation of (a) and (b) is represented in (c) and (d). (B) BLM co-localized with BrdU and p53 in BS A-15 cells. The same as (A) except that BS A-15 cells were used. Immunofluorescence was carried out with antibodies against (a) BrdU/BLM (C-18) and (b) BLM (C-18)/p53 (588). Quantitation of (a) and (b) is represented in (c) and (d). (C) Ser15-phosphorylated p53 interacted with RAD51 in BS A-15, but not in BS cells. BS (lanes 1 and 2) or BS A-15 (lanes 6 and 7) cells were either asynchronous and left untreated (lanes 1 and 6) or contact inhibited and subsequently treated with HU (lanes 2 and 7) for 16 h. Lysates (400 µg) were immunoprecipitated with antibodies against p53 (DO-1) (lanes 3, 4, 8 and 9) or against the corresponding IgG (lanes 5 and 10). Input (lanes 1, 2, 6 and 7) indicates 5% of the lysate used for immunoprecipitation (IP). The efficiency of IP was verified with (a) DO-1 and 1801 antibodies. Antibodies used for detection of other proteins in p53 IP were (b) anti-phospho-p53 (Ser15) and (c) anti-RAD51 (Ab-1). The position of the 46 kDa molecular weight marker is indicated in (a) and (b). Lanes 1, 3, 6 and 8, minus HU; lanes 2, 4, 5, 7, 9 and 10, plus HU.
To determine whether transport of p53 to the sites of stalled DNA replication forks resulted in enhanced interaction of p53 with RAD51, immunoprecipitations were carried out with lysates from BS and BS A-15 cells with p53 DO-1 antibody. Induction of total p53 (DO-1 and Pab 1801 antibodies) was similar in both BS and BS A-15 cells (Figure 3C, a). However, BS A-15 cells had an 8- to 10-fold higher level of induction of Ser15 phosphorylated p53 (Figure 3C, b). The blot for Figure 3C, b is overexposed to reveal the small amount of p53 Ser15 phosphorylated in untreated BS cells. In addition, RAD51 could be detected only in p53 immunoprecipitates of BS A-15 cells and not BS cells (Figure 3C, c), indicating that BLM is critical for efficient accumulation of p53 in RAD51 complexes at the sites of stalled DNA replication forks.
Ser15 phosphorylation of p53 is not obligatory for its transport to the sites of stalled DNA replication forks
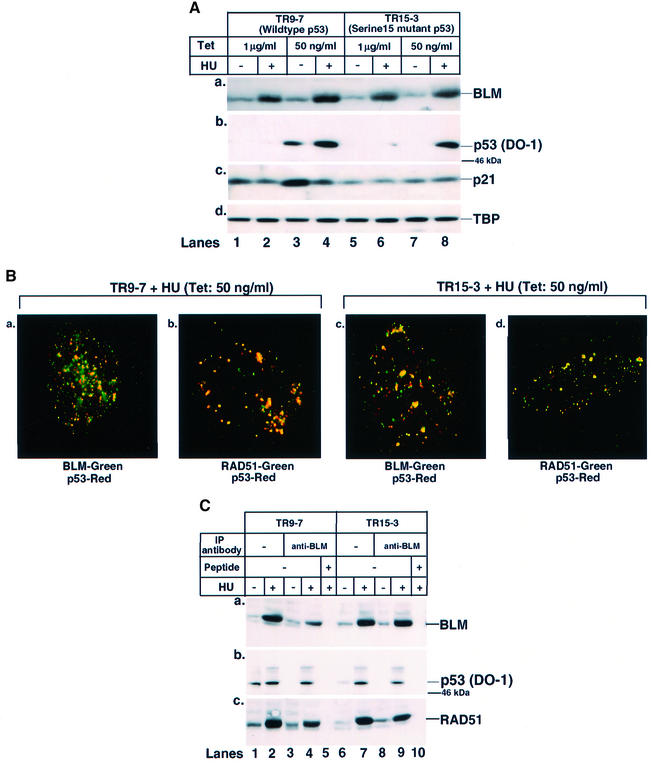
To determine whether Ser15 phosphorylation of p53 was required for its transport to the sites of stalled DNA replication, a tetracycline-regulated isogenic pair of cells expressing either wild-type p53 (TR9-7) or its mutant (Ser15Ala) form (TR15-3) was used (Bean and Stark, 2001). These cells normally are grown in the presence of tetracycline (1 µg/ml). A very low level of tetracycline (50 ng/ml), along with concomitant HU treatment was used to arrest cells in S phase. The low level of tetracycline resulted in the accumulation of transcriptionally active wild-type p53 probably due to leaky expression (Figure 4A). In contrast, concomitant HU treatment resulted in the stabilization of transcriptionally inactive wild-type p53. This, along with co-expression of BLM due to HU treatment, indicated that TR9-7 cells were arrested efficiently in S phase. In TR15-3 cells, mutated (Ser15Ala) p53 did not accumulate when grown in 50 ng/ml tetracycline. However, both mutant p53 and BLM accumulated after HU treatment (Figure 4A).
Fig. 4. Wild-type and mutant (Ser15Ala) p53 both co-localize and interact with BLM. (A) BLM accumulated in TR9-7 and TR15-3 cells in S phase. TR9-7 (lanes 1–4) and TR15-3 (lanes 5–8) cells were grown for 16 h in the presence of either 1 µg/ml (lanes 1, 2, 5 and 6) or 50 ng/ml (lanes 3, 4, 7 and 8) tetracycline and in the simultaneous presence or absence of HU. Cell lysates (75 µg) were run on SDS–polyacrylamide gels and western blotted with (a) anti-BLM, (b) anti-p53 (DO-1), (c) anti-p21WAF1 (Ab-1) and (d) anti-TBP antibodies. The position of the 46 kDa molecular weight marker is indicated. Lanes 1, 3, 5 and 7, minus HU; lanes 2, 4, 6 and 8, plus HU. (B) p53 co-localized with BLM and RAD51 in TR9-7 and TR15-3 cells. The same as (A) except that TR9-7 (a and b) or TR15-3 (c and d) were grown in the presence of 50 ng/ml tetracycline and HU. Immunofluorescence was carried out with antibodies against (a and c) BLM (C-18)/p53 (588) and (b and d) RAD51 (Ab-2)/p53 (588). (C) p53 and RAD51 co-immunoprecipitated with BLM in both TR9-7 and TR15-3 cells. TR9-7 (lanes 1 and 2) or TR15-3 (lanes 6 and 7) cells grown in the presence of 50 ng/ml tetracycline, in the absence (lanes 1 and 6) or presence of HU (lanes 2 and 7) for 16 h. Lysates (400 µg) were immunoprecipitated with antibodies against BLM (C-18) in the absence (lanes 3, 4, 8 and 9) or presence (lanes 5 and 10) of the peptide against which anti-BLM antibody was raised. Input (lanes 1, 2, 6 and 7) indicates 5% of the lysate used for immunoprecipitation (IP). The efficiency of IP was verified with anti-BLM antibody (a). Antibodies used for detection of other proteins in BLM IP were (b) anti-p53 (DO-1) and (c) anti-RAD51 (Ab-1). The position of the 46 kDa molecular weight marker is indicated. Lanes 1, 3, 6 and 8, minus HU; lanes 2, 4, 5, 7, 9 and 10, plus HU.
Immunofluorescence experiments were carried out in TR9-7 and TR15-3 cells grown in 50 ng/ml tetracycline, in the presence of HU. Both wild-type and mutated p53 accumulated at the sites of stalled DNA replication forks as stained by BrdU (data not shown), and co-localized with BLM and RAD51 (Figure 4B). This was confirmed further in immunoprecipitation experiments using anti-BLM antibody (Figure 4C). Wild-type and mutant p53 were detectable at equal levels in BLM immunoprecipitates in the presence of HU (Figure 4C, b). Moreover, the immunoprecipitates also contained RAD51 in both cell lines (Figure 4C, c), thereby indicating that Ser15 phosphorylation was not obligatory for transport of p53 to the sites of stalled DNA replication.
BLM is transported to stalled DNA replication forks in the absence of p53
To determine whether BLM co-localization at sites of stalled DNA replication forks is p53 dependent, isogenic NHF and NHF E6 (lacking detectable p53, data not shown) cell lines were used. In HU-treated cells, the average number of BLM foci was 10–52 per cell. As in NHF, a majority (70%) of BLM signal co-localized with BrdU, indicating that p53 was not obligatory for BLM localization at stalled DNA replication forks (Figure 5A). BLM immunoprecipitate from NHF E6 cells also contained RAD51 during S phase arrest. Reciprocally, BLM was also detected in the RAD51 immunoprecipitate (Figure 5B), indicating that the BLM interaction with RAD51 was p53 independent. Consistent with these findings, RAD51 co-localized with BLM in NHF E6 cells (Figure 5C).
Fig. 5. BLM co-localizes and physically interacts with RAD51 even in the absence of p53. (A) BLM co-localized with sites of stalled DNA replication forks in NHF E6. Contact-inhibited NHF E6 cells were treated with HU for 16 h, washed and incubation continued for 10 min in the presence of BrdU. Immunofluorescence were carried out with antibodies against BrdU/BLM (C-18). Quantitation of (a) is represented in (b). (B) BLM and RAD51 interacted in NHF E6. Cells were either asynchronous and left untreated (lanes 1 and 6), or contact inhibited and subsequently treated with HU (lanes 2 and 7) for 16 h. Lysates (400 µg) were immunoprecipitated with antibodies against BLM (C-18, lanes 3–5) or RAD51 (Ab-1, lanes 8 and 9). The peptide against which BLM antibody was raised (lane 5) or the corresponding IgG (lane 10) was used as control for BLM and RAD51 immunoprecipitation (IP), respectively. Input (lanes 1, 2, 6 and 7) indicates 5% of the lysate used for IP. The efficiency of IPs was verified with the respective self-antibodies (a and c). Antibodies against RAD51 (Ab-1) and BLM (C-18) were used for their detection in reciprocal IPs. Lanes 1, 3, 6 and 8, minus HU; lanes 2, 4, 5, 7, 9 and 10, plus HU. (C) BLM co-localized with RAD51. The same as (A) except that immunofluorescence was carried out with antibodies against BLM (C-18)/RAD51 (Ab-1). Quantitation of (a) is represented in (b).
p53 and BLM negatively regulated HR
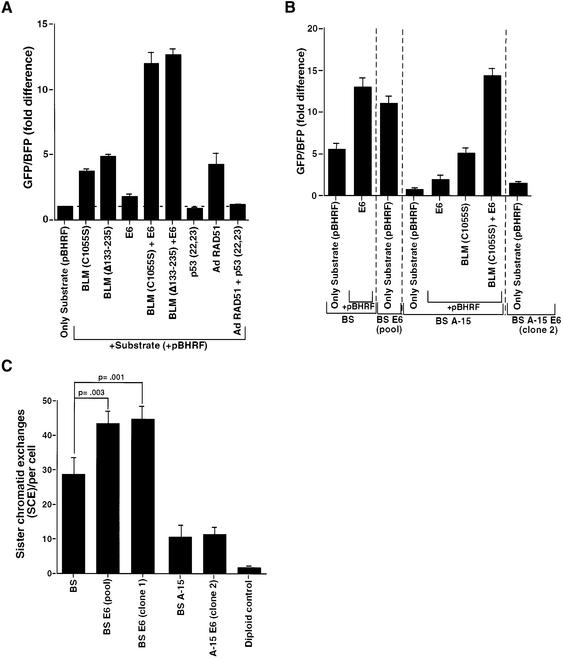
To determine whether the co-localization and physical interaction between p53 and BLM has functional consequences, we undertook two approaches. First, we conducted a plasmid-based host cell reactivation assay (Slebos and Taylor, 2001) in NHF cells to determine if rates of recombination are affected by the selective absence of proteins involved in the process. For this purpose, we used two dominant-negative mutants of BLM, C1055S and Δ133–235, which reduce formation of BLM nuclear foci (Wang et al., 2001). Both dominant-negative mutants were expressed in NHF and could be detected by western blot analysis (data not shown). In accordance with the anti-recombinogenic properties of BLM, each of the two dominant-negative mutants caused an enhancement of HR rate by 4- to 5-fold (Figure 6A). Consistent with previous studies (Susse et al., 2000; Willers et al., 2000; Slebos and Taylor, 2001; Saintigny and Lopez, 2002), reduction in p53 levels through E6 expression also had a modest effect (1.5- to 2-fold) on the HR rate. However, when dominant-negatives of BLM were co-expressed with E6, the increase in rate of HR was greater (11- to 13-fold) than that obtained when either of them was expressed alone (Figure 6A). Increases in the rates of HR similar to those achieved with reduction of p53 levels by E6 were also obtained with two dominant-negative mutants of p53, R175H and R248W (data not shown). Co-expression of these mutants with the dominant-negative BLM mutants affected HR similarly to E6 plus the BLM mutants (data not shown). These results indicate that p53 and BLM act in concert and in interconnected pathways during HR.
Fig. 6. p53 modulates BLM function during HR and SCE. (A and B) The absence of wild-type p53 and BLM synergistically enhanced the rate of HR in (A) NHF and (B) BS, BS E6, BS A-15 and BS A-15 E6 cell lines. Cells were transfected with either pBHRF alone or with pBHRF with different expression plasmids, as indicated. At 36 h post-transfection, cells were harvested and analyzed by flow cytometry in a FACS Vantage (Becton Dickinson) machine. The fold difference of the GFP/BFP ratio for different sets of transfections is represented. (C) The absence of wild-type p53 further enhances the rate of SCE in BS cells. SCE measurements (mean ± SD) were carried out in BS, BS E6 (pool), BS E6 (clone 1), BS A-15 and BS A-15 E6 (clone 2) fibroblasts, and a diploid control. The P-values were obtained by Student’s t-test.
To determine whether p53 can have a direct effect on RAD51/RAD54-mediated HR, overexpression of human RAD51-encoded adenovirus was carried out with or without co-expression of transcriptionally inactive p53, p53 (Leu22Ala, Trp23Ala) (Lin et al., 1994). This mutant of p53 was used to minimize the transcription-dependent functions of wild-type p53. Overexpression of RAD51 increased the HR rate by 3- to 4-fold, while p53 (Leu22Ala, Trp23Ala) did not affect the basal level of HR. However, when RAD51 and p53 (Leu22Ala, Trp23Ala) were co-expressed, the RAD51-dependent increase in HR was no longer obtained (Figure 6A), indicating that p53 modulated HR by direct interaction to govern the pro-recombinogenic function of RAD51.
The possibility that BLM and p53 have a coordinated effect on HR was investigated further in a group of four isogenic cell lines that differ only in their p53 or/and BLM status. To accomplish this, we derived new cell lines from related BS and BS A-15 cell lines in which p53 was inactivated by expression of E6 (BS E6 and BS A-15 E6, respectively). The level of p53 in these cells was extremely low and no DNA damage-induced increase in p53 or its downstream effector p21WAF1 was observed (Supplementary figure 3). As expected, BS cells have a high rate of spontaneous HR, which was enhanced a further 2.5-fold either by inactivation of p53 due to transfection or stable integration of E6. As expected, restoration of BLM in BS A-15 cells led to a lowering of spontaneous HR. Inactivation of p53 or BLM again enhanced the rate of HR from the basal level in BS A-15 by 2- and 5-fold, respectively. As in NHF, when both p53 and BLM were made functionally absent in BS A-15, the rate of HR was synergistically enhanced by 15-fold and reached the levels attained by BS E6 cells (Figure 6B).
Effect of p53 on spontaneous SCE is dependent on BLM
The host cell reactivation assay measures extrachromo somal recombination with artificial plasmid DNA substrates. It has been demonstrated that SCEs are mediated by HR in vertebrate cells (Sonoda et al., 1999). In addition, elevation in SCE rate is a hallmark of BLM deficiency. To ascertain whether p53 affects HR under physiological conditions, we determined the effect of p53 on SCE in the group of four isogenic cell lines described above.
In agreement with a previous report (Kuhn and Therman, 1986), BS cells exhibited a pronounced increase in spontaneous SCE compared with normal cells (Figure 6C). In accordance with another study (Ouellette et al., 2000), correction of BS cells with chromosome 15 containing the BLM gene (BS A-15 cells) resulted in a 3- to 4-fold decrease in SCE frequency to near basal level. However, inactivation of p53 in BS cells (BS E6) resulted in a statistically significant 1.5-fold increase in SCE rate (Figure 6C). This indicates that p53 has a cooperative effect with BLM with respect to SCE frequency. As reported in the literature (Bunz et al., 2002), we found that targeted inactivation of p53 alone (BS A-15 E6) did not lead to an enhancement of spontaneous SCE. Hence, both p53 and BLM modulate SCE (and HR).
Discussion
Intranuclear trafficking of BLM and p53
DNA repair, replication and recombination at the replication fork traditionally are thought to be diverse processes. However, it is likely that these processes are coordinated and that stalled replication foci can be viewed as regions containing a large number of repair and recombination factors coordinated by mutual interactions. Co-localization of BLM with RAD50 and MRE11 at the sites of stalled DNA replication forks (Y.Wang et al., 2000) and with RAD51 (Wu et al., 2001) has been described recently. In this report, we demonstrate that both BLM and p53 foci co-localized and physically associated with each other, along with RAD51 at sites of stalled DNA replication forks (Figures 1A and B, and 2).
Previously, we demonstrated that p53 function is required for BLM to be transported into PML NBs (Wang et al., 2001). Here, we show that BLM can be transported independently to sites of stalled DNA replication forks in the absence of p53 (Figure 5). In contrast, transport of p53 to these sites and its physical interaction with RAD51 is BLM dependent (Figure 3), indicating that BLM may be required for certain S phase-specific functions of p53.
Many kinases can phosphorylate p53 in vivo, often depending on the cell type and stress involved. Ser15 of p53 is phosphorylated in response to treatment with cisplatin, UV radiation, ionizing radiation or HU, but not actinomycin D (Appella and Anderson, 2001; Gottifredi et al., 2001). Previous studies have shown that the phosphatidylinositol-3 kinases ATM and ATR can phosphorylate p53 (Appella and Anderson, 2001), and they are present at RAD51 sites during DNA damage (Gatei et al., 2001). BLM also undergoes radiation-induced phosphorylation, dependent on the presence of a functional ATM (Ababou et al., 2002). Hence, both BLM and p53 are possible targets of ATM at the sites of stalled DNA replication forks. Here, we provide evidence that both wild-type and mutated (Ser15Ala) p53 can be transported to the sites of stalled DNA replication forks and physically interact with BLM and RAD51 (Figure 4). This result, along with the increase in the level of Ser15 phosphorylated p53 in BS A-15 cells (Figure 3C, c), indicates that phosphorylation can occur at the stalled DNA replication fork itself. The phosphorylation at Ser15 of p53 may be required for its stabilization or interaction with other proteins present at the sites.
Role of p53 and BLM in HR
Recent work has demonstrated multiprotein complexes present at stalled replication foci (Y.Wang et al., 2000). p53 was not detected in any of these complexes, possibly because the HeLa cells used were p53 deficient. The present work, conducted on NHFs under physiological conditions, indicates that p53 is also a component of the multiprotein complex(es) that regulates repair, replication and recombination.
p53 can be viewed as an anti-recombinogenic protein and has been shown to maintain genetic stability by suppressing excessive HR. Several studies have shown that p53 inactivation results in elevated rates of recombination (Susse et al., 2000; Willers et al., 2000; Slebos and Taylor, 2001; Saintigny and Lopez, 2002). Using a host cell reactivation assay of HR, we confirm these findings in NHFs (Figure 6A) and in four isogenic cell lines that differed only in p53 and/or BLM status (Figure 6B). In addition, we show here that p53 co-localizes and physically interacts with the pro-recombinogenic protein RAD51 (Figures 2 and 6A). Overexpression of RAD51 increases the rate of HR, whereas co-expression of transcriptionally inactive p53 reduces HR to basal level (Figure 6A). HR rates are also elevated in BLM-deficient cells (Figure 6B). Two BLM mutants act in a dominant-negative fashion, leading to a hyper-recombinogenic phenotype similar to that in BS cells. In addition, we show that simultaneous inactivation of both p53 and BLM results in an HR rate greater than that when either gene is inactivated separately (Figure 6A and B). Hence, these two proteins are cooperating in complementary pathways during HR.
Given the somewhat artificial nature of the short-term recombination assay, we also conducted an SCE assay. Consistent with HR assay results, inactivation of both p53 and BLM led to an increase in the spontaneous SCE rate over that observed with BLM deficiency alone (Figure 6C). Interestingly, the increase in spontaneous SCE rate resulting from p53 inactivation is dependent on BLM, as no increase is observed in cells deficient only in p53. This was consistent with a recent study showing that p53 deficiency alone does not elevate SCE (Bunz et al., 2002).
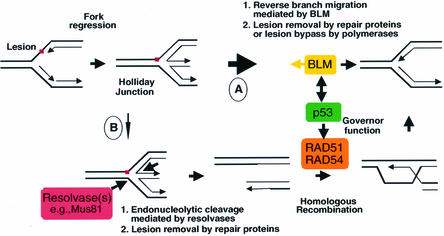
It was demonstrated recently that a considerable amount of SCEs in BLM-deficient DT40 cells were dependent on RAD54 function (i.e. via HR) (W.Wang et al., 2000). However, the RAD51 pathway participates only partially in SCE formation, and alternative pathways should exist (Lambert and Lopez, 2001). We propose that BLM directly regulates the principal pathway by acting as a 3′–5′ helicase to cause reverse branch migration, and it indirectly regulates a secondary pathway by transporting p53 to sites of potential HR, where p53 governs the activity of the pro-recombinogenic protein RAD51.
In the absence of significant stress, such as the conditions under which the SCE assay was conducted, the number and complexity of the sites requiring processing may be handled predominantly by BLM-dependent reverse branch migration (Figure 7, pathway A). In this case, loss of p53 alone (as in BS A-15 E6 cells, Figure 6C) does not have a pronounced effect. When BLM function is compromised (as in BS cells, Figure 6C), the balance could shift towards the HR pathway. However, when BLM and p53 are both inactivated, the equilibrium is shifted towards HR and the absence of inhibition of RAD51 by p53 and/or BLM may elevate the rate of HR further (as seen in BS E6 cells, Figure 6C).
Fig. 7. Mechanistic model for the restoration of stalled DNA replication forks. A regressed replication fork is restored by either reverse branch migration mediated by BLM helicase in a non-recombinogenic pathway (indicated by a thick arrow, pathway A) or a recombinogenic pathway that involves endonucleolytic cleavage by a resolvase(s) (e.g. Mus81), followed by RAD51/RAD54-mediated HR (indicated by a thin arrow, pathway B). p53 functions as a ‘molecular governor’ of HR. The simplified model shows a single DNA lesion at a replication fork that could represent a carcinogen–DNA adduct, a UV photoproduct or a base damaged by a free radical.
The constitutive inhibitory associations between p53 with RAD51 on the one hand and BLM with RAD51 on the other in untreated cells (Figure 2A–C; Supplementary figure 2a and c) may partially suppress HR. The lack of significant co-localization (Supplementary figure 2b) or detectable physical interaction (Figure 2A) between p53 and BLM in untreated cells indicates the presence of two distinct subpopulations of RAD51, one associated with p53 and the other with BLM. Therefore, apart from the predominant role of BLM in unstressed cells, both p53 and BLM are also involved in the modulation of HR due to their respective interactions with RAD51.
When cells are exposed to various stresses, the number of stalled replication forks requiring processing is significantly elevated, which may saturate the proposed BLM-dependent reverse branch migration pathway. In addition, treatment with different carcinogens, UV or ionizing radiation, which are known to induce recombination (Li et al., 1997). The above treatments could lead to more complex lesions on the DNA which are more difficult or impossible to process only by BLM-dependent reverse branch migration. In such situations, the HR pathway involving RAD51/RAD54 is likely to play a more important role (Figure 7, pathway B).
Elevated levels of RAD51 have been observed in various tumor cell lines, suggesting that hyper-recombination may play an important role during tumorigenesis (Raderschall et al., 2002). Hence, under conditions of stress, stalled DNA replication forks are regions where recombination is favorable, but in need of tight control. The inhibitory action of p53 on RAD51 (Figure 6A) may assist in governing inappropriate recombination. Hence, the tumor-derived p53 mutant (R273H) exhibits reduced binding to RAD51 (Buchhop et al., 1997) and fails to suppress RAD51-mediated strand exchange (Susse et al., 2000). This indicates that p53 can ‘govern’ the pro-recombinogenic function of RAD51 through direct binding and by modulating its assembly.
The error-prone non-homologous end joining (NHEJ) DNA repair pathway is also critical for maintaining genomic stability and cell survival (Hoeijmakers, 2001). BLM and p53 have also been implicated in this process. For example, p53–/– mice exhibit attenuated function of the NHEJ factor XRCC4 (Gao et al., 2000). BLM is involved in the alignment of microhomology elements during NHEJ (Langland et al., 2002). Therefore, it would be interesting to determine whether BLM is also involved in the transport of p53 during the end-joining process. Similar roles for p53 and BLM are also possible during mismatch, nucleotide excision and base excision repair.
In conclusion, we have presented evidence that both p53 and BLM are components of the RAD51 complex involved in the resolution of stalled DNA replication forks during S phase. BLM is required for the presence of p53 at stalled DNA replication forks. The p53–BLM interaction has a functional in vivo consequence. p53 and BLM cooperatively affect HR and SCE, providing direct evidence for an S phase-specific and transcription- independent function of p53.
Materials and methods
Recombinants
Recombinants used were: pRCp53 (22,23) (Lin et al., 1994); BLM (C1055S) and BLM (Δ133–235) (Wang et al., 2001); and pTL2 E6 (IGBMC core facility). The recombinant adenovirus Ad-hRad51 was made by inserting hRad51 cDNA as a BamHI fragment from pCRII-HsRad51 into the adenovirus transfer plasmid pZEROTG-CMV. Replication-defective adenovirus genomes were then generated as described previously (Valerie, 1999).
Antibodies
Anti-p53 antibodies were the monoclonals DO-1 and 1801 (Santa Cruz), and the polyclonal 588, raised against GST–human p53 (320–393) (Sengupta and Wasylyk, 2001). Both polyclonal and monoclonal 16G8 (Cell Signaling) were used as anti-phospho-p53 (Ser15) antibodies. Antibodies against BLM were goat polyclonal C-18 (Santa Cruz) and rabbit polyclonal ab476 (Novus). Anti-BrdU monoclonal antibodies were from Amersham. Polyclonal Ab-1 and monoclonal Ab-2 (both Oncogene Research) were used against RAD51, and the anti-p21WAF1 antibody was monoclonal Ab-1 (Oncogene Research). Secondary antibodies were purchased from Jackson Laboratories and Southern Biotechnology Associates.
Cells, culture conditions, treatments
NHF strain GM07532 was obtained from Coriell. Transformed NHF-hTERT cells were generated by transduction of NHF with CLXSN-hTERT-SN retrovirus, which was produced by subcloning the hTERT gene from pGRN145 into the pCLXSN retroviral vector, followed by transfection into the Phoenix-A producer line. GM07532 E6 (NHF E6) cells were generated by transduction of NHF with LXSN-E6 retrovirus harvested from the PA317 producer line. Standard retrovirus protocols were followed, and pools of NHF or NHF E6 cells were selected with 400 µg/ml G-418.
hTERT-transformed BS fibroblasts and chromosome 15-corrected BS fibroblasts, BS A-15 (GM03509 A-15), were described previously (Ouellette et al., 2000). To generate GM03509 E6 (BS E6) and GM03509 A-15 E6 (BS A-15 E6) cell lines, LXSH-E6 retroviral vectors were transfected into 293 packaging cells. Standard retrovirus protocols were followed, and stably transfected cells were selected with 25 µg/ml hygromycin B. All results from NHF, NHF E6, BS, BS A-15, BS E6 and BS A-15 E6 were obtained with hTERT-transformed lines.
Cells were maintained in 10% Dulbecco’s modified Eagle’s medium supplemented with glutamine and antibiotics. For HU treatment, cells were allowed to grow to confluency to attain G1 synchronization. They were then plated out at low density for 24 h so that most of the cells are in S phase. Cells then were either left untreated or treated with 1 mM HU for 16 h (or 100 µM PALA, 6 h) to stop the cells within S phase. The sites of DNA replication were detected with anti-BrdU antibodies after DNase treatment (Kennedy et al., 2000). After HU treatment, cells were washed and incubated with BrdU (150 µM) for 10 min, followed by processing of the samples for immunofluorescence. Asynchronous populations of cells were plated in parallel and represented as untreated cultures.
TR9-7 and TR15-3 cells normally were grown in the presence of 1 µg/ml tetracycline, for 24 h (Bean and Stark, 2001). Cell growth was continued either in the same medium or in the presence of 50 ng/ml tetracycline, in the absence or presence of HU for 16 h, followed by processing for further experiments.
Host cell reactivation assay and flow cytometry
The host cell reactivation assay to determine the HR rate was carried out as described (Slebos and Taylor, 2001). Transfections were carried out with Lipofectamine 2000, in the absence of serum for 6 h. pBHRF alone or with different combinations of expression plasmids was used. After transfection, incubation was continued for 36 h in growth medium, after which cells were harvested and flow cytometry carried out. pBHRF encoded an intact, emission shifted, ‘blue’ variant of green fluorescent protein (i.e. BFP), with a 300 nucleotide stretch of homology to a non-functional copy of GFP. In the absence of HR, only BFP is present, while HR can also create a functional GFP. Green and blue fluorescence were examined simultaneously by exciting the cells using a 488 nm argon laser (GFP) and a UV (350–360 nm) laser (BFP) in a FACS Vantage machine (Becton Dickinson). Each experiment was carried out at least three times.
Western blots and immunoprecipitations
Cells were lysed in a modified RIPA buffer [1 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF) and protease inhibitors]. Western analysis and immunoprecipitations were carried out by standard protocols. The experiments were repeated at least twice, and representative blots are shown.
Microscopy
The cells were grown on coverslips in 6-well plates. Immunofluorescence was carried out as described previously (Balajee et al., 1998) and cells were visualized in a Zeiss Axioskop fluorescence microscope equipped with a high performance CCD imaging system (IP Lab Spectrum). Alternatively, confocal fluorescent images were collected with a Bio-Rad MRC 1024 confocal scan head mounted on a Nikon Optiphot microscope with a 60× objective. Images were analyzed by Bio-Rad software.
The term co-localization refers to the coincidence of green and red fluorescence, as measured by either of the above microscopic processes. Co-localization in quantitation figures indicates the percentage of the proteins that co-localize with each other. For example, in Figure 1A, c, 48% of BrdU foci co-localized with BLM, while 67% of BLM foci co-localized with BrdU. At least 100 cells were analyzed for each co-localization experiment, and 5–8 typical cells per experiment were quantitated either microscopically or by Bio-Rad LaserSharp software. The experiments were repeated at least twice.
Sister chromatid exchange
SCE was carried out according to standard protocols (Perry and Wolff, 1974). The cultures were divided and 3 µg/ml BrdU was added to the medium. The cultures were kept in the dark and harvested after 48 h by treating with 0.05 µg of colcemid for 3 h. Metaphase spreads were made after standard hypotonic treatment and fixed in 3:1 methanol:glacial acetic acid. The slides were kept at 45°C for 18 h and were stained with a solution of Hoechst 33258 (2 mg/ml) in 2× SSC for 10 min. The slides were then mounted with Sorensen’s phosphate buffer pH 7, exposed to a black light for 45 min, subsequently washed with distilled water to remove the coverslip and incubated in 2× SSC at 65°C for 1 h. After washing with distilled water, the slides were stained in 4% Giemsa solution (Sorenson’s buffer pH 6.8) for 30 min, before being washed again in distilled water, dried and mounted. Twenty-five metaphase spreads were imaged for each cell line. The whole experiment was carried out twice, and for each cell line SCE was scored blind. The P-values were obtained by Student’s t-test. The conditions are two-tailed, unpaired data with unequal variance.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors thank Denise Galloway, Robert Naviaux, Inder Verma, Garry Nolan, H.W.Sturzbecher, George Stark, Robbert Slebos and Geron Corp for cells and recombinants, Roscoe Stanyon for SCE analysis, and Dorothea Dudek for editorial assistance.
References
- Ababou M., Dumaire,V., Lecluse,Y. and Amor-Gueret,M. (2002) Bloom’s syndrome protein response to ultraviolet-C radiation and hydroxyurea-mediated DNA synthesis inhibition. Oncogene, 21, 2079–2088. [DOI] [PubMed] [Google Scholar]
- Appella E. and Anderson,C.W. (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem., 268, 2764–2772. [DOI] [PubMed] [Google Scholar]
- Balajee A.S., May,A., Dianova,I. and Bohr,V.A. (1998) Efficient PCNA complex formation is dependent upon both transcription coupled repair and genome overall repair. Mutat. Res., 409, 135–146. [DOI] [PubMed] [Google Scholar]
- Bean L.J. and Stark,G.R. (2001) Phosphorylation of serines 15 and 37 is necessary for efficient accumulation of p53 following irradiation with UV. Oncogene, 20, 1076–1084. [DOI] [PubMed] [Google Scholar]
- Bischof O., Kim,S.H., Irving,J., Beresten,S., Ellis,N.A. and Campisi,J. (2001) Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol., 153, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G., Kipnis,J., Leal,J.F., Yu,C.E., Schellenberg,G.D. and Oren,M. (1999) Physical and functional interaction between p53 and the Werner’s syndrome protein. J. Biol. Chem., 274, 29463–29469. [DOI] [PubMed] [Google Scholar]
- Buchhop S., Gibson,M.K., Wang,X.W., Wagner,P., Sturzbecher,H.W. and Harris,C.C. (1997) Interaction of p53 with the human Rad51 protein. Nucleic Acids Res., 25, 3868–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Fauth,C., Speicher,M.R., Dutriaux,A., Sedivy,J.M., Kinzler,K.W., Vogelstein,B. and Lengauer,C. (2002) Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res., 62, 1129–1133. [PubMed] [Google Scholar]
- Chakraverty R.K. and Hickson,I.D. (1999) Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. BioEssays, 21, 286–294. [DOI] [PubMed] [Google Scholar]
- Collister M., Lane,D.P. and Kuehl,B.L. (1998) Differential expression of p53, p21waf1/cip1 and hdm2 dependent on DNA damage in Bloom’s syndrome fibroblasts. Carcinogenesis, 19, 2115–2120. [DOI] [PubMed] [Google Scholar]
- Franchitto A. and Pichierri,P. (2002) Bloom’s syndrome protein is required for correct relocalization of RAD50/MRE11/NBS1 complex after replication fork arrest. J. Cell Biol., 157, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. et al. (2000) Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature, 404, 897–900. [DOI] [PubMed] [Google Scholar]
- Gatei M., Zhou,B.B., Hobson,K., Scott,S., Young,D. and Khanna,K.K. (2001) Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. In vivo assessment using phospho-specific antibodies. J. Biol. Chem., 276, 17276–17280. [DOI] [PubMed] [Google Scholar]
- Gottifredi V., Shieh,S., Taya,Y. and Prives,C. (2001) From the cover: p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl Acad. Sci. USA, 98, 1036–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J.E. (2000) Partners and pathways repairing a double-strand break. Trends Genet., 16, 259–264. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. (2001) Genome maintenance mechanisms for preventing cancer. Nature, 411, 366–374. [DOI] [PubMed] [Google Scholar]
- Karow J.K., Constantinou,A., Li,J.L., West,S.C. and Hickson,I.D. (2000) The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA, 97, 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie,D.A., Classon,M., Dyson,N. and Harlow,E. (2000) Nuclear organization of DNA replication in primary mammalian cells. Genes Dev., 14, 2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn E.M. and Therman,E. (1986) Cytogenetics of Bloom’s syndrome. Cancer Genet. Cytogenet., 22, 1–18. [DOI] [PubMed] [Google Scholar]
- Lambert S. and Lopez,B.S. (2001) Role of RAD51 in sister-chromatid exchanges in mammalian cells. Oncogene, 20, 6627–6631. [DOI] [PubMed] [Google Scholar]
- Langland G., Elliott,J., Li,Y., Creaney,J., Dixon,K. and Groden,J. (2002) The BLM helicase is necessary for normal DNA double-strand break repair. Cancer Res., 62, 2766–2770. [PubMed] [Google Scholar]
- Lee S., Cavallo,L. and Griffith,J. (1997) Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem., 272, 7532–7539. [DOI] [PubMed] [Google Scholar]
- Li J., Ayyadevera,R. and Shmookler Reis,R.J. (1997) Carcinogens stimulate intrachromosomal homologous recombination at an endogenous locus in human diploid fibroblasts. Mutat. Res., 385, 173–193. [DOI] [PubMed] [Google Scholar]
- Lin J., Chen,J., Elenbaas,B. and Levine,A.J. (1994) Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev., 8, 1235–1246. [DOI] [PubMed] [Google Scholar]
- Lu X. and Lane,D.P. (1993) Differential induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell, 75, 765–778. [DOI] [PubMed] [Google Scholar]
- Oren M. (1999) Regulation of the p53 tumor suppressor protein. J. Biol. Chem., 274, 36031–36034. [DOI] [PubMed] [Google Scholar]
- Ouellette M.M., McDaniel,L.D., Wright,W.E., Shay,J.W. and Schultz,R.A. (2000) The establishment of telomerase-immortalized cell lines representing human chromosome instability syndromes. Hum. Mol. Genet., 9, 403–411. [DOI] [PubMed] [Google Scholar]
- Perry P. and Wolff,S. (1974) New Giemsa method for the differential staining of sister chromatids. Nature, 251, 156–158. [DOI] [PubMed] [Google Scholar]
- Raderschall E., Stout,K., Freier,S., Suckow,V., Schweiger,S. and Haaf,T. (2002) Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res., 62, 219–225. [PubMed] [Google Scholar]
- Saintigny Y. and Lopez,B.S. (2002) Homologous recombination induced by replication inhibition, is stimulated by expression of mutant p53. Oncogene, 21, 488–492. [DOI] [PubMed] [Google Scholar]
- Sengupta S. and Wasylyk,B. (2001) Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev., 15, 2367–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slebos R.J. and Taylor,J.A. (2001) A novel host cell reactivation assay to assess homologous recombination capacity in human cancer cell lines. Biochem. Biophys. Res. Commun., 281, 212–219. [DOI] [PubMed] [Google Scholar]
- Sonoda E., Sasaki,M.S., Morrison,C., Yamaguchi-Iwai,Y., Takata,M. and Takeda,S. (1999) Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol., 19, 5166–5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillare E.A., Robles,A.I., Wang,X.W., Shen,J.C., Yu,C.E., Schellenberg,G.D. and Harris,C.C. (1999) p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev., 13, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susse S., Janz,C., Janus,F., Deppert,W. and Wiesmuller,L. (2000) Role of heteroduplex joints in the functional interactions between human Rad51 and wild-type p53. Oncogene, 19, 4500–4512. [DOI] [PubMed] [Google Scholar]
- Valerie K. (1999) Viral vectors for gene therapy. In Wu-Pong,S. and Rojanasakul,Y. (eds), Biopharmaceutical Drug Design and Development. Humana Press, Totowa, NJ, pp. 69–105.
- van Brabant A.J., Stan,R. and Ellis,N.A. (2000) DNA helicases, genomic instability and human genetic disease. Annu. Rev. Genomics Hum. Genet., 1, 409–459. [DOI] [PubMed] [Google Scholar]
- Wang W., Seki,M., Narita,Y., Sonoda,E., Takeda,S., Yamada,K., Masuko,T., Katada,T. and Enomoto,T. (2000) Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J., 19, 3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W. et al. (2001) Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem., 276, 32948–32955. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cortez,D., Yazdi,P., Neff,N., Elledge,S.J. and Qin,J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev., 14, 927–939. [PMC free article] [PubMed] [Google Scholar]
- Willers H., McCarthy,E.E., Wu,B., Wunsch,H., Tang,W., Taghian,D.G., Xia,F. and Powell,S.N. (2000) Dissociation of p53-mediated suppression of homologous recombination from G1/S cell cycle checkpoint control. Oncogene, 19, 632–639. [DOI] [PubMed] [Google Scholar]
- Wu L., Davies,S.L., Levitt,N.C. and Hickson,I.D. (2001) Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J. Biol. Chem., 276, 19375–19381. [DOI] [PubMed] [Google Scholar]
- Yang Q. et al. (2002) The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J. Biol. Chem., 277, 31980–31987. [DOI] [PubMed] [Google Scholar]