Abstract
Objectives: The authors designed and implemented a clinical data mart composed of an integrated information retrieval (IR) and relational database management system (RDBMS).
Design: Using commodity software, which supports interactive, attribute-centric text and relational searches, the mart houses 2.8 million documents that span a five-year period and supports basic IR features such as Boolean searches, stemming, and proximity and fuzzy searching.
Measurements: Results are relevance-ranked using either “total documents per patient” or “report type weighting.”
Results: Non-curated medical text has a significant degree of malformation with respect to spelling and punctuation, which creates difficulties for text indexing and searching. Presently, the IR facilities of RDBMS packages lack the features necessary to handle such malformed text adequately.
Conclusion: A robust IR+RDBMS system can be developed, but it requires integrating RDBMSs with third-party IR software. RDBMS vendors need to make their IR offerings more accessible to non-programmers.
The clinical patient record represents the confluence of numerous heterogeneous data sources. At one extreme, it includes highly structured, columnar data such as complete blood counts and chemistry panels; at the other extreme is unstructured narrative text, such as operative reports and admission notes, which captures human-interpretable nuances that numbers and codes cannot. Simultaneously managing structured and unstructured data, however, remains an ongoing challenge. To the health care worker, no artificial distinctions are made between a patient’s serum potassium level and the narrative description of the resulting dysrhythmia. At the physical data model level, however, these two facts differ sharply in how they are represented and manipulated by computer information systems. The challenge is to hide these complexities for users who access the information.
We present our experiences with the design and implementation of an integrated system, built using primarily “commodity” software, which provides storage, retrieval, and interactive query of a large collection of structured and textual data. Our efforts, part of the Veteran’s Administration Medical Center, Connecticut (VAMC CT) Clinical Data Warehouse Initiative, represent the first phase in the creation of a clinical data mart.
Background
The prevalence of relational database management systems (RDBMSs) for most clinical patient record systems (CPRSs) reflects their overall dominance of the database market. RDBMSs have undergone considerable evolution since E. F. Codd’s pioneering work in the late 1960s.1 Nonetheless, until recently they have provided only rudimentary storage and retrieval support for arbitrary-sized text. Typically, such text is stored as “binary large objects” (BLOBs) that are composed of a chain of 2–8 KB-sized “pages.” In most RDBMSs, BLOBs cannot, by themselves, be searched for specific content or used for record selection criteria. Furthermore, pages in a BLOB’s chain are not necessarily contiguous on disk. Large BLOBs are often fragmented, with degradation of query performance when the BLOB is reassembled on demand during retrieval.
Information retrieval (IR)2 is the field of computer science concerned with general methods of processing and searching text. In IR terminology, individual units of text, such as clinical notes, are termed documents, and a database of documents is called a collection. The pre-processing of documents by IR software generates indexes, which are structures that facilitate efficient search. At least two indexes are created. The document frequency index records the number of documents in the entire collection in which a particular term occurs. The term frequency index records how often a term occurs in a particular document. The data in these indexes are used to determine how “relevant” a particular document is to a user’s search, which is specified in the form of one or more search terms. General relevance-ranking algorithms in IR include the classic document-vector algorithms,3,4 which rank search results based on the frequency of a search term within a given document and its rarity within the document collection (inverse document frequency).
Until the 1990s, IR systems developed essentially independently of RDBMSs, with little integration between the two. For systems needing both IR and RDBMS functionality, such as CPRSs, designers often built the IR component from first principles. An example is the Medical Archival Retrieval System (MARS),5,6 an IR-based CPRS that was developed by Yount and Vries in the early 1990s at Pittsburgh and is still used actively. Such systems, however, represent formidable software engineering. To avoid duplicating such efforts, there have been various proposals for implementing IR within an RDBMS framework.7 Some IR systems, such as INQUERY,8 are sold as RDBMS add-ons. However, the explosive growth of the World Wide Web, where much information is textual, has prompted all major RDBMS vendors to offer built-in IR support. The potential advantage of IR+RDBMS integration is that one may simultaneously query both columnar and textual data through a common mechanism, structured query language (SQL).
The latest International Standards Organization (ISO) version of SQL, SQL-99,9 has draft recommendations for providing “standard” IR support through the newly introduced CONTAINS function.10 CONTAINS introduces an entire sublanguage with a syntax almost as complex as the data-manipulation subset of the rest of SQL. Despite its seemingly large scope, however, the ISO draft standard does not address numerous important implementation details, such as dealing with documents in their source formats. RDBMS vendors compete either by going beyond the standard (e.g., adding enhancements to CONTAINS) or by providing greater ease of use.
Motivation for the Present Work
The Decentralized Hospital Computer Program (DHCP),11 which is an M (formerly MUMPS) database that is used nationally across all VAMCs, stores clinical narrative along with columnar patient data such as lab test results. DHCP does not, however, support any form of computing with the narrative text (e.g., searching by keywords); the text is merely displayed to the user who is looking up an individual patient’s data.
As part of the Clinical Data Warehouse Initiative, we decided to create a computable resource from the narrative text—specifically, an efficiently searchable document collection for general institutional use. In collaboration with the VAMC Radiology Depart-ment, we also decided to add functionality so that this collection could serve as part of a specialized teaching resource. The Radiology Department had assembled a large teaching collection of radiographic images but lacked sufficient follow-up information on many cases. We needed to allow the radiologist, given a patient and an imaging study date, to search the document collection forward in time, highlighting documents that appeared relevant to the patient’s final diagnosis or outcome. Furthermore, for patients with a diagnosis known to be challenging, the system would facilitate identification of diagnostically similar, but possibly more straightforward, cases for purposes of comparison by enabling access to the VAMC picture archiving system, which is linked to document IDs via Patient ID. An additional goal of the system was to facilitate institutional review board (IRB)-approved clinical investigation.
The VAMC CT data warehouse uses Microsoft SQL Server (MS-SQL) as its production database engine, and we were committed to using this RDBMS. In a first iteration toward implementing an IR system, we loaded extracts of individual DHCP tables into MS-SQL and indexed them using MS-SQL’s built-in facilities. When testing the system by searching with various terms, however, we observed repeatedly that MS-SQL sometimes failed to return certain known documents that should have matched the query. Closer inspection revealed that these failures were due to malformed text, such as incorrect punctuation (e.g., omission of a space after a period or comma) or misspellings. We realized that such failures would be quite common in the context of dictated medical text, which is minimally proofread. A subsequent detailed study of the documentation showed that MS-SQL offered very few options for robust searching in the presence of malformed text. We therefore needed to explore alternative IR solutions.
At the same time, we could not abandon RDBMS technology. The document collection is part of an integrated solution to identifying patients of interest. Search terms in narrative text are only one means of identifying such patients: identification by other criteria, such as demographics and disease codes, is best done through traditional RDBMS technology. We therefore needed to explore whether other integrated RDBMS+IR engines were up to the task. We performed a limited evaluation of Oracle 9i’s IR offering (Oracle Text), using a small subset of notes, as well as a review of the documentation of IBM’s DB2 Text Extender. While DB2 and its documentation are freely downloadable for academic researchers who enroll, at no cost, in the IBM Scholars Program, the software bundle does not currently include the optional Text Extender.
Other significant issues were cost and compatibility with existing VAMC systems. The existing institutional investment in MS-SQL diminished the possibility of replacing it with an alternative RDBMS at an institution-wide level for sake of the IR engine alone. A third option presented itself: the use of a reasonably low-cost “pure” IR package that could integrate with an RDBMS. We therefore evaluated such an IR package, dtSearch (dtSearch, Inc, www.dtSearch.com), currently the most widely used product on the Windows platform. dtSearch is also distributed with several commercial products, notably the Physicians’ Desk Reference. While very easy to use by nonprogrammers in stand-alone mode, it also supports programmatic access to, and indexing of, data stored in RDBMSs. dtSearch’s functionality can be accessed through any programming language that supports COM (Component Object Model), a Microsoft standard that allows programs on Windows platforms to access programming libraries in a language-neutral manner.
We first describe the objectives that our proposed system had to meet, followed by an evaluation of various commercial software alternatives in helping meet these objectives. We then describe a brief benchmarking study to determine the viability of our proposed approach and follow with a description of the system as finally implemented.
System Objectives
To allow attribute-centric queries about textual data.
DHCP, like all systems intended for real-time decision support, is transaction-oriented: it allows only patient-centric query based on demographic criteria such as name, social security number (SSN), or medical record number (MRN). In contrast, attribute-centric queries allow identification of patients who match criteria based on clinical attributes or keywords. This type of searching is better suited to research activities, which tend to focus on groups of patients or diseases.
To allow filtration of the document collection by arbitrary relational criteria (i.e., to allow integrated query of relational and textual data).
Users need to perform queries based on both narrative-text criteria and arbitrarily complex relational criteria, and one must consider how best to support integrated queries when the complexity of the relational criteria cannot be fully anticipated. Our proposed solution to this problem took a two-fold approach. The task of filtering on certain criteria, such as patient identifiers (name, SSN or MRN), document date, or type of report, was considered common enough to warrant direct support. Beyond this, our application would allow use of any query tool, such as Microsoft Access’s Query Design interface, to compose and execute arbitrarily complex relational queries that would create a temporary table of patient IDs. These IDs would then be used to filter the text search results or, in suitable circumstances, to limit the extent of text search.
To provide domain-appropriate relevance ranking of search results.
The built-in relevance-ranking algorithms of IR engines mentioned earlier proved to be of limited usefulness for two reasons. First, we found that term frequencies tended to be quite low: a random selection of documents revealed that keywords signifying positive clinical findings, procedures, or diagnoses rarely occurred more than twice in a document because of the brevity of most documents and the avoidance of unnecessary repetition. The resulting small differences in term frequency reduced its value as a discriminant. Second, every document in this collection always describes a particular patient; any ranking algorithm must take this association into account.
We devised ranking algorithms based on two observations.
For any given search, the greater the number, diversity of type, and occurrence over time of documents belonging to a particular patient, the more likely it was that the patient represented an “interesting case.” In good case summaries, significant positive elements tend to be carried over as “past history,” whereas a keyword occuring in only one document might be mentioned only in passing (e.g., as a condition to be ruled out).
Keywords in certain types of documents, such as operative notes or pathology reports, tended to represent “true positive cases.” In radiology notes, by contrast, the keyword often indicated suspicion rather than confirmation of a condition.
The ranking algorithm that we devised groups documents by patient and ranks, matching patients by total number of documents per patient and document type weighting. By default, all document types are given equal weight (of 1), but the user can change these weights. Thus, types with a weight of zero would be eliminated from a search.
Our use of a problem-specific algorithm is by no means unique. Frisse, in creating a hypertext version of the Washington University Manual of Medicine,12 used a modified document-vector algorithm that weighted the hyperlinks associated with a document.
To support robust search in the presence of malformed text.
Whereas “structured dictation” software, along with voice recognition, is used to record standardized information such as chest x-rays or mammograms, much dictated text is still transcribed off-line before being reviewed by the dictating clinician. The transcription task is often outsourced to service bureaus. Persons who are not professional transcriptionists, such as residents, may enter other text, such as progress notes, directly into the system. In any case, medical text within a hospital record system contains many more misspellings and incorrect punctuation than written text intended for publication, which is vigorously proofed by editorial staff.
Comparative Evaluation of IR/RDBMS Software
This section reports the results of our comparative evaluation of the IR features of MS-SQL, dtSearch, Oracle Text and DB2 Text Extender.
Indexing
Ease of Indexing
Creating IR indexes (“Full-Text Catalogs”) with MS-SQL is exceptionally easy. A “wizard” makes this a trivial point-and-click operation.
As stated before, dtSearch can be used in two modes: as a stand-alone IR engine and integrated with an RDBMS. In the former mode, indexing and searching can be done easily and interactively through a GUI that is geared toward nonprogrammers. In contrast, when accessing data that is physically stored in an RDBMS, the indexing and searching interface must be programmed. For the production system that we ultimately created, however, this merely entailed making minor modifications to the code examples in the dtSearch documentation, which constitute less than 100 lines of code.
Indexing in Oracle and DB2 Text Extender is done through the command line.
Indexing Options
MS-SQL provides little control over index creation beyond defining the index name, location, and which tables and columns are indexed.
dtSearch provides better control over index creation. Options include whether numbers are indexed, whether the index is accent-sensitive or case-sensitive, how hyphens are handled, maximum word length, and which configuration files are used to specify stop-words, and so forth. (Stop-words are very common words like “the” and “and,” which are not useful for searching.)
Oracle allows fine-grained control of indexing through a “Preferences” system, which is invoked by calling special subroutines (“stored procedures”). Although not particularly intuitive, the default options are reasonable for most situations. Oracle has numerous options for non-English languages; for example, compound words in German and Dutch are divided into their constituent words during indexing.
With DB2 Text Extender, when designating a column for indexing, one must define an alias (“handle”) for it: a query that invokes CONTAINS must use the handle rather than the column name itself. Although not a major barrier to use, this is a significant deviation from the ISO standard. DB2 allows less flexibility than dtSearch or Oracle in that one must decide whether to allow inflectional search, in which variations in tense or person are ignored; precise search for exact phrases; or fuzzy search to identify words despite misspellings. A different index type supports each of the three search categories. One cannot, for example, perform inflectional or fuzzy search with a precise-search index. Changing the index type requires re-indexing of the documents, because a text column cannot be simultaneously indexed with two different types of indexes.
Comparison of Searching Features
The feature sets of MS SQL Server, dtSearch and Oracle, are summarized in Table 1▶. We discuss details of each feature below. Features that are particularly relevant to medical text are marked with asterisks, with a brief explanation as to why they are relevant.
Table 1 .
Summary of the IR Capabilities of the Four Software Packages Evaluated*
| Feature | MS SQL Server 2000 | dtSearch Engine 6.0 | Oracle 9i | IBM DB2 Text Extender |
|---|---|---|---|---|
| Exact term and phrase matching | Yes | Yes | Yes | Yes |
| Boolean searches | Yes | Yes | Yes | Yes |
| Natural Language searches | Yes | Yes | Yes | Yes |
| Prefix searches | Yes | Yes | Yes | Yes |
| Word stemming (inflectional searches) | Yes | Yes | Yes | Yes |
| Term weighting | Yes | Yes | Yes | Yes |
| Proximity searches | Very limited | Yes | Yes | Limited to sentences and paragraphs only |
| XML-section-aware searches | No | Yes | Yes | Yes |
| Wildcard searches | Yes | Yes | Yes | Yes |
| Regular expression searches | No | Yes | No | No |
| Fuzzy searches | No | Yes | Yes, set at indexing time | Yes, set at indexing time |
| Phonic searches | No | Yes | Yes | Yes |
| Synonym expansion | No | Yes | Yes, | Yes |
| Built-in thesaurus | No | Wordnet | No | No |
| Case-sensitive searches (*) available? | No | Yes | Yes, set at indexing time | Yes, set at indexing time |
| Individually Customizable Stop-word lists | No | Yes | Yes | No |
| Availability of Word List | No | Yes | No | No |
*A detailed explanation of each feature that is listed is provided in the text.
All of the packages provide basic functionality, such as exact term and phrase searching, searching for a word by prefix, inflectional searching, and variable term weighting, in which different terms in a query are given different weights to indicate their relative importance to the user. They support complex Boolean search as well as “natural language” search. In the latter, a sentence or passage can be supplied to the IR engine, which then strips off all stop-words, and then relevance-ranks documents in the collection to the remaining words in the passage. The differences among the packages are described below.
Robust Handling of Variations in Punctuation Style*
As stated previously, the punctuation within dictated, nonreviewed medical text often departs from strict rules of style. Of the packages, only MS-SQL falls short. When a space is omitted after a period, MS-SQL treats the construct <token-1><period> <token-2> as though it were a new word. The instances of token-1 and token-2 are not recognized as individual words: this is a significant bug. Improper recognition of word and sentence boundaries not only results in missed searches, but also tends to swell the IR indexes with spurious “compound words.”
Fuzzy Search*
Spelling errors in medical text tend to be especially numerous in collections in which a significant fraction of the documents consists of brief notes (e.g., follow-up notes) that are reminders to oneself rather than meant for public review. We encountered 68 different variants of “orchiectomy,” of which only one other form (the British “orchidectomy”) could be considered correct. The polysyllabic medical terms that are most likely to be misspelled are also most likely to be keywords of interest. Fuzzy search deals with this situation. Existing implementations tend to be based on the well-known agrep (approximate grep) algorithm of Wu and Manber.13 DB2 uses a special type of index (N-gram index) for fast fuzzy search.
MS-SQL lacks fuzzy search capability entirely. dtSearch allows the user to control the level of “fuzziness” on a per-search basis, in terms of number of mismatched letters, in specifying a search. Oracle and DB2 allow fuzzy search if the index has been set up accordingly: with Oracle, enabling fuzzy search is the default. The latter two packages, however, require one to specify the level of mismatch in terms of a fuzzy “score” or “level” whose interpretation is not intuitive, and whose use requires experimentation. Oracle’s default setting matched “hyperpituitarism,” also a commonly misspelled word, to “hypopituitarism,” which has a mismatch of two letters.
Proximity Searching with Distance Specification
Proximity search lets one specify that the keywords of interest should occur within a certain range (e.g., a certain number of words from each other). MS-SQL provides a very limited form of proximity (X NEAR Y), but the interpretation of “NEAR” is not documented. A conference with Microsoft developers indicated that it used a threshold of 50 words. Because proximity information is not indexed in MS-SQL, searches using NEAR run relatively slowly. Oracle and dtSearch allow precise specification of distance, in terms of number of words or words within the same sentence or paragraph. Queries specifying proximity in terms of word distance utilize proximity indexes for speed. DB2 allows specification of proximity within the same sentence or paragraph, but not in terms of number of words.
XML-Aware Searching
Many documents have their content structured by divisions into sections. Currently, such structure is increasingly provided using standards based on XML (Extensible Markup Language).14 For example, the title of a document may be enclosed within tags such as <TITLE>....</TITLE>. The names of the tags that are used for a given document are determined by another document called an XML schema15 (or its older counterpart, the Document Type Definition16). The latter document specifies what tags are permissible and how they are used; for example, certain tags can be nested within others. XML is too vast a topic to introduce here; a suitable reference is Means and Harold.17 In the medical field, the proposed HL-7 Patient Record Architecture (PRA) standards18 may eventually yield clinical documents in tagged XML format.
All of the IR engines discussed here, except MS-SQL, are XML-aware and allow the user to specify that the terms of interest must occur within a given “field” indicated within the span of particular XML tags. Such search may be regarded as a special type of proximity search. Currently, however, none of the engines appear to be XML-Schema–aware. Because different documents in the same collection may be based on different XML schemas, such engines “play it safe” and do not take advantage of an existing schema to create indexes that are optimized for such searches. Although specialized database engines, termed native XML databases,19 exist for the purpose of efficiently storing and manipulating XML documents, they currently lack the advanced features necessary for IR capability.
Ability to Index Formatted Documents
This is an important feature when source documents are either created or stored with formatting information. dtSearch and Oracle recognize a wide variety of formats: Oracle, through third-party software, can recognize up to 100 formats, although many of these are rarely encountered. MS-SQL recognizes Word, Excel, PowerPoint, and HTML. DB2 supports the fewest. For practical purposes, only HTML and XML are recognized; the supported versions of word processor formats have long been obsolete.
Regular Expression Search*
Regular expressions20 provide a powerful syntax for searching for complicated patterns in text, such as one of several possible sequences of letters followed by a sequence of numbers. In the medical context, the use of regular expressions is typically the first step in information extraction from structured medical notes that are generated by assisted dictation systems, which use boilerplate formats with numerous standardized headings and sections. Regular expressions can also be used to express spelling variations of individual terms through a compact notation (e.g., Latinized vs. U.S. English, such as “haemoglobin” vs. “hemoglobin,” or British vs. U.S. English, such as “odour” vs. “odor”). Lexical analyzers,21 which are used in compilers or interpreters for programming languages, use regular expressions extensively, as do ad hoc text mining programs. By definition, regular expression patterns are arbitrary (i.e., supplied by the user dynamically) and cannot be pre-indexed. However, algorithms for regular expression pattern search have been extensively refined over the years and are very efficient. dtSearch is the only package that supports regular expression search.
Controlling Case-sensitivity of Search*
Medical text contains a significant number of acronyms, many of which would be common words if they were written in lower case (e.g., “AIDS”). Therefore, although one generally needs to locate keywords regardless of case, the ability to specify case-sensitive search can be important when needed. MS-SQL does not allow any control. The behavior of Oracle and DB2 is dictated by how a particular column’s index was created; this cannot be changed without re-indexing. dtSearch allows maximal flexibility. Two different indexes, one case-sensitive and the other case-insensitive, can be created on the same data, and the user can switch from one to the other. Alternatively, if a case-insensitive index was created, case-sensitive keywords can still be searched using regular expressions.
Ability to Specify Search Across All Fields, or Limiting Search to Specific Fields
Searching across all fields is of value when a table or tables possess numerous string columns, any or all of which have been indexed. The user may wish to search across all indexed fields or limit the search to a specified subset of fields. This is MS-SQL’s strong point. The CONTAINS implementation allows one to specify, with a wildcard, that all columns in a table that are IR-indexed should be searched. In Oracle and DB2, the CONTAINS function allows only a single column to be specified: this is a weakness that limits their power for some applications, as discussed below. For dtSearch, one can implement the equivalent of MS-SQL programmatically; at index creation time, all columns of interest in a table are indexed.
Support for an External Thesaurus*
Although support for synonym-based query expansion can benefit search of documents in any knowledge domain, it may arguably have a greater impact in medicine, where both Greco-Latin as well as common (Anglo-Saxon) forms exist for the same concept (e.g., emesis vs. vomiting). External thesauri provide powerful query term expansion capability. Thesauri generally provide concepts with several types of relationships, including synonyms, related terms, “parent-child” (general/specific), and “part-of” relationships. MS-SQL lacks thesaurus capability. dtSearch uses a customized version of the WordNet lexical and semantic network22 and also allows supply of external thesauri. Oracle and DB2 are a shade more powerful than dtSearch, in that they allow search of many other types of hierarchical relationships. However, they do not come with built-in thesauri.
Displaying a List of Words in the Index, with their Frequency Counts*
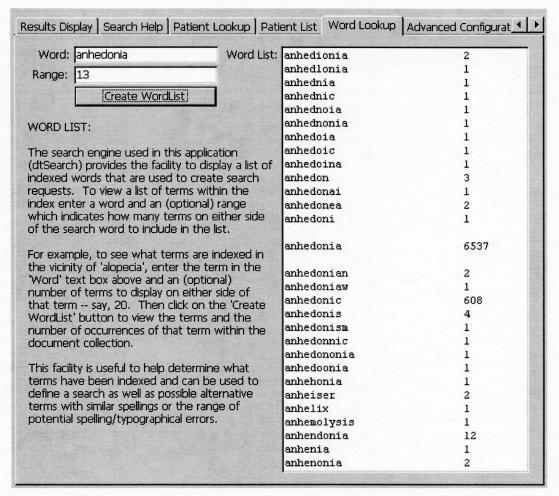
Medicine is well known for numerous compound words that are derived from several Greco-Latin roots (e.g., chole-docho-jejun-ostomy [hyphens added]). Such words are often misspelled. Fuzzy search and, to a lesser extent, regular expressions are useful for dealing with misspellings, but it is clearly impossible to anticipate every possible misspelling. Ability to interrogate the alphabetically sorted document frequency index, within a “window” centered on a particular term, can help in this regard, because misspellings for such words tend to occur in the latter part of a word rather than the first part. A query’s recall is then improved when the user can specify the multiple alternative forms that are discovered in the list. dtSearch is the only package that allows this, as shown in the Word List of Figure 1▶.
Figure 1.
The “Word list” for a search term of interest, which is accessed through one of the pages in the multi-tabbed control of the left half of Figure 2▶. This page lets the user access the Document Frequency Index for all terms in the collection that are centered around a particular term. The list shows the word and the number of documents in which it occurs. This screen shot illustrates the numerous ways in which the term “anhedonia” and its adjectival form “anhedonic” have been misspelled.
In summary, MS-SQL is the weakest of the packages in all aspects except two: ease of use and search across multiple text fields in a table. For dictated medical text, dtSearch has a significant edge over Oracle and DB2. For multi-language document collections, such as those that might be gathered by a Web search, Oracle and DB2 are more powerful than dtSearch. Their features in this regard have not been discussed because they are not directly relevant to this paper. However, Oracle and DB2, especially the former, have a significant learning curve because of their command-line orientation, which makes much of their power needlessly hard to access.
For our proposed system, we decided to use dtSearch for the reasons of feature set and cost. It is less expensive than the RDBMSs by more than an order of magnitude. We had to consider one final issue: integration with MS-SQL. This issue is discussed below.
One Software Package or Two?
As we have stated, our proposed system needs both IR and RDBMS functionality. A presumed advantage of using a single software package to perform both IR and RDBMS tasks, as opposed to using two separate programs that must communicate through code written by the developer, is a higher level of integration, with possibly much better performance.
The MS-SQL and DB2 IR engines are external programs that happen to be architecturally distinct from the RDBMS engines: MS-SQL, for example, uses the Microsoft Full-Text Indexing and Searching services. The RDBMS engines merely act as brokers for text search requests. The consequences of this are illustrated by using the following simple query as an example. Suppose we wish to identify all documents of type “surgery note” with the report date 5/1/ 2001, which contain the keywords “lung” and “cancer.” The relational criteria (report type and date) match 8 documents in the VA CT collection; the IR criterion matches about 16,000 documents. One would expect the RDBMS to use an evaluation of relational criteria, which is very fast because both report type and report date are indexed, to limit the IR search and return matching results rapidly. Instead, when the IR is performed by an external service, the search is actually performed as follows23:
The RDBMS passes the CONTAINS part of the query to the IR service.
The service processes the request and returns all 16,000 matching document IDs into an internal table.
The query is rewritten as a standard SQL statement that joins the results of the relational query to the internal table on the common field (document ID).
The rewritten query is executed, the results are returned to the user, and the internal table is truncated, to yield the final results (2 documents).
Oracle Text integrates the IR engine more closely with the RDBMS; as described later, however, this integration is far from complete.
The importance of such “loose coupling” is that replacement of the built-in IR engine by manually integrating a third-party IR engine may not incur significant additional performance penalty. In fact, our benchmark in the next section shows that this particular query runs almost twice as fast when dtSearch is used in conjunction with MS-SQL. The major issue to be addressed is ease of software development. Some programming is required in any case. The richness and complexity of SQL-99’s CONTAINS function currently make it necessary to build a GUI for non-expert users to access its features. The question is whether having to send the search commands that result from the user’s actions to an external IR engine, instead of to the RDBMS, constitutes a significant hurdle, or whether such an effort is justified in terms of yielding additional functionality.
Comparative Benchmarking of MS SQL Server and dtSearch
The benchmarks described below compare MS-SQL with dtSearch. Development and testing were performed on a dual-processor microcomputer with two Intel Pentium III 1.0 GHz CPUs, 512 MB of RAM, with two 40-GB hard drives in a RAID-0 configuration. All software ran locally and included a MS-SQL 2000 database back-end and a Microsoft Access 2000 front-end client connected to MS-SQL via ODBC. The client accessed the dtSearch Engine (version 6.04) via its COM interface.
Document Procurement and Storage
We used a commercial ODBC driver for DHCP (KB-SQL, Seattle Systems) to bulk-import selected DHCP tables into multiple Microsoft Access files and thence into the MS-SQL database. Because DHCP often stores reports as multiple 80-character-per-line records, we converted such data from a sequence of records to a single string that was then stored in a TEXT column. All document texts were stored in a single table, one row per document, together with basic descriptors such as unique document ID, patient ID, report type, and report date. Merging of the various report types in this way allowed us to avoid unnecessary and potentially expensive join operations across multiple tables and simplified the program code that performed document indexing and searching.
Document Collection Characteristics
The document collection totaled 2,809,339 reports for 70,328 patients and spanned a 5-year period. The number of documents per patient ranged from 1 to 1,642 with an average of 39 and a median of 12; many patients had reports of multiple types. The significance of this is that the performance of our ranking algorithms improves as the number and diversity of documents per patient increase. We also noted the brevity of most reports. Although length varied considerably, 85% contained fewer than 300 words, and 96% had fewer than 600 words. All pathology and cytology reports were shorter than 300 words. Tables 2▶ and 3▶ summarize the document collection characteristics.
Table 2 .
Summary of Descriptive Statistics on Various Types of Reports in the VAMC CT Collection*
| Report Type | Document Count | % of Documents | # of Distinct Patients | Word Count | Avg. Word Count | Median Word Count | Range (rounded) | Report Period | Byte Count |
|---|---|---|---|---|---|---|---|---|---|
| Medical Report | 1,966,600 | 70.00 | 61,192 | 384,358,313 | 195 | 116 | 100–6000 | 10/1/1994–9/1/2001 | 2,594,725,432 |
| Radiology Report | 403,929 | 14.38 | 43,185 | 34,693,902 | 85 | 68 | 50–2200 | 5/1/1996–8/1/2001 | 230,897,633 |
| Radiology Impression | 399,342 | 14.21 | 43,063 | 9,478,025 | 23 | 15 | 10–1000 | 5/1/1996–8/1/2001 | 65,036,534 |
| Surgical Note | 13,491 | 0.48 | 7,495 | 5,529,147 | 409 | 356 | 150–3400 | 4/1/1996–7/1/2001 | 36,888,138 |
| Pathology Report | 19,194 | 0.68 | 10,655 | 380,097 | 19 | 5 | 3–100 | 4/1/1996–7/1/2001 | 3,034,330 |
| Cytology Report | 6,515 | 0.23 | 3,418 | 92,896 | 14 | 12 | 5–100 | 4/1/1996–7/1/2001 | 779,143 |
| Autopsy Report | 268 | 0.01 | 268 | 284,024 | 1,054 | 1,331 | 150–3400 | 1/1/1997–2/1/2001 | 1,959,049 |
| TOTALS: | 2,809,339 | 434,816,404 | 2,933,320,259 |
*The distribution of words in all types of documents is highly non-Gaussian, as indicated by a significant difference between the average and median word counts.
Table 3 .
Number of Patients with More Than One Type of Note*
| Report Types | # of Distinct Patients |
|---|---|
| Medical | 61,192 |
| Medical + Radiology | 34,177 |
| Medical + Radiology + Surgery | 7,158 |
| Medical + Radiology + Pathology/Cytology | 11,047 |
| Medical + Radiology + Surgery + Pathology/Cytology | 5,196 |
| Medical + Radiology + Surgery + Pathology/Cytology + Autopsy | 67 |
*This information is useful in determining how useful our patient-ranking algorithm, which ranks patients by the number of documents containing a keyword, will be in practice.
Indexing Speed and Index Size
In this test, dtSearch operated on data stored within the MS-SQL database, although its indexes were created as disk files outside the database. dtSearch ran considerably faster than MS-SQL. Both indexers were run repeatedly: dtSearch took from 3.5–4 hours, whereas MS-SQL took from 20–28 hours. MS-SQL indexer runs only as a “background” process, although we attempted to improve performance by increasing its job priority and shutting down all other applications and services. The index created by dtSearch was significantly larger than that created by MS-SQL (2.25 GB vs. 1.1 GB), because it stores finer-grained proximity information. The proximity index records the exact position of an individual term within a document. Position is typically recorded by word offset from the start of the document, but other measures, such as sentence and paragraph offsets, may also be recorded.
Search Performance
In these benchmarks, we compared the performance of dtSearch to that of MS-SQL after indexing was complete. We automated the testing through a routine that executed queries against both engines in tandem, with results being saved to separate temporary tables. A second routine identified rows in one table that were absent in the other. Analysis required careful review of each document in the difference set. The observations, apart from helping to diagnose the reasons for failure, also enriched our understanding of the document collection itself. Table 4▶ shows benchmarks for comparison of MS-SQL vs. dtSearch for a variety of queries, both simple and complex. Several queries involved relational and IR criteria. In this scenario, dtSearch operated in conjunction with MS-SQL, with the latter program performing the relational part of the query. The mechanism of communication between the two programs is discussed in the System Description section.
Table 4 .
Comparison of MS-SQL vs. dtSearch for 14 Queries*
| Search Time (secs) |
Number of Matches |
Additional Matches |
||||
|---|---|---|---|---|---|---|
| Query | dtSearch | MS-SQL | dtSearch | MS-SQL | dtSearch | MS-SQL |
| “craniopharyngioma” | 4 | 4 | 71 | 71 | 0 | 0 |
| “angioplasty” | 215 | 303 | 17,187 | 16,994 | 193 | 0 |
| “hypopituitarism” | 2 | 6 | 142 | 142 | 0 | 0 |
| “nerve sheath tumor”* | 9 | 1 | 42 | 42 | 0 | 0 |
| “degenerative disk disease” | 9 | 13 | 3,365 | 3,365 | 0 | 0 |
| “guillain barre syndrome” | 5 | 10 | 127 | 127 | 0 | 0 |
| “seminoma” AND “orchiectomy” | 2 | 3 | 56 | 53 | 3 | 0 |
| “esophageal varices” AND “cirrhosis” | 9 | 38 | 1,783 | 1,729 | 54 | 0 |
| “tumor” OR “mass” OR “malignancy” [limit to radiology report] | 473 | 893 | 59,211 | 59,183 | 28 | 0 |
| (“squamous cell carcinoma” OR “SCC” OR “SCCa”) AND “laryngectomy” | 4 | 10 | 180 | 178 | 2 | 0 |
| “aorta” AND “aneurysm” [limit to date 7/1/2000–7/5/2000] | 9 | 19 | 18 | 18 | 0 | 0 |
| “hepatic” AND “cirrhosis” [limit to date 9/1/2000–9/7/2000] | 9 | 19 | 4 | 3 | 1 | 0 |
| “lung” AND “cancer” [limit to surgery notes, date 5/1/01] | 11 | 20 | 2 | 2 | 0 | 0 |
| “colon NEAR adenocarcinoma”* [limit to date 1/1/2000–1/1/2001] | 18 | 11 | 80 | 73 | 7 | 0 |
*dtSearch is faster in all except two, indicated by asterisks. The columns show the respective times for the query (in seconds), the total number of documents returned by both programs, and the documents returned by one program that were not returned by the other. The documents returned by dtSearch are always equal to, or a superset of, the documents returned by MS-SQL. MS-SQL missed some documents due to malformed punctuation, e.g., no space after a period, so that a term of interest is “conjoined” to the first word of the next sentence.
dtSearch was faster in all of the queries except two, which are indicated by asterisks. The timing benchmarks represent the average of two values, which were obtained each time after shutting down the machine and starting the software again. This was done to eliminate cache effects, which are well-known with disk-based operations. Sometimes running the same query a second time shortly after the first will return results instantaneously, because the previous results are still in memory. The timing figures show that search times increase with the number of matching documents. Although we have no explanation for the two cases in which dtSearch ran slower, the results suggest that the IR implementation of dtSearch is more optimized than that of MS-SQL. Certainly, any performance penalty that might have been expected for dtSearch, due to the need for two programs to communicate with each other through our code, appears to be absent.
For result sets of less than 200 documents, we visually inspected each document through the interface, which facilitates rapid visual evaluation by highlighting individual words in the search terms where they occur in the text. There were no false positives, but in a few cases, dtSearch returned more documents than MS-SQL. When inspected, these documents showed malformed punctuation in the vicinity of the word (e.g., absence of spaces after commas or periods). This set of documents, therefore, constituted false negatives with respect to MS-SQL. No documents identified by MS-SQL were missed by dtSearch. Based on the above benchmarks, we elected to use the dtSearch engine for text indexing and searching of data stored in MS-SQL rather than using MS-SQL’s built-in IR features.
System Implementation and Description
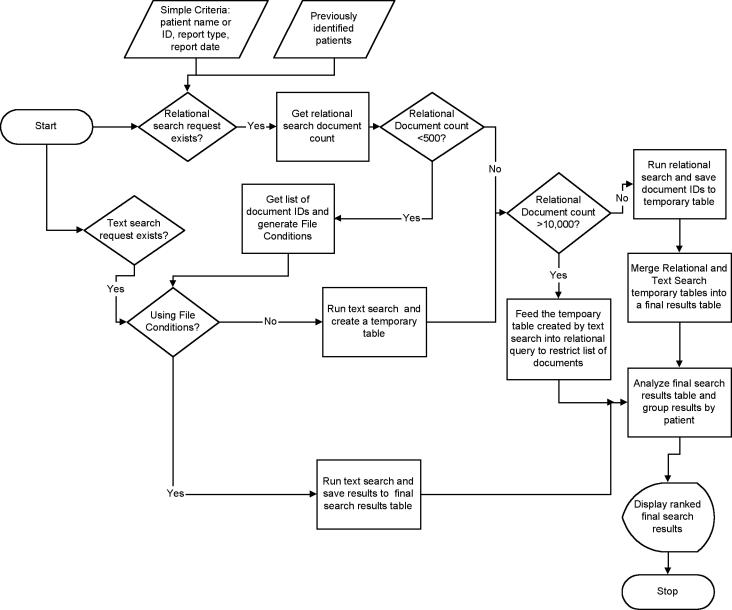
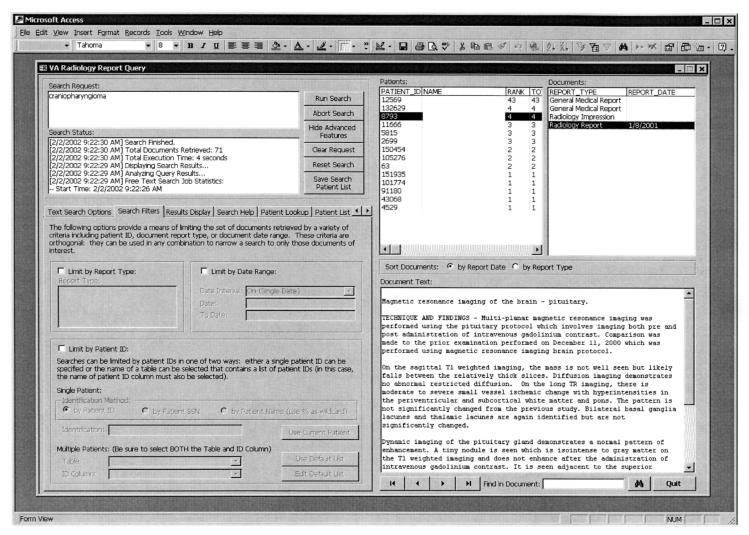
Figure 2▶ outlines the algorithm used to couple text and relational searches. Figure 3▶ shows the user interface of our application in operation. The salient features of the interface, and the algorithm, are described below.
The relational part of the query is generated automatically through the user’s interactions with the program. The user can search for documents related to a single patient or provide a table containing a list of patient IDs of interest. This table may have been generated by any external power-user-oriented query application, such as Microsoft Access’s own query builder, that is capable of creating a list of patient IDs (e.g., patients seen in a particular department within a date range) and matching a particular set of ICD-10 codes. The database executes the generated SQL, returning the results for display and/or further use.
The IR search request is input using the dtSearch “query language,” which is partially, but not entirely, composed through the user’s actions. Online help assists query formulation. In addition, a “word list” (see Figure 1▶) allows the user to retrieve a list of terms in the index that are in the lexically ordered vicinity of a term of interest, with the number of occurrences of each term in the document collection. This list is useful to detect alternative misspellings of a term in the collection.
- In brief, the algorithm analyzes the search request and partitions it into its relational and text search components. The two search operations are coupled as follows. A count of the number of rows matching the relational criterion is first determined by using SQL’s COUNT() function.
- If this count is less than 500, the relational search is executed, the matching rows are retrieved, and the resulting document IDs are fed into the IR search to limit the latter and produce a final results table (scenario 1).
- If the count is greater than 10,000, the IR search is executed first, a temporary results table is created, and the original relational query is rewritten by incorporating this temporary table. The rewritten query is then executed to yield the final results table (scenario 2).
- If the number of rows is intermediate, then both relational and IR queries are executed independently, and the resulting temporary tables are merged in a second step to yield the final results table.
For scenario 1, we use a dtSearch feature called File Conditions, which allows a textual search to be limited based on properties of the documents that are being searched, such as document name or creation date. During indexing time, we specify the primary key of the Reports table (the Report ID) as the document name and the date of the report as its creation date. Thus, one can limit the text search by date range of the reports. Similarly, if one is searching for keywords in documents applicable only to a single patient, one can retrieve the relatively few Report IDs for this patient and pass them in the File Condition along with the search command proper. File conditions work well when the number of documents matching the specified file properties is reasonably small (i.e., less than 500). Tests showed that, beyond this threshold, File Conditions do not help and may even affect the search adversely.
-
Scenario 2 is used to preclude situations in which the relational criterion by itself is so broad (e.g., all radiology notes) as to return a significant fraction of the database’s contents if executed directly. Executing a rewritten query limits the final number of documents in a more efficient manner.
The File Conditions feature was used in the penultimate benchmark of Table 4▶, described in the previous section. The search time decreased from 20 seconds with MS-SQL alone to 11 seconds with dtSearch and MS-SQL working cooperatively.
By default, search results are organized by patient and presented in descending rank by the total number of documents per patient. Selecting a patient displays a list of associated documents, sorted by time to give a sense of the tempo of an illness or condition. Alternatively, the user can rank search results by report type weighting. Initially, all document types, such as pathology report or surgical operative note, are given a weight of 1. Giving a report type a higher weight results in higher ranking for patients for whom the desired report type exists. This is valuable, for example, if the user had performed a search for “pancreatic adenocarcinoma” and was particularly interested in cases for which a surgical note was available.
Figure 2.
Search and ranking algorithm used by the application. The search request is separated into its relational and IR components, either of which, but not both, may be omitted. If a relational criterion exists, the count of records that match it are determined, and the result is used to determine whether the list of documents that match the criteria can be supplied to the IR engine to restrict the degree of search. If the number of documents matching the relational criterion is less than 500, one can save time by feeding the list of document IDs to the IR engine.
Figure 3.
The user interface of our application. The user can search for one or more keywords or interest. Various search options, such as weighting of terms and regular expressions, are available under the “Text Search Options” tab. The search results can be limited to one or more report types or to within a range of report dates. One can also search for reports of a single patient or for multiple patients who have been previously identified and recorded in a temporary table, which is identified to the application in the lower-left part of the screen. The “Search Status” displays search progress—the most recent message is at the top—and a ranked list of matching patients is shown on the top right. Patient names have been removed by editing of this screen shot. Selecting a particular patient displays a list of reports available for the patient, and clicking on a particular report illustrates its contents. The terms of interest are highlighted in the document where they occur. Here the term “craniopharyngioma” is off-screen.
Present Status
The application that we have described has been developed primarily for limited deployment within the VAMC, chiefly within the Information Technology Group and the Department of Radiology. In general, applications built using Microsoft Access clients are not suitable for large-scale deployment in an institution. The logistics of updating the client software on dozens of machines each time enhancements are made to the program are formidable. “Three-tier” solutions that provide access through a Web browser are more appropriate to such scenarios, but these require a significant degree of software engineering.
We regard the present application as only a first step in providing text-mining facilities for the VAMC CT collection.
Limitations of the Present Work and Future Work
Several limitations of this present study are worth noting. First, we have evaluated only the three most popular high-end RDBMSs in terms of number of licenses, Oracle, MS-SQL and DB2. We have not evaluated Informix, which is now an IBM acquisition whose technology is expected to be merged with DB2 in the long term, or Sybase. We were interested primarily in determining the feasibility of using commodity software components to provide the framework for an IR+RDBMS system and what issues need to be addressed in this undertaking. For developers using RDBMS packages other than the ones we have evaluated, however, the features listed in Table 4▶ can serve as the basis of an evaluation worksheet.
A well-known problem with indexing and searching by words alone is that, because medicine has multiple synonyms, the user must specify alternative forms of a keyword to avoid missing potentially relevant documents. Although thesaurus-based synonym expansion can address this issue partially for single-word synonyms, it is vulnerable to word order when multi-word concepts are involved. A more robust approach for addressing this issue is to preprocess the documents and match phrases in the text to concepts in a controlled medical vocabulary. This technique, concept indexing, is potentially powerful, but not foolproof, because of the presence of polysemy, as pointed out in an editorial by Masys.24 Polysemy refers to the existence of terms with multiple meanings that cannot be readily disambiguated by the concept-matching algorithm (e.g., “anesthesia” can refer to a procedure ancillary to surgery or to the clinical finding of loss of sensation). Concept matching is also complicated by the presence of non-standard abbreviations, grammatical errors and neologisms. For these reasons, concept indexing cannot replace word indexing but can only be ancillary to it.
Another problem with both word and concept indexing is that of negation. Negated concepts come principally from two sources: pertinent negatives recorded during medical history taking and the formulation of a differential diagnosis that lists conditions that are to be ruled out. The mere presence of a term or concept in a document does not necessarily make the document relevant for that keyword. The ranking algorithm used with our search application partially addresses this issue. If a patient has only one document matching a keyword of interest, we found that it frequently was either a pertinent negative or part of a differential diagnosis. In contrast, the patient for whom multiple matching documents existed, particularly if the documents occurred over a span of time, nearly always represented a relevant case. This is admittedly a half-measure, and more robust recognition and indexing of concept negation need to be incorporated. We envision future work in at least two areas.
-
Our group has previously studied the problems of both concept indexing25 as well as concept indexing with detection of negation26 using the National Library of Medicine’s Unified Medical Language System.27 We have implemented negation detection through a natural-language processing technique using a parser that recognizes a large number of negation and affirmation word forms and concepts in the vicinity of these forms.
We hope eventually to implement full concept indexing with negation on the VAMC CT collection. One practical problem is that the work cited above has focused on feasibility rather than efficiency. Negation indexing of 3 million notes would take about 1 year of continuous CPU time on a 600 MHz PC using our existing concept/negation-matching algorithm, which takes about 1 minute on an 800-word note. Much of this time represents network bandwidth latency, because we are indexing on a client CPU that makes requests to a separate server CPU on which the UMLS database resides. In any case, we need to consider seriously ways of optimizing both the algorithm and the hardware infrastructure that supports it.
The ability to add thesaurus-driven query term expansion is potentially attractive. dtSearch supports external thesauri via several possible mechanisms, and subsets of the UMLS can be loaded and accessed for use. Although the term expansion algorithm is vulnerable to word order, as stated earlier, this approach still has value for single-word terms and synonyms, as well as standard acronyms and abbreviations. The use of the thesaurus is, of course, subject to the caveat of polysemy, which is seen with both abbreviated and nonabbreviated terms.
Conclusions
IR and RDBMS System Integration: Impact on Query Execution Speed
In our benchmark results, dtSearch and MS-SQL typically perform better in conjunction than MS-SQL running by itself. This seems even more surprising considering that in the former scenario, dtSearch and MS-SQL communicate through code written by us in an interpreted language (MS-Access’s Visual Basic for Applications), in the latter scenario, a single compound SQL statement containing Boolean criteria and a CONTAINS() function is passed to MS-SQL, with subsequent execution of the statement running uninterrupted. These results might appear counter-intuitive, until one considers the several reasons for suboptimal performance of the IR add-ons of RDBMSs.
Although RDBMS query optimizers have undergone considerable evolution, the RDBMS, in the case of MS-SQL and DB2, merely functions as a passive broker for IR requests that are passed off to an architecturally distinct search engine, which returns a set of matching records to the RDBMS that are filtered afterward. Currently, this is no better, from the standpoint of query optimization, than the “manual” approach we have taken using a third-party IR component.
Installation or purchase of the add-on is typically optional; therefore, the RDBMS optimizer’s designers sometimes spend little effort in accommodating an add-on that may not be installed.
In cases in which closer coupling between the IR and RDBMS engines has been attempted, as in Oracle 9i, these efforts are relatively preliminary. Thus, when the relational part of a mixed relational+ IR query is determined to return few rows, Oracle 9i treats CONTAINS as a function call, invoking the IR engine repeatedly for each row. In reality, it should not need to call the IR engine more than once.
The choice of IR data structure may not be the most appropriate. Although inverted files,2 which are used by both Oracle and MS-SQL, are generally used to implement IR indexes, they allow rapid search only by term/keyword. They cannot efficiently answer queries that wish to limit search to a particular subset of documents, however small that subset may be. (dtSearch goes beyond traditional inverted files by storing extra information that permits the use of File Conditions.) De Vries28 argues that inverted files are a “black box” to RDBMS optimizers and advocates structures other than inverted files for systems that must perform relational + IR search. For example, one can store IR indexes as relational tables, which are then indexed using B-trees.29 Such an approach has been described by Hersh et al.30 A cost-based RDBMS optimizer can then generate statistics that use both the term and document ID frequency distributions to make reasonable decisions.
Inverted-file implementations in RDBMSs are possibly suboptimal. Dynamic Information Systems Corporation made a large sale of Omnidex, a “pure” IR engine that also uses inverted files, to Lawrence Livermore Labs after reducing the run-time of queries against a large Oracle database of scientists’ notes from up to 13 minutes with a previous version of Oracle’s IR engine to less than 4 seconds with Omnidex.31 For this application, multiple columns in a table needed to be searched. Oracle’s implementation of CONTAINS, as stated earlier, does not allow search across all columns; multiple calls must be issued, with resulting inefficiency.
We anticipate considerable improvement in this situation in the future. The gap between “pure” IR vendors and RDBMS vendors has shrunk rapidly in the past two years, in part through IR software acquisitions by the latter. The “pure” IR vendors will have to innovate significantly to remain financially viable as independent entities. In fact, if we had been working, for example, with MEDLINE documents instead of clinical text, the differences between the packages and the reasons for using a “pure” IR package would have been much less clear-cut.
The Malformed Text Problem with Dictated Medical Text
The VAMC’s CPRS (DHCP), although eminently capable, is a legacy system that lacks contemporary industry-standard features such as a word processor with integrated spell checking of medical terms, and a significant proportion of notes in the VAMC CT collection—notably, progress notes that are intended for internal use only—reflect its use as a “computer instead of paper” receptacle. Such notes are little more than medical shorthand—intelligible to the author but difficult to compute with. The problem is compounded by obvious typing errors. The list of problems includes errors in spelling, punctuation, spacing, capitalization, and use of abbreviations and acronyms, some of which are clearly idiosyncratic to the author and of questionable interpretability to others. A particularly “pathological” note illustrating all of these problems is reproduced in Figure 4▶. It is not certain that modern software alone will remedy the situation: it is also necessary for policies regarding the quality of notes’ content to be formulated and enforced.
Figure 4.
A follow-up note from the VAMC CT collection, reproduced verbatim. Although not typical of the collection, this note illustrates the practical difficulties in applying IR to medical text: The note has numerous abbreviations that are idiosyncratic to the author, and indifferent transcription has introduced errors in spelling, punctuation, spacing, and capitalization.
There is no way to assess the magnitude of search failure (i.e., false negatives) because of malformed text that the search engines simply miss. Based on our limited testing, the spelling error rate in our collection appears to be around 4%. On searches with multiple terms and large potential result sets, this would result in a nontrivial degradation of performance in terms of recall. If, however, we were dealing with a collection of MedLine documents, in which spelling errors and malformed punctuation are rare, the distinction between the search engines would have been much less.
Currently, the only way to deal with malformed medical text appears to be through an interactive search-refinement process; an entirely automated approach that eliminates human intervention is not conceivable. Although one cannot guarantee 100% recall in the presence of malformed or misspelled text, software that provides robust handling of punctuation variants, fuzzy search, regular expression capability, and inspection of a window within the term frequency index can help significantly. Of note, the only evaluated software that supports the last two features, as well as the most intuitive implementation of fuzzy search, is dtSearch, a non-RDBMS package.
Implementation Issues with IR and RDBMS Integration
Existing commercial clinical information systems, many of which are built on top of RDBMSs, have not yet taken full advantage of IR functionality. Although many of them provide limited text processing for cross-patient queries, their features fall far short of what full-fledged IR can offer. In many legacy systems, such capability has been based on string-processing capabilities of the host programming language; for example, DEC-RAD 32 (now IDXrad) relies on the features within the M language.
In general, existing clinical data repositories (CDRs) support collection of all clinical data, including text, in a single database but leave the task of devising means of analyzing textual data up to the repository’s computing support team. The variable extent of IR support by individual RDBMS vendors complicates the task of analyzing/mining textual CDR data. It is important to know the limitations of individual RDBMSs for the task at hand, but the choice of RDBMS is often preordained by what the repository uses. Switching RDBMS vendors is a delicate and expensive operation that is also fraught with political issues, and integration with a relatively low-cost “pure” IR tool may be more attractive for researchers performing text mining on a limited budget.
Although it was not particularly difficult for us to build the system described above, most RDBMS users do not wish to spend time in software-integration programming chores. Such programming, as stated earlier, is partly necessary simply to make the rich and complex IR features of a package more readily accessible to nonprogramming users. There is no reason why RDBMS vendors should not provide civilized user interfaces for integrated relational and textual query out of the box. Currently, however, the vendors have just barely managed to get IR functionality in place, and even this functionality is sometimes incomplete. The vendors’ present decision to leave the responsibility of interface-building to third-party software developers leaves non-SQL experts in a bind. Commercial generic end-user query tools are still too rooted in the relational world and do not yet take advantage of RDBMS IR functionality.
The latter situation exists partly because ISO’s work on the SQL-99 standard has moved, as it typically does, at a very leisurely pace, so that it is still in a draft stage. Meanwhile, the commercial pressure on RDBMS vendors to introduce IR functionality has caused them to devise their own divergent approaches. The individual IR implementations differ much more than the “relational” SQL implementations. Although the respective feature sets differ significantly, the syntactic differences are far greater, with little commonality beyond the word “CONTAINS.” As such, it is much harder to devise a lingua franca form of IR-functional SQL analogous to the Open Database Connectivity (ODBC) standard. (This standard, originally developed by Microsoft but voluntarily relinquished by them to standards bodies such as ANSI and ISO, was aimed at shielding developers from the syntactic idiosyncrasies of particular SQL dialects for standard relational operations, thereby facilitating software portability greatly.) Conse-quently, generic-query-tool vendors have shied away from supporting IR functionality. In addition, IR/RDBMS applications with a large body of code are subject to vendor lock-in, because all of the IR-specific code would need to be rewritten if the RDBMS vendor was changed.
It may be expecting too much of RDBMS vendors to give up the advantages of lock-in voluntarily. RDBMS users will benefit greatly, however, if the IR component of RDBMS software is made both more robust to deal with malformed text and more accessible to nonprogrammers. Doing this needs to be a high priority in future releases of RDBMS software.
Availability of Software.
The software described in this article will be distributed freely on making a request to Dr. Nadkarni. While the database storage component can use any RDBMS that can store text, the dtSearch engine must be licensed from dtSearch, Inc.
Acknowledgments
The authors thank IBM and the IBM Scholar’s Program for providing the senior author with a free copy of DB2. The work reported in this article was supported by NIH Grants U01 ES10867, R01 LM06843-01, and U01 CA78266.
Bibliography
- 1.Codd E. A Relational Model for Large Shared Data Banks. Commun ACM 1970; 13(6):377–387. [PubMed] [Google Scholar]
- 2.Baeza-Yates R, Ribeiro-Neto B. Modern Information Retrieval. Harlow, UK, Addison-Wesley Longman, 1999.
- 3.Sparck Jones K. A statistical interpretation of term specificity and its application in retrieval. J Document 1972; 28(1):11–21. [Google Scholar]
- 4.Salton G, Wu H, Yu CT. Measurement of Term Importance in Automatic Indexing. J Am Soc Inform Sci 1981; 32(3):175–186. [Google Scholar]
- 5.Yount R, Vries J, Councill C. The Medical ARchival System: an Information Retrieval System Based on Distributed Parallel Processing. Inform Process Manage 1991; 27(4):1–11. [Google Scholar]
- 6.Giuse D, Mickish A. Increasing the availability of the computerized patient record. In AMIA Annual Fall Symposium, 1996. Philadelphia, Hanley & Belfus, 1996, pp 633–637. [PMC free article] [PubMed]
- 7.Vasanthakumar SR, Callan JP, Croft WB. Integrating INQUERY with an RDBMS to support text retrieval. Bulletin of the IEEE Technical Committee on Data Engineering. 1996; 19(1):24–33. [Google Scholar]
- 8.Callan J, Croft W, Harding S. The INQUERY Retrieval System. In Proceedings of the International Conference on Database and Expert Systems Applications, 1992. 1992, pp 347–356.
- 9.Melton J, Simon AR, Gray J. SQL 1999: Understanding Relational Language Components. San Mateo, CA, Morgan Kaufman, 2001.
- 10.International Standards Organization. SQL Multimedia and Application Packages. Part 2: Full-Text. Geneva, 2000. Report Number ISO/IEC JTC 1/SC 32/WG 4.
- 11.Department of Veterans Affairs. Decentralized Hospital Computer System Version 2.1: Programmer’s Manual. San Francisco, Information Systems Center, 1994.
- 12.Frisse ME. Searching for information in a hypertext medical handbook. Commun ACM 1988; 31(7):880–886. [Google Scholar]
- 13.Wu S, Manber U. Fast text searching allowing errors. Commun ACM 1992; 35(10):83–91. [Google Scholar]
- 14.World Wide Web Consortium. Extensible Markup Language. 2002. http://www.w3.org/XML/Schema. Accessed on February 20, 2002.
- 15.World Wide Web Consortium. XML Schema. 2001. http://www.w3.org/XML/Schema. Accessed on February 20, 2002.
- 16.World Wide Web Consortium. Guide to the W3C XML Specification DTD, Version 2.1. 1998. http://www.w3.org/XML/1998/06/xmlspec-report-v21.htm. Accessed on October 12, 2001.
- 17.Means WS, Harold ER. XML in a Nutshell: A Desktop Quick Reference, 2nd ed. Sebastopol, CA, O’Reilly & Associates, 2002.
- 18.Health Level Seven. HL7 PRA Level 2 Proposal. 2002. http://www.hl7.org/Special/dotf/docs/Proposal_to_HL7_on_Mayo_DTD_final.doc. Accessed on June 28, 2002.
- 19.Staken K. Introduction to native XML databases. 2002. http://www.xml.com/pub/a/2001/10/31/nativexmldb.html. Accessed on June 27, 2002.
- 20.Friedl JEF. Mastering Regular Expressions. Sebastopol, CA, O’Reilly & Associates, 1997.
- 21.Levine JR, Mason T, Brown D. lex & yacc. Sebastopol, CA, O’Reilly & Associates, 1992.
- 22.Miller GA, Fellbaum C, Tengi R, Wakefield P. Wordnet: A lexical database for the English Language. 2002. http://www.cogsci.princeton.edu/~wn/.
- 23.Bain T. SQL Server Full Text Search Optimizaiton. 2001. http://www.sql-server-performance.com/tb_search_optimization.asp. Accessed on February 21, 2002.
- 24.Masys D. Linking microarray data to the literature [editorial]. Nature Genetics 2001; 27(6):9–10. [DOI] [PubMed] [Google Scholar]
- 25.Nadkarni PM, Chen RS, Brandt CA. UMLS Concept Indexing for Production Databases: A Feasibility Study. J Am Med Inform Assoc 2001; 8(1):80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mutalik P, Deshpande A, Nadkarni P. Use of general-purpose negation detection to augment concept Indexing of medical documents: a quantitative study using the UMLS. J Am Med Inform Assoc 2001; 8(6):598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindberg DAB, Humphreys BL, McCray AT. The Unified Medical Language System. Meth Inform Med 1993; 32:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vries A. Challenging Ubiquitous Inverted Files. In First DELOS Workshop on Information Seeking, Searching and Querying in Digital Libraries. Zurich, 2000.
- 29.Garcia-Molina H, Ullman J, Widom J. Database Systems Implementation. Upper Saddle River, NJ, Prentice Hall, 2000.
- 30.Hersh W, Hickam D, Leone T. Words, concepts, or both: optimal indexing units for automated information retrieval. In Proceedings of the Annual Symposium on Computer Applications in Medical Care, 1992, 1992, pp 644–648. [PMC free article] [PubMed]
- 31.Dynamic Information Systems Corporation. LLNL Chooses OMNIDEX to Speed Oracle Full Text Searches. 2002. http://www.disc.com/prllnl.html. Accessed on February 20, 2002.
- 32.Niedner C. Use of SQL with an entity-attribute-value database. MUG Q 1991; 21(3):40–45. [Google Scholar]