Abstract
In Saccharomyces cerevisiae, the 60S ribosomal subunit assembles in the nucleolus and then is exported to the cytoplasm, where it joins the 40S subunit for translation. Export of the 60S subunit from the nucleus is known to be an energy-dependent and factor-mediated process, but very little is known about the specifics of its transport. To begin to address this problem, an assay was developed to follow the localization of the 60S ribosomal subunit in S. cerevisiae. Ribosomal protein L11b (Rpl11b), one of the ∼45 ribosomal proteins of the 60S subunit, was tagged at its carboxyl terminus with the green fluorescent protein (GFP) to enable visualization of the 60S subunit in living cells. A panel of mutant yeast strains was screened for their accumulation of Rpl11b–GFP in the nucleus as an indicator of their involvement in ribosome synthesis and/or transport. This panel included conditional alleles of several rRNA-processing factors, nucleoporins, general transport factors, and karyopherins. As predicted, conditional alleles of rRNA-processing factors that affect 60S ribosomal subunit assembly accumulated Rpl11b–GFP in the nucleus. In addition, several of the nucleoporin mutants as well as a few of the karyopherin and transport factor mutants also mislocalized Rpl11b–GFP. In particular, deletion of the previously uncharacterized karyopherin KAP120 caused accumulation of Rpl11b–GFP in the nucleus, whereas ribosomal protein import was not impaired. Together, these data further define the requirements for ribosomal subunit export and suggest a biological function for KAP120.
INTRODUCTION
Although eukaryotic ribosomes function in the cytoplasm, the synthesis, processing, and assembly of the ribosomal subunits in Saccharomyces cerevisiae and higher eukaryotes occur in the nucleolus. The entire ribosome is composed of four rRNA species and ∼75 ribosomal proteins (r-proteins) distributed between two subunits. The 18S, 5.8S, and 25S rRNAs are derived from a single 35S rRNA precursor that is synthesized by RNA polymerase I and then processed by a series of endonucleolytic and exonucleolytic cleavages (reviewed by Kressler et al., 1999; Venema and Tollervey, 1999). The 5S rRNA is synthesized separately by RNA polymerase III and associates with the 60S preribosomal subunit early in assembly. The mature 40S ribosomal subunit contains the 18S rRNA and ∼32 r-proteins, whereas the 60S subunit is composed of the 5S, 5.8S, and 25S rRNAs and ∼45 r-proteins. Proper assembly of each ribosomal subunit requires the coordination of several events, including the synthesis and import of r-proteins, the synthesis and processing of rRNA, and the concomitant assembly of r-proteins into the preribosomal subunits. Although a pathway for 35S rRNA maturation has been well defined through both genetic and biochemical approaches (reviewed by Kressler et al., 1999; Venema and Tollervey, 1999), less is known about the association of r-proteins with the rRNA and the export of the assembled subunits out of the nucleus.
All nucleocytoplasmic transport occurs through the nuclear pore complex (NPC). The yeast NPC is composed of multiple copies of ∼30 different nuclear pore proteins referred to as nucleoporins (Rout et al., 2000). Together, these nucleoporins form the overall structure of the NPC, which consists of a membrane-embedded central core with fibrils protruding from its cytoplasmic face and a basket-like structure that extends out on the nuclear side (Yang et al., 1998; Stoffler et al., 1999; Allen et al., 2000). The active nuclear pore can accommodate transport of large macromolecules, including export of the ribosomal subunits.
Transport of cargo into and out of the nucleus requires not only interactions with the NPC but also soluble transport factors. The best defined transport path is the import of proteins containing a classic nuclear localization signal (NLS) by the importin α/β receptor (reviewed by Corbett and Silver, 1997; Gorlich and Kutay, 1999; Wente, 2000). Importin α/Srp1 recognizes and binds to proteins containing a NLS and together with importin β/Rsl1 travels through the NPC. Thirteen importin β homologues termed karyopherins were identified in S. cerevisiae by sequence comparison to importin β (Gorlich et al., 1997). These karyopherins act as receptors for specific import and export cargoes. Transport substrates for several of the karyopherins have already been identified (reviewed by Gorlich and Kutay, 1999). The nucleotide-bound state of the small GTPase Ran/Gsp1 imparts directionality to transport (Gorlich et al., 1996; Izaurralde et al., 1997). Dissociation of the karyopherin–cargo complex in the nucleus is mediated by RanGTP association (Rexach and Blobel, 1995; Chi et al., 1996; Gorlich et al., 1996), whereas GTP hydrolysis in the cytoplasm results in the release of export cargo (Bischoff and Gorlich, 1997; Floer et al., 1997; Lounsbury and Macara, 1997). In yeast, the high concentration of RanGTP in the nucleus is maintained by the guanine nucleotide exchange factor Prp20 (Amberg et al., 1993; Kadowaki et al., 1993), and the cytoplasmic pool of RanGDP is generated by the GTPase-activating protein Rna1 (Becker et al., 1995). The mechanism by which the transport complexes travel through the NPC is not as clear.
Microinjection experiments in Xenopus oocytes demonstrated that ribosomal subunit export is an energy-dependent, factor-mediated, and unidirectional process that requires components of the NPC (Bataille et al., 1990). In addition, export of microinjected 40S ribosomal subunits does not compete with export of tRNA out of the nucleus, indicating separate pathways for export (Pokrywka and Goldfarb, 1995). A role for the GTPase Ran in ribosome assembly or export was proposed based on the observation that rRNA processing is delayed in yeast strains bearing conditional alleles of either of the Ran regulators PRP20 or RNA1 (Traglia et al., 1989; Kadowaki et al., 1993). These early observations were recently confirmed for both the 40S and 60S ribosomal subunits with the use of two independent assays (Hurt et al., 1999; Moy and Silver, 1999). Mutation of the Ran regulators Rna1, Prp20, and Yrb1 caused the 40S ribosomal subunit to accumulate in the nucleus of yeast, as determined by in situ hybridization to 20S rRNA (Moy and Silver, 1999). In addition to the Ran regulators, a subset of nucleoporin mutations and a temperature-sensitive mutation of the nuclear export sequence receptor Xpo1/Crm1 also blocked export of the 40S ribosomal subunit (Moy and Silver, 1999). The 60S ribosomal subunit also accumulated in the nucleus of yeast strains bearing conditional alleles of RNA1 and PRP20, as determined by the localization of a fusion between the ribosomal protein L25 (Rpl25) and the green fluorescent protein (GFP) (Hurt et al., 1999). In addition, temperature-sensitive mutations of the nucleoporins Nsp1, Nup49, and Nic96 also caused mislocalization of the 60S ribosomal subunit (Hurt et al., 1999). It is likely that the factors identified to date represent only a subset of the transport factors required for ribosome export.
Here we describe an assay to follow the localization of the 60S ribosomal subunit in S. cerevisiae with the use of a fusion between the ribosomal protein L11b (Rpl11b) and GFP. This assay differs from the Rpl25–GFP export assay described previously (Hurt et al., 1999) in that nuclear accumulation of Rpl11b–GFP can be detected under conditions of logarithmic growth. A large panel of mutant yeast strains were screened for defects in 60S ribosomal subunit assembly and transport with the use of this assay. Surprisingly, deletion of KAP120, one of the nonessential karyopherins, caused a strong accumulation of the 60S ribosomal subunit in the nucleus. Further characterization revealed that kap120Δ cells have a slight delay in rRNA processing and a significant reduction in free 60S ribosomal subunits.
MATERIALS AND METHODS
Yeast Strains
Table 1 lists the yeast strains used in this study. The strains PSY2083 and PSY2084 were constructed by replacing the ORF of RPL11A and RPL11B with HIS3 by means of a PCR-based method (Baudin et al., 1993). The haploid strain PSY685 was transformed with a HIS3 PCR product targeted to the RPL11A locus to generate PSY2083, and PSY581 was transformed with a HIS3 PCR product targeted to the RPL11B locus to create PSY2084. Disruptions of RPL11A and RPL11B were verified by Southern blot analysis (Southern, 1975).
Table 1.
Yeast Strains
| Strain | Genotype | Reference |
|---|---|---|
| PSY580 | MATa ura3-52 leu2Δ1 trp1Δ63 | Winston et al., 1995 |
| PSY581 | MATα ura3-52 leu2Δ1 his3Δ200 | F. Winston (Harvard Medical School) |
| PSY685 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | A.H. Corbett and P.A. Silver, unpublished results |
| PSY2083 | MATa rpl11a∷HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | This study |
| PSY2084 | MATα rpl11b∷HIS3 ura3-52 leu2Δ1 his3Δ200 | This study |
| PSY2085 | MATa rpl11a∷HIS3 rpl11b∷HIS3 ura3-52 leu2Δ1 his3Δ200 + YCp50L11A URA3 | This study |
| PSY2086 | MATα RPL11A RPL11B ura3-52 leu2Δ1 his3Δ200 + YCp50L11A URA3 | This study |
| PSY2087 | MATa RPL11A RPL11B ura3-52 leu2Δ1 trp1Δ63 his3Δ200 + YCp50L11A URA3 | This study |
| PSY2088 | MATα rpl11a∷HIS3 rpl11b∷HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 + YCpL11A URA3 | This study |
| PSY713 | MATα prp20-1 ura3-52 leu2Δ1 trp1Δ63 | Koepp et al., 1996 |
| PSY714 | MATa rna1-1 ura3-52 leu2Δ1 trp1 | Corbett et al., 1995 |
| PSY1038 | MATα Δyrb1∷HIS3 trp1 his3Δ200 lys ade2 ade3 + yrb1-1 LEU2 CEN | Schlenstedt et al., 1995 |
| PSY1105 | MATα xpo1∷LEU2 ura3-1 trp1-1 his3 ade2-1 can1-1 + PKW457 xpo1-1 HIS3 | Stade et al., 1997 |
| PSY1654 | MATα nup82∷HIS3 ura3-52 leu2-3,112 trp1-1 his3Δ200 lys2-801 + nup82Δ108 URA3 | Hurwitz and Blobel, 1995 |
| PSY1634 | MATa nup116-5∷HIS3 ura3 leu2 trp1 his3 | Wente and Blobel, 1993; M. Damelin and P.A. Silver, unpublished |
| PSY413 | MATα Δnup49∷TRP1 ura3 leu2 trp1 his3 ade2 ade3 + pUN100-nup49-313 LEU2 | Doye et al., 1994 |
| PSY466 | MATα nup1-2∷LEU2 ura3-52 leu2-3,112 trp1-1 his3Δ200 + nup1-8 TRP1 | Loeb et al., 1993 |
| PSY851 | MATα nup2-4∷URA3∷HIS3 ura3-52 leu-2,3,112 trp1Δ63 his3Δ200 ade 2 | Loeb et al., 1993 |
| PSY1658 | MATa nup84∷HIS3 ura3 leu2 trp1 his3Δ200 ade2 | Siniossoglou et al., 1996 |
| PSY889 | MATa Δnup100-3∷TRP1 ura3 leu2 his3 ade2 | Wente et al., 1992 |
| PSY205 | MATa rat2-2 ura3-52 leu2Δ1 trp1Δ63 | Heath et al. 1995 |
| PSY888 | MATa rat3-1 ura3-52 leu2Δ1 trp1Δ63 | Li et al., 1995 |
| PSY1646 | MATα nup170-1∷HIS3 ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 can1-100 | Aitchison et al., 1995 |
| PSY1659 | MATa nic96∷HIS3 ura3 leu2 trp1 his3 ade2 + pUN100-nic96-1 LEU2 | Zabel et al., 1996 |
| PSY1077 | MATα gle2-1 ura3-1 leu2-3,112, trp1-1 his3-11 ade2-1 | Murphy et al., 1996 |
| PSY1169 | MATα rip1∷HIS3 ura3-52 leu2Δ1 his3Δ200 | Saavedra et al., 1997 |
| PSY1645 | MATa nup157-2∷URA3 ura3-52 leu2-3,112 trp1-1 his3Δ200 lys2-801 | Aitchison et al., 1995b |
| PSY635 | MATα rat7-1 ura3-52 leu2Δ1 his3Δ200 | Gorsch et al., 1995 |
| PSY1661 | MATa seh1∷HIS3 ura3 leu2 trp1 his3 ade2 | Siniossoglou et al., 1996 |
| PSY1040 | MATα cse1-1 ura3-52 trp1Δ901 his3-11,15 ade2-101 | Xiao et al., 1993 |
| PSY1643 | MATα gle1-4 ura3-1 leu2-3,112 his3-11,15 ade2-1 | Murphy and Wente, 1996 |
| PSY1133 | MATa mtr10-1 ura3-52 lys2-301 | Kadowaki et al., 1994 |
| PSY1201 | MATa pse1-1 ura3-52 leu2Δ1 trp1Δ63 | Seedorf and Silver, 1997 |
| PSY688 | MATα srp1-31 ura3 leu2 trp1 his3 ade2 | Loeb et al., 1995 |
| PSY1103 | MATa rsl1-4 ura3-52 leu2Δ1 trp1Δ63 | Koepp, 1997 |
| PSY1234 | MATa mtr2-1 ura3-52 his3Δ200 | Kadowaki et al., 1994 |
| PSY1024 | MATa npl3-17 ura3-52 leu2-3 trp1-1 his3 lys1-1 ade2-1 ade8 can1-100 | Lee et al., 1996 |
| PSY1034 | MATa npl3-27 ura3-52 leu2-3 trp1-1 his3 lys1-1 ade2-1 ade8 can1-100 | Lee et al., 1996 |
| PSY825 | MATa npl4-1 ura3-52 leu2 | DeHoratius and Silver, 1996 |
| PSY636 | MATα rat8-1 ura3-52 leu2Δ1 trp1Δ63 | Snay-Hodge et al., 1998 |
| PSY1233 | MATa rat8-2 ura3-52 leu2Δ1 trp1Δ63 | Snay-Hodge et al., 1998 |
| PSY1664 | MATa kap104∷ura3∷HIS3 leu2 lys trp1 + pRS314-kap104-16 TRP1 | Aitchison et al., 1996 |
| PSY967 | MATa kap123∷HIS3 ura3-52 leu2Δ1 his3Δ200 | Seedorf and Silver, 1997 |
| PSY1082 | MATa kap120∷HIS3 ura3-52 leu2Δ1 his3Δ200 | M. Damelin and P.A. Silver, unpublished results |
| PSY1083 | MATα KAP120 ura3-52 leu2Δ1 his3Δ200 lys2 | M. Damelin and P.A. Silver, unpublished results |
| PSY1138 | MATa msn5∷HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | J.A. Kahana and P.A. Silver, unpublished results |
| PSY1200 | MATa sxm1∷HIS3 ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | Seedorf and Silver, 1997 |
| PSY412 | MATa nsp1-10Ats∷URA3 ura3 leu2 lys1 his3 ade2 ade8 | Nehrbass et al., 1990 |
| PSY1799 | MATa mex67∷HIS3 ura3 leu2 trp1 his3 ade2 + pHT4667-mex67-5 URA3 | Santos-Rosa et al., 1998 |
| PSY2101 | MATα los1∷HIS3 ura3 leu2 trp3 his3 ade2 | Simos et al., 1996 |
| PSY1784 | MATa kap114∷HIS3 ura3-52 leu2Δ1 trp1 his3Δ200 | Morehouse et al., 1999 |
| PSY1198 | MATa nmd5∷HIS3 ura3Δleu2Δ1 his3Δ200 ade2 ade8 | Ferrigno et al., 1998 |
| Strain | Genotype | Reference |
| PSY1171 | MATa fal1∷HIS3MX6 ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 + pRS316-fal1-1 LEU2 | Kressler et al., 1997 |
| PSY410 | MATa nop1-3∷URA3 ura3 leu2 lys1 ade2 ade8 | Tollervey et al., 1993 |
| PSY374 | MATa nsr1∷URA3 ura3 leu2 trp1 his3 ade8 can1 | Kondo and Inouye, 1992 |
| PSY1252 | MATa npl2-1 cyt1∷HIS3 ura3-1 leu2-3,112 trp1-1 his3- 11,15 ade2-1 can1-100 | S.R. Valentini and P.A. Silver, unpublished results |
| PSY1243 | MATα npl2-2 cyt1∷HIS3 ura3-1 leu2-3,112 trp1-1 his3- 11,15 ade2-1 can1-100 | S.R. Valentini and P.A. Silver, unpublished results |
| PSY1131 | MATα mtr4-1 ura3-52 lys2-801 pep4∷HIS3 prb1-Δ1.6R | Liang et al., 1996 |
| PSY204 | MATα rat1-1 ura3-52 leu2Δ1 his3Δ200 | Amberg et al., 1992 |
| PSY637 | MATα rat9-1 ura3-52 leu2Δ1 trp1Δ63 | Goldstein et al., 1996 |
| PSY1652 | MATα dob1-1 ura3-1 leu2-3,112 trp1-1 his3-11,15 ade2-1 | de la Cruz et al., 1998 |
| PSY216 | MATα rat6-1 ura3-52 leu2Δ1 his3Δ200 | Amberg et al., 1992 |
Yeast strains lacking wild-type copies of both RPL11A and RPL11B were generated by crossing PSY2083 carrying a wild-type copy of RPL11A on a URA3 CEN plasmid to PSY2084. The diploid was sporulated, and tetrads were analyzed. HIS+ and 5-FOA–sensitive (FOAS) spores in tetrads that segregated 2:2 HIS+:his− and 2:2 FOAS:FOAR (5-FOA–resistant) were identified as the double mutant strain rpl11a::HIS3 rpl11b::HIS3.
Plasmids
The LEU2 CEN (pPS2167) and TRP1 CEN (pPS2168) RPL11B–GFP plasmids were constructed as follows. Yeast genomic DNA was amplified by PCR with a 5′ primer that annealed 891 base pairs 5′ of the ORF for RPL11B (5′GCATCTACTAGTGCAGGATTACGAAGACTTC3′) and a 3′ primer that annealed to the 3′ end of RPL11B excluding the stop codon (5′CAGTCACTCGAGCTTTATCGAGCACATCAGCG3′). The PCR product was digested with SpeI and XhoI and cloned into the SpeI–XhoI site of either a LEU2 or a TRP1 CEN vector that contained the ORF of GFP followed by the 3′ untranslated region of NUF2 (Kahana and Silver, 1998). To generate the URA3 CEN RPL11B–GFP plasmid (pPS2169), a NotI–XhoI fragment of pPS2167 was cloned into the NotI–XhoI site of a URA3 CEN plasmid that contained GFP and the 3′ untranslated region of NUF2 (Kahana and Silver, 1998). The wild-type RPL11A clone YCp50L16A (RPL11A URA3 CEN) was provided by J. Woolford (Carnegie Mellon University, Pittsburgh, PA) (Rotenberg et al., 1988).
Separation of Ribosomal Subunits
Yeast cells expressing Rpl11b–GFP from pPS2167 were grown in 100 ml of synthetic complete medium lacking leucine (leu−) to a density of 1 × 107 cells/ml at 25°C. Cells were harvested by centrifugation at 2000 rpm at room temperature and washed with buffer B (50 mM Tris-Cl, pH 7.5, 50 mM NaCl, 1 mM DTT [Foiani et al., 1991]). Lysate was prepared by glass-bead disruption of cell pellets resuspended in buffer B containing protease inhibitors (1 mM PMSF and 2.5 μg/ml each pepstatin A, leupeptin, chymostatin, and aprotinin). Lysate (200 μg of total RNA) was loaded onto a 10 ml 7–30% sucrose gradient prepared in buffer B and centrifuged at 39,000 rpm for 2.5 h at 4°C. Sucrose gradient profiles were recorded by absorption at 254 nm, and 0.5-ml fractions were collected.
Total protein was precipitated from each 0.5 ml sucrose gradient fraction by the addition of ice-cold trichloroacetic acid to a final concentration of 12% and incubation overnight at −20°C. The fractions were pelleted by centrifugation at 14,000 rpm for 15 min at 4°C, and pellets were washed two times with 200 μl of ice-cold 100% acetone. Dried pellets were resuspended in 90 μl of 1× gel sample buffer (10% glycerol, 2% SDS, 0.1 M DTT, 0.1% bromphenol blue), and 10 μl of each fraction was separated on a 10% SDS-polyacrylamide gel followed by transfer to a nitrocellulose membrane by standard methods (Sambrook et al., 1989). For GFP detection, anti-GFP was used at a 1:2500 dilution in PBST (PBS, 0.25% Tween 20) plus 2.5% milk for 1 h at 25°C. HRP-conjugated anti-rabbit secondary antibody (Jackson Immunoresearch, West Grove, PA) was used at a dilution of 1:5000 in PBST plus milk, and ECL (Amersham Pharmacia Biotech, Uppsala, Sweden) was used to detect immunoreactive bands.
Rpl11b–GFP Localization Assay
Yeast strains expressing Rpl11b–GFP were grown in selective medium (12 ml) to 1–2 × 107 cells/ml at 25°C. Strains that were ade2 were grown in selective medium supplemented with 20 μg/ml adenine sulfate. Before shifting the culture to the nonpermissive growth temperature, a 3-ml aliquot was removed and cells were fixed by the addition of formaldehyde to 3.7% and incubation with agitation at 25°C. After 30 min, the fixed cells were collected, washed two times with 1 ml of 0.1 M potassium phosphate, pH 6.5 (KPi), and stored in 1 ml of solution P (0.1 M KPi, pH 6.5, 1.2 M sorbitol) at 4°C. The remaining culture was shifted to either 37°C for temperature-sensitive strains or 15°C for cold-sensitive strains. The length of the shift depended on the yeast strain being tested and the onset of its growth defect as determined from the literature. At the end of the temperature shift, a 3-ml aliquot was fixed as described above with incubation at the shift temperature (37 or 15°C). The culture was then shifted back to 25°C, and aliquots were removed at 30 and 60 min and fixed at 25°C as described. After permeabilization with 0.5% Triton X-100 for 10 min at room temperature, cell nuclei were stained with DAPI at a final concentration of 1 μg/ml for 3 min at room temperature. Cells were washed two times with 1 ml of 1× PBS and stored at 4°C in 100–200 μl of 1× PBS. Rpl11b–GFP was visualized by fluorescence microscopy as described previously with the use of a Nikon (Garden City, NY) microscope (Ferrigno et al., 1998). Images of cells were captured with a Princeton Instruments (Trenton, NJ) Micromax camera and Metamorph Imaging software (Universal Imaging, Westchester, PA).
Pulse-Chase Labeling of rRNA
Yeast cultures (100 ml) were grown in synthetic complete medium lacking methionine (met−) to 1 × 107 cells/ml at 25°C. Cells were harvested by centrifugation at 2000 rpm and resuspended in 3 ml of met− medium. To label rRNA specifically, 250 μCi of [3H-methyl]-methionine (specific activity, 70–85 Ci/mmol; Amersham Pharmacia Biotech) was added, and cells were incubated for 3 min at 25°C. At 3 min, unlabeled methionine in met− medium was added to a final concentration of 5.1 mM, and at chase times of 15 s, 2 min 15 s, and 9 min 15 s, 1 ml aliquots of cells were removed from the reaction. Cells were collected by brief centrifugation, the supernatant was removed, and the pellets were frozen on dry ice. Processing of each sample took ∼45 s, so this time was included in the time of chase, giving 1-, 3-, and 10-min chase time points.
Total RNA was extracted from labeled cells by hot acid phenol treatment as described previously (Lundblad, 1997), and 10,000 cpm of each sample was separated on a 1.2% formaldehyde agarose gel. RNA was transferred to a Hybond-N+ membrane (Amersham Pharmacia Biotech) by vacuum transfer (VacuGene XL, Amersham Pharmacia Biotech), and the membrane was sprayed with EN3HANCE (New England Nuclear, Boston, MA) before exposure to film for 3 d at −80°C.
RESULTS
Rpl11b–GFP Serves as a Marker for the 60S Ribosomal Subunit in S. cerevisiae
Rpl11 is one of the ∼45 ribosomal proteins that together with 5S, 5.8S, and 25S rRNA forms the 60S ribosomal subunit in S. cerevisiae. Like many of the r-proteins in yeast, Rpl11 is expressed from two gene copies, RPL11A and RPL11B (Woolford et al., 1979; Leer et al., 1984). These two copies code for 99% identical proteins; however, the level of expression from each copy differs significantly (Rotenberg et al., 1988). Two-thirds of Rpl11 expressed in yeast is from the B copy and one-third is from the A copy. Deletion of both RPL11A and RPL11B is lethal, indicating that Rpl11 is an essential component of the 60S subunit in yeast (Rotenberg et al., 1988). Rpl11 associates with the preribosomal subunit late in the assembly pathway, presumably after the 35S rRNA precursor is cleaved into its 40S and 60S components (Kruiswijk et al., 1978; Kressler et al., 1999). We chose to use Rpl11b as a marker for the 60S subunit because it is an essential component of the 60S subunit, it binds specifically to the 60S preribosomal subunit, and it is expressed at a higher level than Rpl11a. Because a fusion between Rpl11a and β-galactosidase was functional in yeast (Tsay et al., 1994), we predicted that the addition of the much smaller GFP to the carboxyl terminus of Rpl11b would also produce a functional fusion protein. In addition, the GFP tag would allow visualization of the 60S ribosomal subunit in living cells.
For Rpl11b–GFP to serve as a reporter for the 60S ribosomal subunit, it must meet three criteria. First, Rpl11b–GFP must be able to functionally replace endogenous Rpl11; second, the localization of Rpl11b–GFP in the cell must reflect that of the 60S ribosomal subunit; and third, the majority of Rpl11b–GFP expressed in the cell must be incorporated into the 60S ribosomal subunit.
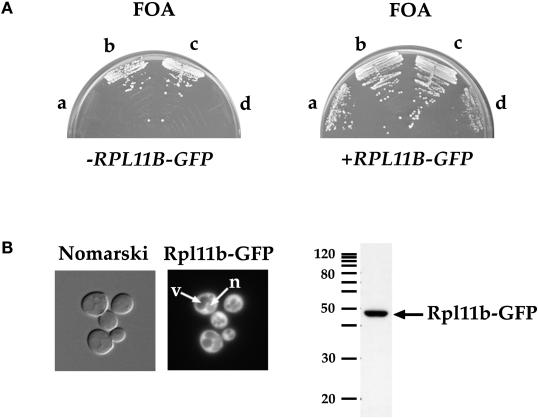
A low-copy LEU2 plasmid bearing GFP fused in frame to the 3′ end of RPL11B was constructed. The 5′ promoter of RPL11B was used to drive expression of the fusion protein. A genetic background in which RPL11B is required for viability was generated to verify that RPL11B–GFP was functional in yeast. A diploid strain disrupted for one copy of each RPL11A and RPL11B by replacement with HIS3 and carrying a wild-type copy of RPL11A on a URA3 CEN plasmid was constructed. The diploid was sporulated, and the resulting tetrads were analyzed to identify tetrads that segregated 2:2 his−:HIS+ and 2:2 FOAR:FOAS, indicating the presence of two wild-type (RPL11A RPL11B) and two double mutant spores (rpl11a::HIS3 rpl11b::HIS3), respectively. An example tetrad is shown in Figure 1A (left), where spores a and d do not grow on synthetic complete medium containing FOA and thus require the RPL11A URA3 CEN plasmid for cell viability, and spores b and c, the wild-type spores, are FOA-resistant. Spores a and d were no longer FOA-sensitive when transformed with the RPL11B–GFP LEU2 CEN plasmid (Figure 1A, right), indicating that RPL11B–GFP is a functional gene fusion.
Figure 1.
The RPL11B–GFP gene fusion is functional in yeast. (A) Four spores of a representative tetrad from the cross between PSY2083 (rpl11a::HIS3) covered by YCp50L11A URA3 CEN and PSY2084 (rpl11b::HIS3) streaked on synthetic complete medium containing FOA (left panel). Spores a and d are HIS+ and FOAS and spores b and c are his− and FOAR. Spores a–d were transformed with pPS2167 (RPL11B–GFP LEU2 CEN) and streaked on synthetic complete medium containing FOA (right panel). (B) Rpl11b–GFP expressed from a CEN plasmid in wild-type yeast was visualized by fluorescence microscopy of living cells (left panel). Arrows point to a vacuole (v) and the nucleus (n). Nomarski, phase-contrast image of cells; Rpl11b–GFP, fluorescence signal. Yeast lysate prepared from wild-type yeast cells expressing Rpl11b–GFP was probed with anti-GFP to detect Rpl11b–GFP (right panel). Five micrograms of total protein of lysate was loaded. Migration of a 10-kDa ladder of molecular mass markers is shown.
The localization of Rpl11b–GFP in wild-type yeast cells grown to early log phase was determined by fluorescence microscopy. Rpl11b-GFP is predominantly cytoplasmic and for the most part excluded from both the vacuoles (v) and the nucleus (n) (Figure 1B, arrows). This localization is consistent with the steady-state cytoplasmic location of 60S ribosomal subunits active in translation. To verify that the GFP signal seen in the cytoplasm is due to intact Rpl11b–GFP, lysate prepared from wild-type yeast cells expressing Rpl11b–GFP was run on a 10% SDS-polyacrylamide gel and immunoblotted with anti-GFP (Figure 1B, right). A single polypeptide migrating just under 50 kDa was present, consistent with the predicted molecular mass of ∼47 kDa for a fusion between ∼20-kDa Rpl11b and 27-kDa GFP.
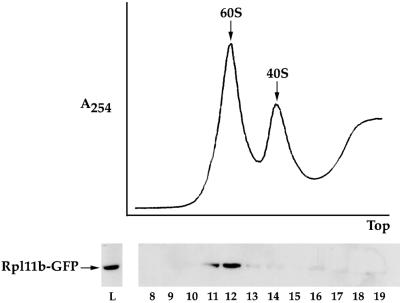
The incorporation of Rpl11b–GFP into the 60S ribosomal subunit was verified by preparing lysate from wild-type yeast cells expressing Rpl11b–GFP and separating the individual ribosomal subunits by high-speed sucrose gradient centrifugation. Fractions from the sucrose gradient were assayed for the presence of Rpl11b–GFP by separation on a 10% SDS gel and immunoblotting with anti-GFP. As seen in Figure 2, Rpl11b–GFP sedimented with the 60S ribosomal subunit and only trace amounts of Rpl11b–GFP were detectable in the 40S subunit and soluble fractions of the sucrose gradient. The gradient profile and distribution of Rpl11b–GFP were similar for a yeast strain that has GFP integrated at the 3′ end of the genomic copy of RPL11B (our unpublished results).
Figure 2.
Rpl11b–GFP is incorporated in the 60S ribosomal subunit. Yeast lysate from wild-type cells expressing Rpl11b–GFP from a CEN plasmid was separated on a 7–30% sucrose gradient. The sucrose gradient profile was recorded by absorbance at 254 nm (upper panel). Fractions 8–19 collected from the sucrose gradient and lysate (5 μg of total protein) were probed with anti-GFP to detect Rpl11b–GFP (bottom panel).
60S Ribosomal Subunit Localization Assay
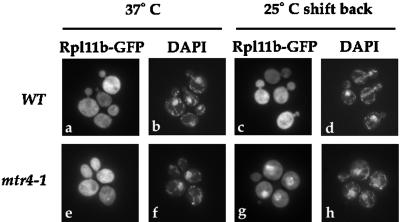
An assay was developed to screen for factors involved in ribosome subunit synthesis or export with the use of Rpl11b–GFP as a marker for the 60S ribosomal subunit. Mutant yeast cells expressing Rpl11b–GFP were grown to early log phase in liquid culture at 25°C, shifted to the nonpermissive growth temperature to induce the mutant phenotype, and then shifted back to the permissive growth temperature to induce new ribosomal protein synthesis. Yeast cells were formaldehyde fixed, their nuclei were stained with DAPI at each step of the assay, and the localization of Rpl11b–GFP was determined by fluorescence microscopy. The location and distribution of Rpl11b–GFP in cells was not affected by the fixing procedure. The shift back to the permissive temperature was required because synthesis of ribosomal proteins, and thus of the reporter Rpl11b–GFP, is reduced at 37°C in yeast (Gorenstein and Warner, 1976; Kim and Warner, 1983; Hurt et al., 1999). A yeast strain that is defective in 60S ribosomal subunit assembly or transport should accumulate Rpl11b–GFP in the nucleus upon shifting back to the permissive temperature if the rate of ribosome synthesis is faster than the rate at which the mutant phenotype reverses. The 60S ribosomal subunit should remain cytoplasmic in wild-type yeast cells throughout this assay. As seen in Figure 3, Rpl11b–GFP is cytoplasmic in a wild-type strain after a 1-h shift to 37°C (Figure 3a) and after shifting the cells back to 25°C for 30 min (Figure 3c). The GFP signal was not as bright at 37°C, consistent with a reduction in r-protein synthesis at this temperature. Because this assay depends on the synthesis, import, and assembly of Rpl11b–GFP into the 60S subunit, mutants that slow the import of r-proteins, inhibit rRNA synthesis or processing, or reduce import of any factor required for ribosome transport may also affect Rpl11b–GFP localization.
Figure 3.
Rpl11b–GFP localization assay. Wild-type (PSY580) or mtr4-1 cells expressing Rpl11b–GFP from a CEN plasmid were grown to early log phase, shifted to 37°C for 1 h, and then shifted back to 25°C for up to 1 h. Yeast cells were fixed and incubated with DAPI to stain the nuclei. Rpl11b–GFP was visualized by fluorescence microscopy. Panels a, b, e, and f show cells shifted to 37°C for 1 h, and panels c, d, g, and h show cells shifted back to 25°C for 30 min.
Yeast Strains Defective in Ribosome Assembly Accumulate Rpl11b–GFP in the Nucleus
Proper assembly of 60S ribosomal subunits is required for their export from the nucleus (Gorlich and Kutay, 1999). Therefore, yeast mutants that are defective in 60S ribosomal subunit assembly may alter the steady-state localization of Rpl11b–GFP. To test this prediction, several ribosomal assembly mutants, including mtr4-1/dob1-1, nop1-3, fal1-1, nsr1Δ, and rat1-1, were screened for effects on Rpl11b–GFP localization. Mtr4p/Dob1p is involved in the processing of 7S rRNA to mature 5.8S rRNA, and mutations cause defects in 60S ribosomal subunit assembly (de la Cruz et al., 1998). The location of Rpl11b–GFP in mtr4-1 cells grown to early log phase, shifted to 37°C for 1 h, and then shifted back to 25°C was determined by fluorescence microscopy. As predicted for an assembly mutant, Rpl11b–GFP accumulated in the nucleus of >70% of mtr4-1 cells as early as 30 min after shifting back to the permissive temperature (Figure 3g). The fluorescent signal overlaps with the DAPI signal, indicating that Rpl11b–GFP is in the nucleus of mtr4-1 cells (Figure 3h). Similar results were obtained with the dob1-1 mutation, although the defect was even more pronounced in this strain. Rpl11b–GFP accumulated in the nucleus of >90% of dob1-1 cells after the shift back to 25°C.
The data for all of the ribosomal assembly mutants tested are summarized in the fourth column of Table 2. The temperature-sensitive nop1-3 strain that has defects in rRNA methylation and 60S subunit assembly mislocalized Rpl11b–GFP to the nucleus of 20–25% of cells after shifting back to the permissive temperature. Unexpectedly, fal1-1 and to a greater extent nsr1Δ cells also accumulated Rpl11b–GFP in the nucleus. Both mutations inhibit cleavage of the 35S rRNA at sites that divide the precursor into the 40S (20S RNA) and 60S precursor RNAs (27SA2 RNA). These strains have a reduced level of mature 18S rRNA and therefore are considered to be primarily defective in 40S ribosomal subunit assembly (Kondo and Inouye, 1992; Kondo et al., 1992; Lee et al., 1992; Kressler et al., 1997). The localization of Rpl11b–GFP was not altered in yeast cells bearing a mutation of the 5′ to 3′ exonuclease Rat1 that is involved in the conversion of the 7SS RNA precursor to 5.8SS rRNA (Amberg et al., 1992; Henry et al., 1994).
Table 2.
Summary of Rpl11b–GFP localization data
| β subunits | Nucleoporins | Transport factors | Ribosomal processing | Other |
|---|---|---|---|---|
| Strains that accumulate Rpl11b–GFP in the nucleusa | ||||
| rsl1-4b | nup159/rat7-1b | srp1-31b | mtr4-1/dob1-1b | npl2-1/npl2-2 |
| kap120Δb | nup116-5b | yrb1-1b | nsr1Δc | |
| nmd5Δb | nup1-8b | rat8-1b/rat8-2b | nop1-3 | |
| kap104-16 | nup120/rat2-2b | prp20-1 | fal1-1 | |
| kap123Δ | nup82Δ108b | rna1-1 | ||
| xpo1-1 | nup49-313b | npl3-27/npl3-17 | ||
| cse1-1c | nic96-1 | mtr2-1 | ||
| mtr10-1b | nsp1 | |||
| nup85/rat9-1b | ||||
| nup133/rat3-1 | ||||
| nup40/gle2-1 | ||||
| gle1-4b | ||||
| Strains that do not accumulate Rpl11b–GFP in the nucleusd | ||||
| msn5Δ | nup100Δ | mex67-5 | rat1-1 | WT |
| kap114Δ | nup170-1 | npl4-1 | rat6-1 | |
| sxm1Δ | nup42/rip1Δ | |||
| los1Δ | nup2-4 | |||
| pse1-1 | nup157-2 | |||
| nup84Δ | ||||
| seh1Δc | ||||
Strains in boldface type accumulated Rpl11b–GFP in >40% of cells, and strains in lightface type accumulated Rpl11b–GFP in 10–40% of cells.
Strains accumulated Rpl11b–GFP at 25°C.
Cold-sensitive strains were shifted to 15°C.
Ten percent or fewer cells accumulated Rpl11b–GFP in the nucleus.
Several Nucleoporin Mutations Cause Rpl11b–GFP to Accumulate in the Nucleus
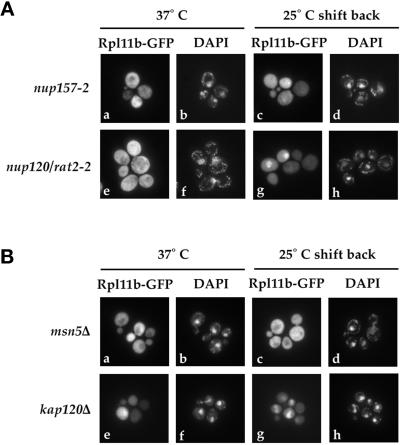
Eighteen nucleoporin mutants were screened for their effect on Rpl11b–GFP localization, and the results for two of these mutants are shown in Figure 4A. Nup157 is a nonessential nucleoporin that is one of the core nuclear pore proteins (Aitchison et al., 1995b). Rpl11b–GFP remained cytoplasmic in nup157-2 cells throughout the localization assay (Figure 4A, a and c). In contrast, mutation of NUP120 (rat2-2), a nucleoporin implicated in mRNA export, accumulated Rpl11b–GFP in the nucleus of >50% of cells 30 min after shifting back to the permissive temperature (Figure 4A, g). The change in Rpl11b–GFP localization in nup120 cells was not due to accumulation of free Rpl11b–GFP because Rpl11b–GFP remained in the 60S ribosomal subunit throughout the assay, as determined by sucrose gradient analysis (our unpublished results).
Figure 4.
Rpl11b–GFP localization in nucleoporin and karyopherin mutants. Yeast strains nup157-2, rat2-2, msn5Δ, and kap120Δ expressing Rpl11b–GFP from a CEN plasmid were grown to early log phase, shifted to 37°C for 1 h, and then shifted back to 25°C for up to 1 h. Cells were fixed, and their nuclei were stained with DAPI. (A) Fluorescence microscopy images of nup157-2 cells shifted to 37°C (a and b), nup157-2 cells shifted back to 25°C for 30 min (c and d), rat2-2 cells shifted to 37°C (e and f), and rat2-2 cells 30 min after shifting back to 25°C (g and h). (B) Fluorescence microscopy images of msn5Δ cells shifted to 37°C (a and b), msn5Δ cells shifted back to 25°C for 30 min (c and d), kap120Δ cells shifted to 37°C (e and f), and kap120Δ cells 30 min after shifting back to 25°C (g and h).
The results for all of the nucleoporin mutants screened for mislocalization of the 60S ribosomal subunit are summarized in Table 2. Temperature-sensitive alleles of the essential nucleoporins NSP1, NUP1, NUP49, NUP82, NIC96, and NUP159 all caused a strong accumulation of Rpl11b–GFP in the nucleus after shift back to the permissive temperature (Table 2). In addition, several of these strains accumulated Rpl11b–GFP in the nucleus at the permissive temperature (Table 2). Interestingly, nup85 cells accumulated Rpl11b–GFP in the nucleus at the permissive temperature of 25°C before shifting to 37°C and appeared to concentrate Rpl11b–GFP in the nucleolus after 1 h at 37°C. Not all of the nucleoporin mutants tested accumulated Rpl11b–GFP in the nucleus (Table 2), indicating that there is some degree of specificity for the 60S ribosomal subunit.
Transport Factors
Several known transport factors, including mutations of the RAN regulators PRP20 and RNA1, were screened for accumulation of Rpl11b–GFP in the nucleus, and the results are shown in Table 2. Both prp20-1 and rna1-1 cells accumulated Rpl11b–GFP in the nucleus after shift back to the permissive temperature; however, the defect was present in only ∼25% of the cells. A temperature-sensitive mutation of the Ran-binding protein Yrb1 caused a strong accumulation of Rpl11b–GFP in the nucleus. Mutations in the hnRNP-like protein Npl3 and its importer Mtr10 also caused accumulation of Rpl11b–GFP in the nucleus. Rpl11b–GFP also accumulated in the nucleus of cells bearing mutant alleles of RAT8, a gene that codes for a helicase that is implicated in mRNA export (Snay-Hodge et al., 1998; Tseng et al., 1998). Not all mutations that cause defects in mRNA export mislocalized Rpl11b–GFP, because mutation of the RNA export factor Mex67 did not block export of Rpl11b–GFP from the nucleus.
Karyopherins
Mutations of importin α (SRP1), importin β (RSL1), and 12 additional karyopherins were screened for their effect on 60S ribosomal subunit localization (Table 2). Temperature-sensitive mutations of the NLS receptor SRP1 (srp1-31) and of the importin α receptor RSL1 (rsl1-4) both caused Rpl11b–GFP to accumulate in the nucleus. A cold-sensitive mutation of CSE1 (cse1-1), the export receptor for Srp1, also caused Rpl11b–GFP to accumulate in cells shifted to 15°C. The results for two nonessential karyopherins, MSN5 and KAP120, are shown in Figure 4B. Deletion of MSN5 did not alter the cytoplasmic localization of Rpl11b–GFP at either 37 or 25°C (Figure 4B, a–d), whereas deletion of KAP120 caused a surprisingly strong mislocalization of Rpl11b–GFP in the nucleus at 25°C (Figure 4B, g). Rpl11b–GFP was trapped in the nucleus of >90% of kap120Δ cells even before shifting to 37°C, whereas at 37°C, when ribosomal protein synthesis was reduced, Rpl11b–GFP was distributed throughout the cell (Figure 4B, e).
Deletion of NMD5, the yeast homologue of the mammalian ribosomal protein importer RanBP7 (Gorlich et al., 1997; Jakel and Gorlich, 1998), also caused a strong nuclear accumulation of Rpl11b–GFP at 25°C. Deletion of KAP123, one of the ribosomal protein importers in yeast (Rout et al., 1997), and a temperature-sensitive mutant of KAP104, the yeast homologue of the mammalian ribosomal protein importer transportin (Jakel and Gorlich, 1998), also caused the 60S subunit to accumulate in the nucleus to a lesser extent. Although Pse1 has been implicated in the import of ribosomal proteins (Rout et al., 1997), the 60S reporter Rpl11b–GFP remained in the cytoplasm of pse1-1 cells throughout the localization assay. Interestingly, Rpl11b–GFP also accumulated in ∼30% of xpo1-1 cells that were first shifted to 37°C for 1 h and then shifted back to 25°C for 1 h. Xpo1 is the receptor for nuclear export signal–containing proteins (Stade et al., 1997). Deletion of the karyopherins KAP114, SXM1, and LOS1 had no effect on the localization of the 60S ribosomal subunit (Table 2).
Deletion of KAP120 Causes a Delay in rRNA Processing and a Reduction in the Level of 60S Ribosomal Subunits
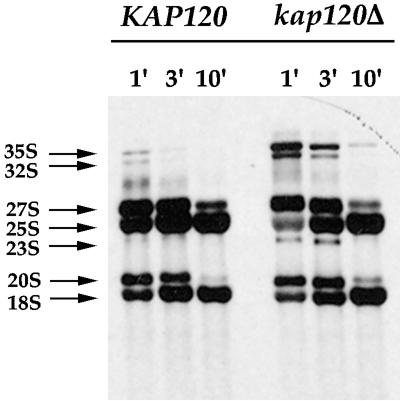
Because Kap120 is one of the few karyopherins whose transport substrate(s) has not yet been identified and whose function is not known, we chose to characterize the 60S export defect in kap120Δ cells in more detail. Processing of the 35S rRNA precursor was analyzed in kap120Δ cells to investigate whether nuclear accumulation of Rpl11b–GFP was due to a defect in ribosomal rRNA maturation. The 35S rRNA precursor can be specifically labeled with [3H]methionine because rRNA is highly methylated in vivo. To follow the processing of the 35S rRNA precursor, KAP120 and kap120Δ strains were grown to 1 × 107 cells/ml at 25°C, pulse labeled with [3H]methionine for 3 min, and then chased with an excess of unlabeled methionine for up to 10 min. Conversion of the 35S rRNA precursor to the mature 25S and 18S rRNAs was nearly complete after 10 min of chase with unlabeled methionine for wild-type KAP120 cells (Figure 5). In contrast, deletion of KAP120 led to an accumulation of the 35S rRNA precursor that was still detectable after 10 min of chase (Figure 5). A 23S aberrant RNA species was also detected in kap120Δ cells at early chase times (Figure 5). Although rRNA processing was delayed in kap120Δ cells, the levels of mature 25S and 18S rRNA were not depleted significantly (Figure 5).
Figure 5.
Processing of rRNA in KAP120 and kap120Δ cells. KAP120 and kap120Δ cells were grown at 25°C to a density of 1 × 107 cells/ml, pulse labeled for 3 min with [3H-methyl]methionine, and chased with an excess of unlabeled methionine. Chase time points of 1, 3, and 10 min for each yeast strain are shown. The 35S rRNA precursor and processing intermediates are labeled.
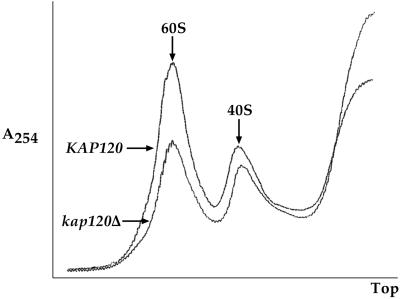
Because a delay in rRNA processing is often indicative of a ribosome assembly defect, the ratio of 60S to 40S ribosomal subunits in kap120Δ and KAP120 cells was determined. Lysates prepared from kap120Δ and KAP120 cells grown at 25°C were subjected to sucrose gradient sedimentation to separate the individual subunits. The subunit peaks were detected by their absorption at 254 nm. As seen in the sucrose gradient profiles in Figure 6, deletion of KAP120 reduced the 60S ribosomal subunit peak significantly compared with the wild-type KAP120 strain. The level of 40S ribosomal subunits, however, was reduced only slightly in kap120Δ cells (Figure 6).
Figure 6.
Deletion of KAP120 causes a large reduction in 60S ribosomal subunit levels. Yeast lysates prepared from KAP120 and kap120Δ cells expressing Rpl11b–GFP from a CEN plasmid were centrifuged through 7–30% sucrose gradients, and gradient profiles were collected by absorbance at 254 nm. An overlay of the gradient profiles for KAP120 and kap120Δ cells is shown to emphasize the difference in peak heights for the 60S and 40S ribosomal subunits. Equal amounts of total RNA (200 μg) were loaded for each gradient.
The accumulation of Rpl11b–GFP in the nucleus of kap120Δ cells was not an indirect effect of trapping mRNA in the nucleus. Polyadenylated RNA was localized throughout the cytoplasm of kap120Δ cells, as determined by in situ hybridization with a digoxigenin-labeled oligo-dT probe (our unpublished results). In addition, nuclear accumulation of Rpl11b–GFP in kap120Δ cells was not due to free Rpl11b–GFP because Rpl11b–GFP was incorporated in the 60S ribosomal subunit, as determined by sucrose gradient analysis (our unpublished results). Although it is possible that Kap120 is another ribosomal protein importer, it does not import either Rpl11 or Rpl25. Rpl11b–GFP and a Rpl25–NLS–GFP–GFP reporter are both localized in the nucleus of kap120Δ cells at 25°C, indicating that their import is not blocked. In contrast, the Rpl25–NLS–GFP–GFP reporter is distributed in both the cytoplasm and the nucleus of cells deleted for KAP123, an Rpl25 importer (our unpublished results).
DISCUSSION
To further our understanding of 60S ribosomal subunit assembly and export, we have designed an assay that uses Rpl11b–GFP to follow 60S ribosomal subunit localization in yeast. Mutant yeast strains that are defective for 60S ribosomal subunit assembly or export accumulate Rpl11b–GFP in the nucleus upon shifting cells from the nonpermissive growth temperature back to the permissive growth temperature. The shift back to the permissive growth temperature is required to increase the nuclear pool of Rpl11b–GFP available for ribosome biogenesis because r-protein synthesis in yeast is reduced under mild stress conditions (Gorenstein and Warner, 1976; Kim and Warner, 1983). Using this assay, we identified mutations in several nucleoporins and transport factors that trap the 60S ribosomal subunit in the nucleus, including mutation of the karyopherin Kap120.
The Rpl11b–GFP localization assay described here differs from the 60S export assay that uses Rpl25–GFP as the tag for the 60S ribosomal subunit (Hurt et al., 1999). Nuclear accumulation of Rpl11b–GFP can be detected in cells during logarithmic growth when they are actively producing ribosomes, whereas Rpl25–GFP accumulation is observed when cells are shifted from conditions of starvation (saturation) to fresh medium (Hurt et al., 1999). The two r-proteins also bind at different points on the ribosome biogenesis pathway (Kruiswijk et al., 1978; Kressler et al., 1999). Rpl25 binds to the 35S rRNA precursor before it is divided into its 40S (20S rRNA) and 60S (27SA2 rRNA) specific components, whereas Rpl11 associates later in the pathway, most likely with the 27SA2 preribosomal particle. Although the differences between these two 60S localization assays may seem minor, they have the potential to identify a different set of factors required for ribosomal subunit assembly and export. Because Rpl25 binds directly to the rRNA, it is likely to be buried within the 60S ribosomal subunit (El-Baradi et al., 1984, 1987; Yeh and Lee, 1998). Rpl11 binds later in ribosome assembly and appears to be located at the surface of the 60S ribosomal subunit (Tsay et al., 1994). Thus, Rpl11 may be available for recognition by the ribosome assembly or export machinery.
The distribution of Rpl11b–GFP in yeast cells is sensitive to defects in ribosome synthesis, as expected. Rpl11b–GFP is predominantly cytoplasmic in wild-type yeast cells at all temperatures assayed, consistent with the cytoplasmic localization of active 80S ribosomes. In contrast, Rpl11b–GFP accumulates in the nucleus of yeast strains bearing temperature-sensitive mutations in the 60S ribosome assembly factors Mtr4/Dob1 and Nop1. Although rat1-1 cells have a defect in 5.8S rRNA processing (Amberg et al., 1992; Henry et al., 1994), Rpl11b–GFP did not accumulate in this strain. It is possible that a defect in rat1-1 cells was not detected because in its absence the exonuclease Xrn1 completes 5.8S rRNA processing (Henry et al., 1994; Petfalski et al., 1998). Rpl11b–GFP did accumulate in fal1-1 and nsr1Δ cells, which are defective in cleavage of the 35S rRNA precursor into the 20S and 27SA2 rRNAs (Kondo and Inouye, 1992; Kondo et al., 1992; Lee et al., 1992; Kressler et al., 1997). Rpl11b–GFP may accumulate in the nucleus of these strains because it binds to the 27SA2 preribosomal particle after this major cleavage event (Kressler et al., 1999). Thus, we were able to detect defects in ribosome assembly mutants with the use of Rpl11b–GFP as a marker for the 60S ribosomal subunit. In contrast, Rpl25–GFP did not mislocalize in cells bearing a mutation in the 60S subunit assembly factor Nop1 (Hurt et al., 1999), and we were unable to detect a defect in mtr4-1 cells with the use of the Rpl25–GFP assay (our unpublished results).
The 60S ribosomal subunit accumulates in the nucleus of several yeast strains bearing mutations in nucleoporins. The nucleoporin mutants nup49-313, nic96-1, nsp1, nup82Δ108, nup116-5, and nup120 cause Rpl11b–GFP to accumulate in the nucleus (Table 2). The 40S ribosomal subunit also accumulates in the nucleus of these strains (Moy and Silver, 1999; T. Moy and P. Silver, unpublished results), suggesting that these nucleoporins may be involved in the export of both ribosomal subunits. In addition, the yeast strains nup85, nup133, nup159, nup1, nup40, and gle1-4 all accumulated Rpl11b–GFP in the nucleus to various degrees (Table 2). These strains have also been shown to accumulate poly(A)+ RNA in the nucleus at nonpermissive growth temperatures (Bogerd et al., 1994; Doye et al., 1994; Gorsch et al., 1995; Li et al., 1995; Goldstein et al., 1996; Murphy et al., 1996; Murphy and Wente, 1996). Some of these nucleoporins may play a role in the export of both poly(A)+ RNA and 60S ribosomal subunits. Rpl11b–GFP accumulation in nup1-8, nup133, gle1-4, and nup159 cells may be due to a delay in ribosome assembly and not an export defect, because the 40S ribosomal subunit accumulated in the nucleolus of nup1-8, nup133, and gle1-4 cells (Moy and Silver, 1999) and rRNA processing was inhibited in nup159 cells (Del Priore et al., 1996; Goldstein et al., 1996).
Mutation of either Nup120 or Nup85, two members of the Nup84 nuclear pore subcomplex, causes nuclear accumulation of Rpl11b–GFP. The Nup84 subcomplex also contains Nup145-C, Seh1, and Sec13 (Siniossoglou et al., 1996, 2000). It has been proposed that this subcomplex plays a role in mRNA export because mutations in Nup120, Nup85, Nup84, and Nup145 trap poly(A)+ RNA in the nucleus (Aitchison et al., 1995a; Heath et al., 1995; Goldstein et al., 1996; Siniossoglou et al., 1996; Dockendorff et al., 1997). The 60S subunit accumulated in nup120 cells after a 1-h shift to 37°C, and a slight defect was also detected at the permissive temperature. It does not appear that nuclear accumulation of Rpl11b–GFP in nup120 cells is due to a defect in ribosome assembly, because the ratio of 60S to 40S subunits is similar to that of wild-type cells in all conditions assayed (our unpublished results). In nup85 cells, Rpl11b–GFP accumulates in the entire nucleus at 25°C and is concentrated in the nucleolus at 37°C, suggesting that both ribosome assembly and export are affected by this mutation. It is possible that the Nup120 and Nup85 portion of the Nup84 subcomplex plays a role in 60S ribosomal subunit export in addition to mRNA export.
Consistent with a role for Ran in ribosomal subunit export, mutation of the Ran regulators Rna1, Prp20, and Yrb1 resulted in nuclear accumulation of Rpl11b–GFP. The 40S ribosomal subunit also accumulates in the entire nucleus of rna1-1, prp20-1, and yrb1-1 cells (Moy and Silver, 1999). It is not clear whether Ran and its regulators play a direct role in ribosomal subunit export or if the defect in these mutants is due to a block in other transport processes. The importin α and importin β mutants srp1-31 and rsl1-4 also caused 60S subunit accumulation, presumably due to a delay in the import of factors required for 60S ribosomal subunit assembly or export.
The mutants rat8-1 and rat8-2 accumulated Rpl11b–GFP in the nucleus. Rat8 is a member of the DEAD box family of helicases that is localized in the cytoplasm and associates with the NPC in yeast cells (Snay-Hodge et al., 1998; Tseng et al., 1998; Hodge et al., 1999). A role for Rat8 in mRNA export has been proposed (Snay-Hodge et al., 1998). The 60S export defect in rat8 mutants was not due to a ribosome assembly defect, because processing of the 35S rRNA in rat8-1 cells was delayed only slightly and the ratio of 60S to 40S subunits as well as the polyribosome profiles of rat8-1 and rat8-2 cells were similar to those of wild-type cells (our unpublished results). We were also able to detect the 60S export defect in rat8-1 cells with the use of the Rpl25–GFP export assay described previously (our unpublished results). Rpl25–GFP accumulated in both the nucleolus and the nucleus of rat8-1 cells within 15 min of shifting cells from the nonpermissive growth temperature back to the permissive temperature (our unpublished results). It is possible that Rat8 is involved in both mRNA and 60S ribosomal subunit export. A role for a helicase in the release of mRNA or 60S ribosomal subunits from the cytoplasmic face of the NPC is an intriguing possibility.
A subset of karyopherin mutants mislocalize Rpl11b–GFP to the nucleus. Mutation of KAP104 and deletions of NMD5 and KAP123 all lead to nuclear accumulation of the 60S ribosomal subunit. Because Kap123 is a putative yeast r-protein importer (Rout et al., 1997) and both Nmd5 and Kap104 are homologous to mammalian r-protein importers (Jakel and Gorlich, 1998), it is possible that mutation of these factors slows r-protein import, which in turn inhibits ribosome subunit assembly. In contrast, mutation of Pse1 and Sxm1, two karyopherins implicated in r-protein import (Rosenblum et al., 1997; Rout et al., 1997), did not affect Rpl11b–GFP localization. Because ribosome biogenesis is essential for cell viability, it is likely that there are multiple r-protein importers in yeast. Mutation of the nuclear export sequence receptor Xpo1/Crm1 also mislocalizes Rpl11b–GFP to the nucleus, although the defect occurs in only 30% of cells. Because Rna1 accumulates in the nucleus of xpo1-1 cells at the nonpermissive temperature (Feng et al., 1999), it is possible that disruption of the RanGTP gradient causes the 60S mislocalization defect in these cells.
We have identified a potential role for the karyopherin Kap120 in the assembly or export of 60S ribosomal subunits in yeast. Deletion of KAP120 causes a strong accumulation of Rpl11b–GFP in the nucleus at 25°C (Figure 4B) and a large reduction in the level of mature 60S ribosomal subunits (Figure 6). The decrease in 60S ribosomal subunits is not simply due to a deficiency in the level of mature 25S rRNA because deletion of KAP120 only delays (does not block) processing of the 35S rRNA precursor (Figure 5). The 60S ribosomal subunit defect is also not caused by a block in mRNA export because poly(A)+ RNA is cytoplasmic in kap120Δ cells (our unpublished results). Deletion of KAP120 appears to affect only the 60S ribosomal subunit. The 40S subunit is not mislocalized in kap120Δ cells (Moy and Silver, 1999), and nearly wild-type levels of 40S ribosomal subunits are present (Figure 6). At both 37°C (Figure 4B) and at saturation (our unpublished results), conditions in which ribosome synthesis in the cell is reduced, Rpl11b–GFP is distributed throughout the cytoplasm of kap120Δ cells. This redistribution of Rpl11b–GFP indicates that deletion of KAP120 does not cause a complete block in assembly or export. There are several potential roles for Kap120 in ribosome biogenesis and export. Kap120 may be an importer for factor(s) involved in ribosomal subunit assembly, a r-protein importer, or a factor that recognizes fully assembled ribosomal subunits for export. We are currently investigating the role of Kap120 in ribosome assembly and export in more detail.
ACKNOWLEDGMENTS
Special thanks to J. Woolford and E. Hurt for providing plasmids used in this study. We also thank Alexander S. Brodsky, for critical input at the initial phase of the project, Terrence Moy for technical assistance and helpful discussions, Karin Anderson and Anne McBride for critical reading of the manuscript, and members of the Silver laboratory for their useful advice. This work was funded by grants from the National Institutes of Health to P.A.S. and a National Institutes of Health postdoctoral fellowship to T.S.-Z
REFERENCES
- Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995a;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science. 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol. 1995b;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Amberg DC, Fleischmann M, Stagljar I, Cole CN, Aebi M. Nuclear PRP20 protein is required for mRNA export. EMBO J. 1993;12:233–241. doi: 10.1002/j.1460-2075.1993.tb05649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Bataille N, Helser T, Fried HM. Cytoplasmic transport of ribosomal subunits microinjected into the Xenopus laevis oocyte nucleus: a generalized, facilitated process. J Cell Biol. 1990;111:1571–1582. doi: 10.1083/jcb.111.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Gorlich D. RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett. 1997;419:249–254. doi: 10.1016/s0014-5793(97)01467-1. [DOI] [PubMed] [Google Scholar]
- Bogerd AM, Hoffman JA, Amberg DC, Fink GR, Davis LI. nup1 mutants exhibit pleiotropic defects in nuclear pore complex function. J Cell Biol. 1994;127:319–332. doi: 10.1083/jcb.127.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJ, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeHoratius C, Silver PA. Nuclear transport defects and nuclear envelope alterations are associated with mutation of the Saccharomyces cerevisiae NPL4 gene. Mol Biol Cell. 1996;7:1835–1855. doi: 10.1091/mbc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Priore V, Snay CA, Bahr A, Cole CN. The product of the Saccharomyces cerevisiae RSS1 gene, identified as a high-copy suppressor of the rat7-1 temperature-sensitive allele of the RAT7/NUP159 nucleoporin, is required for efficient mRNA export. Mol Biol Cell. 1996;7:1601–1621. doi: 10.1091/mbc.7.10.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure Mol. Cell Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baradi TT, de Regt VC, Planta RJ, Nierhaus KH, Raue HA. Interaction of ribosomal proteins L25 from yeast and EL23 from E. coli with yeast 26S and mouse 28S rRNA. Biochimie. 1987;69:939–948. doi: 10.1016/0300-9084(87)90227-6. [DOI] [PubMed] [Google Scholar]
- El-Baradi TT, Raue HA, De Regt CH, Planta RJ. Stepwise dissociation of yeast 60S ribosomal subunits by LiCl and identification of L25 as a primary 26S rRNA binding protein. Eur J Biochem. 1984;144:393–400. doi: 10.1111/j.1432-1033.1984.tb08477.x. [DOI] [PubMed] [Google Scholar]
- Feng W, Benko AL, Lee JH, Stanford DR, Hopper AK. Antagonistic effects of NES and NLS motifs determine S. cerevisiae Rna1p subcellular distribution. J Cell Sci. 1999;112:339–347. doi: 10.1242/jcs.112.3.339. [DOI] [PubMed] [Google Scholar]
- Ferrigno P, Posas F, Koepp D, Saito H, Silver PA. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin beta homologs NMD5 and XPO1. EMBO J. 1998;17:5606–5614. doi: 10.1093/emboj/17.19.5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M. Disassembly of RanGTP-karyopherin beta complex, an intermediate in nuclear protein import. J Biol Chem. 1997;272:19538–19546. doi: 10.1074/jbc.272.31.19538. [DOI] [PubMed] [Google Scholar]
- Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, Snay CA, Heath CV, Cole CN. Pleiotropic nuclear defects associated with a conditional allele of the novel nucleoporin Rat9p/Nup85p. Mol Biol Cell. 1996;7:917–934. doi: 10.1091/mbc.7.6.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorenstein C, Warner JR. Coordinate regulation of the synthesis of eukaryotic ribosomal proteins. Proc Natl Acad Sci USA. 1976;73:1547–1551. doi: 10.1073/pnas.73.5.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Pante N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath CV, Copeland CS, Amberg DC, Del Priore V, Snyder M, Cole CN. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1697. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Hannus S, Schmelzl B, Lau D, Tollervey D, Simos G. A novel in vivo assay reveals inhibition of ribosomal nuclear export in ran-cycle and nucleoporin mutants. J Cell Biol. 1999;144:389–401. doi: 10.1083/jcb.144.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz ME, Blobel G. NUP82 is an essential yeast nucleoporin required for poly(A)+ RNA export. J Cell Biol. 1995;130:1275–1281. doi: 10.1083/jcb.130.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Goldfarb D, Spitz LM, Tartakoff AM, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Silver PA. The uses of green fluorescent protein in yeasts. In: Chalfie M, Kain S, editors. Green Fluorescent Protein: Properties, Applications and Protocols. New York: Wiley-Liss; 1998. pp. 139–152. [Google Scholar]
- Kim CH, Warner JR. Mild temperature shock alters the transcription of a discrete class of Saccharomyces cerevisiae genes. Mol Cell Biol. 1983;3:457–465. doi: 10.1128/mcb.3.3.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM. Microbiology and Molecular Genetics. Cambridge, MA: Harvard University Press; 1997. Analysis of nuclear transport factors in Saccharomyces cerevisiae. [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992;267:16252–16258. [PubMed] [Google Scholar]
- Kondo K, Kowalski LR, Inouye M. Cold shock induction of yeast NSR1 protein and its role in pre-rRNA processing. J Biol Chem. 1992;267:16259–16265. [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk T, Planta RJ, Krop JM. The course of the assembly of ribosomal subunits in yeast. Biochim Biophys Acta. 1978;517:378–389. doi: 10.1016/0005-2787(78)90204-6. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Lee WC, Zabetakis D, Melese T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer RJ, van Raamsdonk-Duin MM, Mager WH, Planta RJ. The primary structure of the gene encoding yeast ribosomal protein L16. FEBS Lett. 1984;175:371–376. doi: 10.1016/0014-5793(84)80771-1. [DOI] [PubMed] [Google Scholar]
- Li O, Heath CV, Amberg DC, Dockendorff TC, Copeland CS, Snyder M, Cole CN. Mutation or deletion of the Saccharomyces cerevisiae RAT3/NUP133 gene causes temperature-dependent nuclear accumulation of poly(A)+ RNA and constitutive clustering of nuclear pore complexes. Mol Biol Cell. 1995;6:401–417. doi: 10.1091/mbc.6.4.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Hitomi M, Hu YH, Liu Y, Tartakoff AM. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol Cell Biol. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JD, Davis LI, Fink GR. NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol Biol Cell. 1993;4:209–222. doi: 10.1091/mbc.4.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JD, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink GR. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin beta and reduces Ran GTPase-activating protein (RanGAP) inhibition by karyopherin beta. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Lundblad V. Saccaromyces cerevisiae. In: Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. Boston: John Wiley & Sons; 1997. pp. 13.12.1–13.12.3. [Google Scholar]
- Morehouse H, Buratowski RM, Silver PA, Buratowski S. The importin/karyopherin Kap114 mediates the nuclear import of TATA-binding protein. Proc Natl Acad Sci USA. 1999;96:12542–12547. doi: 10.1073/pnas.96.22.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy TI, Silver PA. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 1999;13:2118–2133. doi: 10.1101/gad.13.16.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente SR. An RNA-export mediator with an essential nuclear export signal. Nature. 1996;383:357–360. doi: 10.1038/383357a0. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrywka NJ, Goldfarb DS. Nuclear export pathways of tRNA and 40S ribosomes include both common and specific intermediates. J Biol Chem. 1995;270:3619–3624. doi: 10.1074/jbc.270.8.3619. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg MO, Moritz M, Woolford JL., Jr Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 1988;2:160–172. doi: 10.1101/gad.2.2.160. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G, Aitchison JD. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Genes Dev. 1997;11:2845–2856. doi: 10.1101/gad.11.21.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A. Laboratory Manual. Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Pante N, Hurt E. Nuclear mRNA export requires complex formation between Mex67p and Mtr2p at the nuclear pores. Mol Cell Biol. 1998;18:6826–6838. doi: 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M, Silver PA. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, Kern H, Hurt EC. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993;72:443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Traglia HM, Atkinson NS, Hopper AK. Structural and functional analyses of Saccharomyces cerevisiae wild-type and mutant RNA1 genes. Mol Cell Biol. 1989;9:2989–2999. doi: 10.1128/mcb.9.7.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay YF, Shankweiler G, Lake J, Woolford JL., Jr Localization of Saccharomyces cerevisiae ribosomal protein L16 on the surface of 60S ribosomal subunits by immunoelectron microscopy. J Biol Chem. 1994;269:7579–7586. [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Wente SR. Gatekeepers of the nucleus. Science. 2000;288:1374–1377. doi: 10.1126/science.288.5470.1374. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic traffic. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP, Blobel G. A new family of yeast nuclear pore complex proteins. J Cell Biol. 1992;119:705–723. doi: 10.1083/jcb.119.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Woolford JL, Jr, Hereford LM, Rosbash M. Isolation of cloned DNA sequences containing ribosomal protein genes from Saccharomyces cerevisiae. Cell. 1979;18:1247–1259. doi: 10.1016/0092-8674(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Xiao Z, McGrew JT, Schroeder AJ, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- Yeh LC, Lee JC. Yeast ribosomal proteins L4, L17, L20, and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA. Biochim Biophys Acta. 1998;1443:139–148. doi: 10.1016/s0167-4781(98)00202-4. [DOI] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol. 1996;133:1141–1152. doi: 10.1083/jcb.133.6.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]