Abstract
Magnetic resonance imaging (MRI) can provide highresolution 3D maps of structural and functional information, yet its use of mapping in vivo gene expression has only recently been explored. A potential application for this technology is to noninvasively image transgene expression. The current study explores the latter using a nonregulatable internalizing engineered transferrin receptor (ETR) whose expression can be probed for with a superparamagnetic Tf-CLIO probe. Using an HSV-based amplicon vector system for transgene delivery, we demonstrate that: 1) ETR is a sensitive MR marker gene; 2) several transgenes can be efficiently expressed from a single amplicon; 3) expression of each transgene results in functional gene product; and 4) ETR gene expression correlates with expression of therapeutic genes when the latter are contained within the same amplicon. These data, taken together, suggest that MRI of ETR expression can serve as a surrogate for measuring therapeutic transgene expression.
Keywords: CLIO, ETR, transgene, amplicon, MRI
Introduction
Severe side effects of many conventional anticancer agents have driven the search for alternative anticancer drugs and biologics to maximize the therapeutic efficacy while minimizing side effects. Among the strategies attempted, combination of biologics and chemotherapeutic agents in the form of transgene therapy and prodrug administration has shown some promise. Prodrug-activating gene therapy, as it has come to be called, attempts to sensitize cells to prodrugs by delivery of exogenous therapeutic genes. This approach has shown positive results in several different laboratory models [1–6] and these successes have spawned clinical trials [7–10]. For gene therapy, regardless of the transgene used, it would be a major advance to be able to monitor the efficiency of transgene expression in target tissues noninvasively, in real time, and at high spatial resolution. This would allow precise determination of productive transduction of diseased tissues, improve understanding of the temporal expression of the transgene, and eventually result in maximum prodrug activation in situ. Several different imaging modalities (e.g., nuclear and optical) are being using to monitor real-time transgene expression noninvasively and in vivo [11–14] and in clinical applications [15]. However, none of these modalities has the high spatial resolution of magnetic resonance imaging (MRI) and would need to be combined with another imaging modality (such as CT scan or MRI) to obtain anatomical information [15]. Therefore, using a single modality for both transgene monitoring and obtaining high-resolution anatomical information would be highly desirable.
Others have used magnetic resonance spectroscopy (MRS) to monitor transgene expression in vivo [16–18]. We have been developing MRI technology for monitoring gene expression and have demonstrated the utility of MRI for imaging gene expression both in vitro and in vivo as well as its utility for mapping transgene expression in tumor sections at microscopic resolution [19]. MRI at clinical field strengths results in high soft tissue contrast anatomical images with voxel resolutions in the 101 to 102 µm3 range, but, contrasted to the other imaging modalities, MRI suffers from lower sensitivity to reporter probes. We have developed an amplification scheme to overcome this limitation and have pioneered the use of MR for in vivo imaging of transgene expression employing the engineered transferrin receptor (ETR) in cells transfected with transgenes ex vivo [19,20]. Recently, we have synthesized novel ETR-targeted MR contrast agents [21] and further improved the sensitivity of MRI for detection of ETR by 16-fold. Here we present a gene therapy vector that contains several transgenes, one of which is useful for prodrug therapy and the other for MRI of gene expression. The work described demonstrates that HSV amplicons can be used to simultaneously transfer several genes into target cells, that the expression of each of the transgenes is tightly correlated to the expression of the contiguous genes, and that the therapeutic transgene can enable effective prodrug-dependent cell killing, whereas the imaging transgene can allow for effective imaging of gene expression in transduced cells.
Materials and Methods
Cell Lines
The Gli36∂EGFR human glioma cells were generated from parental Gli36 cells (a gift from Dr. D. Louis, MGH/HMS) by retroviral transduction of a cDNA coding for a mutant EGF receptor (a gift from Drs. H.J. Huang and Webster K. Cavenee [22]) and subcutaneous passage in nude mice. The Gli36∂EGFR cell line used in these studies was isolated from ground up tumors and maintained in tissue culture in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 0.5 µg/ml puromycin at 378C in 5% CO2.
HSV-1 Amplicon Vectors
The vectors used in this study were genetically engineered HSV-1 amplicons. Three vectors were used for this study, EZ-ETR-P450 (see Figure 1) and two control vectors derived from EZ-ETR-P450, ETR(-)/P450(+), and ETR(+)/P450(-). The parent vector, EZ-ETR-P450, contained three experimental cDNA, the LacZ and CYP2B1 cDNA and also the engineered transferrin receptor (ETR) cDNA under the control of the CAG promoter [23], a gift from Prof. Jun-ichi Miyazaki at Osaka University (Osaka, Japan). The control vector for imaging, ETR(-)/P450(+), contained the β-galactosidase cDNA, LacZ, under control of the HSV-1 IE4/5 promoter and the cDNA for a P450 enzyme, CYP2B1, under control of the CMV promoter, but lacked ETR cDNA. For the chemosensitivity assay, the control vector, ETR(+)/P450(-), contained the β-galactosidase cDNA, LacZ, under control of the HSV-1 IE4/5 promoter and the ETR cDNA under control of the CAG promoter, but no CYP2B1 gene. Each vector also contained 1) the packaging signal and the oriS sequences of HSV-1, and 2) the EBNA-1 and oriP gene of Epstein-Barr virus. The above HSV-1 vectors were propagated and concentrated as described by Saeki et al. [24,25]. Viral titers were determined by measuring LacZ-transducing units on Gli36∂EGFR cells after X-gal staining. ∂EGFR-transfected Gli36 cells were used for these studies as expression of this protein allows for increased tumorigenicity. These cells have been very well characterized and the inclusion of the EGFR does not alter the experiments or conclusion of the paper [26]. Vectors were stored at -80°C prior to their use. Procedures involving viruses were performed in accordance with guidelines issued by the Harvard Office of Biological Safety.
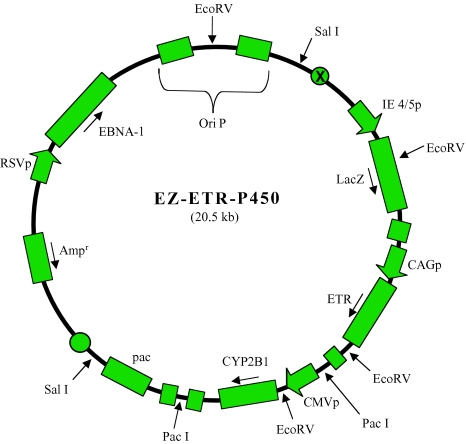
Figure 1.
Schematic of EZ-ETR-P450 amplicon. Three transgene cDNA are contained within the amplicon: 1) the LacZ cDNA under control of an IE 4/5 promoter; 2) the ETR cDNA under control of the CAG promoter; and 3) the cDNA for rat cytochrome c P450 2B1 (CYP) under control of the CMV promoter. Sequences for EBNA-1, to allow for episomal maintenance in mammalian cells, and the pac signal, to allow for packaging into empty HSV virion capsids, are also included (see Materials and Methods).
Analysis of Gene Expression by Western Blotting
Immunoblot analysis was performed with an ECL kit (Amersham, Buckinghamshire, England, UK). Cells were lysed by incubating them for 1 minute on ice in cell lysis buffer (50 mM Tris-HCl, pH 8.0, 250 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% Na deoxycholate, 0.02% sodium azide, and protease inhibitor), and the protein content of the supernatant was quantified with a DC protein assay kit (Bio-Rad, Hercules, CA), according to the manufacturer's instructions. An equal volume of 2x Laemmli sample buffer was added to the supernatant, and this was boiled for 5 minutes. Equal amounts of protein from each extract (50 µg/lane) were separated on 8% SDS polyacrylamide gels and transferred onto nitrocellulose membranes (Hybond-P). After blocking with 5% dry milk in TBS-T (20 mM Tris-HCl, pH7.6, 137 mM NaCl, 0.1% Tween 20), the membranes were incubated with the primary antibody for 1 hour at room temperature. Primary antibodies used were sheep antihuman transferrin receptor antibodies (a gift from Dr. Caroline A. Enns [27]), rabbit anti-β-galactosidase antibody (Chemicon, Temecula, CA), rabbit antirat CYP2B1/2B2 antibody (Chemicon), and antimouse anti-β-actin monoclonal antibody (Sigma, St. Louis, MO) as an internal control. After washing, the membrane was incubated for 1 hour at room temperature with horseradish peroxidase-conjugated secondary antibodies. The secondary antibodies used were swine antigoat IgG (Boehringer Mannheim, Indianapolis, IN), antimouse and antirabbit IgG included with the ECL kit (Amersham). Staining was carried out using the ECL kit according to the manufacturer's instructions. Semiquantitative assessment of Western blot data was performed by scanning the photographic films and then quantifying the intensity of the bands on the film using NIH Image 1.6. Reported data have been corrected for background intensity. Because no preflashing of the film was performed, those data can only be described as semiquantitative.
Immunohistochemical Staining
Fluorescence microscopy was used to confirm the presence or absence of ETR, LacZ, and P450 transgenes overexpression in the infected cells. Cells were plated on 12-mm microscope cover glass (Fisher Scientific, Pittsburgh, PA), placed in 12-well dishes (Falcon; Becton Dickinson, Lincoln Park, NJ), and allowed to grow for 24 hours prior to the start of the experiment. Cells were then infected with amplicon and incubated for another 24 hours, fixed with 2% formaldehyde, rinsed with PBS (0.01 M phosphate buffer, 0.15 M sodium chloride, pH 7.4) and 10% horse serum (Sigma). The expression of LacZ, CYP2B1, or ETR expression was revealed by sequential addition and staining of target-specific primary antibodies followed by fluorescence microscopy at three different wavelengths as follows. First fixed samples were incubated for 1 hour with sheep antirat CYP2B1/2B2 polyclonal antibody (Chemicon) diluted (1:100) in incubation buffer (PBS with 10% horse serum and 0.2% saponin; Sigma). Following incubation, cells were rinsed with the same buffer and incubated with Cy5.5-labeled donkey antisheep IgG (Jackson Immuno Research, West Grove, PA) diluted (1: 200) in incubation buffer. After three washes in incubation buffer, cells were incubated for 1 hour with a mouse antihuman transferrin receptor monoclonal antibody (clone B3/25; Boehringer Mannheim) and a rabbit anti-β-galactosidase polyclonal antibody (Chemicon) diluted (1:200) in incubation buffer (PBS with 10% horse serum and 0.2% saponin; Sigma). Following incubation, cells were rinsed with the same buffer and incubated with FITC-labeled goat antimouse IgG (Southern Biotechnology Associates, Birmingham, AL) and rhodamine-labeled antirabbit IgG (Chemicon) diluted (1:500 and 1:200, respectively) in incubation buffer. After three washes, cover slips were mounted on the microscopy slides (Fisher Scientific) using Fluoromount-G (Southern Biotechnology Associates). Cells were examined with a fluorescence microscope equipped with triple fluorescent filter sets and a SenSys CCD camera connected to a Macintosh computer [20].
In Vitro Therapeutic Efficacy
To show that transduction of the CYP2B1 gene sensitizes cells to cyclophosphamide (CPA), Gli36∂EGFR human glioma cells were seeded in a 24-well plate at a density of 5x104 cells/well, cultured for 24 hours, and then virus administered; EZ-ETR-P450 (MOI: 0, 5) or ETR(+)/P450(x) (MOI: 5) as a control. Twenty-four hours after infection, medium-containing CPA at various concentrations ranging from 1 to 10,000 µM was added. Three days after CPA administration, the percentage of surviving cells was determined to evaluate cell chemosensitivity to CPA. Live cells were quantified using a hemocytometer.
MRI
To determine if we could detect increased cellular uptake of Tf-targeted iron oxide particles by cells infected with ETR(+) amplicon, human glioma cells were infected with EZ-ETR-P450 amplicon or ETR(-)/P450(+) amplicon, exposed to media containing the Tf-S-S-CLIO conjugate for 1 hour at 37°C, washed with HBSS, trypsinized, and rewashed in DMEM supplemented with 10% FBS. Cells were then collected by centrifugation and cell pellets resuspended in culture medium and transferred to 250-µl tubes. To form uniform pellets, the cells were again centrifuged at 700 rpm for 2 minutes (Sorvall 7 RT; Kendro Laboratory Products, Newtown, CT). To prevent drying and susceptibility artifacts, the supernatant was not removed. The tubes were placed into a water bath at room temperature for MRI. MRI was performed with a clinical 1.5-T superconducting magnet (Signa 5.0; GE Medical Systems, Milwaukee, WI) using a 5-in. surface coil. The imaging protocol consisted of a multiecho sequence (SE, TR 3000 milliseconds, variable TE 16-100 milliseconds). The 1.9mm imaging slice was carefully placed to avoid partial volume effects. At a field of view of 8 cm2 and a 256x256 imaging matrix, each voxel had a size of 0.186 mm3. Tf-SS-CLIO imaging agent used for these studies was synthesized as described in Högemann et al. [21]. The stock of Tf-S-S-CLIO contained approximately four Tf molecules per CLIO and the relaxivities of the particles were measured using a 0.47-T table top minispec (R1: 32 [mM s]-1; R2: 146 [mM s]-1).
Results
The goal of these studies was to facilitate noninvasive, real-time MRI of therapeutic gene transfer and expression. For these studies, we constructed a gene therapy vector that would allow for simultaneous expression of multiple cDNA, coding for therapeutic and imaging transgenes that would enable “surrogate” noninvasive real-time imaging of therapeutic transgene expression. The high capacity of HSV amplicons compared to most other viral vectors (allowing up to 150 kb of foreign DNA to be delivered) provides a suitable vector with desirable characteristics for the studies performed here [28]. We thus sought to determine if an imagable marker gene delivered by an HSV amplicon also expressing a therapeutic transgene could serve as a surrogate for monitoring therapeutic transgene expression in glioma cells.
Figure 1 provides a schematic of this construct named EZ-ETR-P450. Three transgene cDNA are expressed from this vector: 1) the β-galactosidase (LacZ) cDNA, 2) the ETR cDNA, and 3) the cDNA for rat cytochrome P450 2B1 (CYP2B1). Sequences for EBNA-1 and OriP from Epstein-Bar virus (EBV), to allow for episomal maintenance in mammalian cells, and the OriS and pac signals from HSV-1, to allow for DNA replication and packaging of the plasmid in the presence of HSV helper functions, are also included [28].
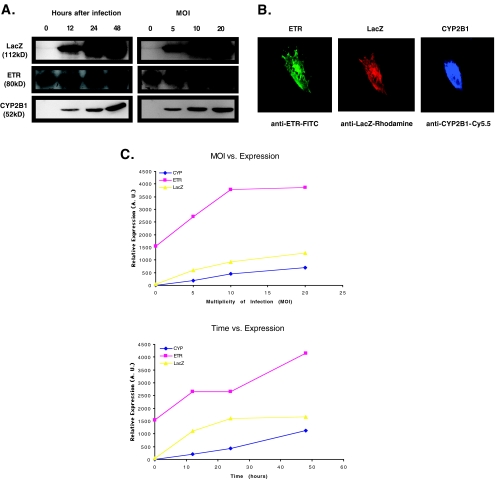
To show that this HSV/EBV hybrid amplicon can express protein from all three cDNA simultaneously, human glioma Gli36∂EGFR cells were infected and expression of each of the transgenes was monitored by Western blot analysis (Figure 2A). Because Gli36∂EGFR cells express the endogenous form of the human transferrin receptor, the blot in Figure 2A also shows a band present in the control lanes (i.e., time point 0 or MOI of 0). However, there is a clear increase in the signal from this band after amplicon infection, indicating increased expression from the ETR cDNA in the amplicon. To determine if variations in gene expression would be reflected similarly by all three gene products, different levels of gene expression were obtained by changing either viral dose (MOI) or time after viral infection. Analysis under these conditions showed that expression of all three gene products was tightly correlated at different levels of gene expression (Figure 2, A and C). Inspection of the semiquantitative analysis of Western blot data in Figure 2C reveals that expression of CYP2B1, ETR, and LacZ increases similarly and correlates with both MOI and time after infection. These results thus indicate that multiple transgenes can be expressed from amplicons infecting glioma cells and that their expression is tightly correlated at different levels of expression.
Figure 2.
(A) Correlation of transgene expression in cells infected with EZ-ETR-P450 amplicon. Gli36∂EGFR cells were infected with amplicon containing three transgenes. At the indicated times or MOI, cells were harvested and lysates subjected to analysis by Western blot to confirm expression of transgenes. Blots were treated and signal-analyzed using NIH image as described in Materials and Methods. These data are plotted in (C). When protein expression was assessed at varying times after infection, a single MOI of 1 was used (representative data, n=3). (B) Expression of three cDNA after infection with EZ-ETR-P450 amplicon in each cell. Cells were fixed and analyzed using three different antibodies specific for each protein product. Complexes were then visualized using a second antibody conjugated to one of three different fluorochromes, each emitting at different wavelengths. All three gene products were expressed in all cells observed. Presented is a representative cell from an experiment (n=3). (C) Graphical analysis of Western blot from (A). The ECL signal from (A) was captured on to photographic film, scanned into the computer, and the resulting electronic figure analyzed using NIH Image.
Infected cells were also analyzed immunohistochemically to determine if expression of all three transgenes occurred in the same cell. Figure 2B shows microscopic analysis of a representative human glioma cell from a culture infected with the amplicon, EZ-ETR-P450. All three transgenes were expressed in the same cell simultaneously. Control cells incubated without primary antibody confirmed specificity of signal. The distribution of transgene targets is in agreement with previous reports describing the subcellular distribution of the transferrin receptor [29,30] and of CYP2B1 [31,32].
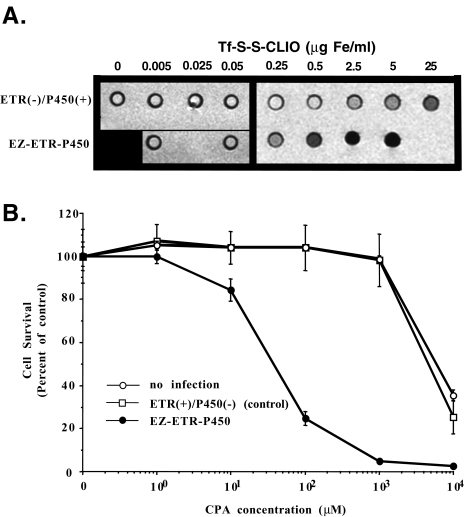
Although we had previously shown that cells stably transfected with ETR could be visualized by MRI [19–21], we have not determined if cells infected by an ETR-expressing viral vector could also be imaged by MRI. Human Gli36∂EGFR glioma cells were infected with either the EZ-ETR-P450 vector [ETR(+)/P450(+)] or with a control vector [ETR(-)/P450(+)], identical to EZ-ETRP450 but lacking ETR cDNA, and then treated in culture with different concentrations of the imaging probe, Tf-S-S-CLIO [21]. It is evident that, at Tf-S-S-CLIO concentrations of 0.5 µg/ml, changes in MRI signal intensity are detectable in the wells containing ETR(+)/P450(+)-infected cells (Figure 3A). Doses of probe that are at least 10 times higher are required to produce the same change in signal intensity in the wells containing control ETR(-)/P450(+)-infected cells. Presumably this is due to the uptake of Tf-S-S-CLIO through the endogenous TfR. Additionally, we demonstrated that increasing the MOI during treatment of the cells with amplicon resulted in greater changes in MR signal intensities. In experiments where cells were infected with amplicons at an MOI of 20 vs an MOI of 4, a 1.8-fold change in T2 was measured and was in good accordance with the difference in ETR expression measured by Western blots (data not shown and Figure 2C). These data thus show that ETR delivery (and relative overexpression) into cells by a gene transfer vector can be imaged using MRI.
Figure 3.
(A) Detection of increased cellular uptake of Tf iron oxide particles by cells infected with EZ-ETR-P450 amplicon. Twenty-four hours after infection, the cells were incubated for 1 hour with increasing concentrations of Tf-S-S-CLIO contrast agent, washed, pelleted into tissue culture tubes, and imaged in a clinical GE 1.5-T MRI. T2-weighted MRI of wells containing cell pellets treated with Tf-S-S-CLIO in culture shows that EZ-ETR-P450-infected cells show a significant signal decrease at 0.5 µg/ml iron compared to cells infected with the ETR-negative control vector, ETR(-)/P450(+). To conserve probe and other reagents, selective informative data points were measured for the EZ-ETR-P450-infected cells. The black square in the lower left corner is a place holder (n=2, representative experiment shown). (B) Infection of Gli36∂EGFR cells with EZ-ETR-P450 confers prodrug sensitivity. Gli36∂EGFR cells were infected with EZETR-P450 or control amplicon [ETR(+)/P450(-)] in tissue culture and cell survival 72 hours after CPA treatment was quantified by cell counting. EZ-ETRP450-infected cells have significantly increased drug sensitivity compared to uninfected and control infected cells (n=3).
To confirm that therapeutic gene expression measured in Western blots yielded functional gene product, the ability of the expressed gene product to metabolize CPA was functionally determined. Figure 3B shows that there is a significant dose-dependent increase in cell killing (chemosensitivity) in cells treated with CPA and infected with amplicons containing the CYP2B1 cDNA compared to uninfected cells or cells infected with control amplicons [(ETR(+)/P450(-)]. Additionally, the β-galactosidase enzyme was functional as the infected cells stained “blue” upon addition of the chromophore, X-gal (data not shown). These data suggest that the LacZ cDNA could be replaced by another therapeutic gene, making it possible to deliver and express multiple prodrug-activating enzymes from a single amplicon, potentially increasing efficacy of the gene therapy.
Discussion
The genomic revolution is enabling a more comprehensive understanding of the molecular basis of disease and, for the first time, molecular therapeutic approaches to correct “defects” can be attempted. Further, other advances have allowed investigators to express xenogenes in cells, imparting additional functions to the cells. Implicit in the evolution of these approaches is the development of efficient gene delivery vehicles for in vivo delivery of “corrective/additional” genes and the need to identify ways of monitoring gene delivery and expression noninvasively and in vivo.
Here we demonstrate that a high-capacity, high-efficiency HSV-based gene therapy vector is amendable for noninvasive imaging of therapeutic transgene expression. We reasoned that if relative expression of the different transgenes expressed from a single vector was tightly correlated and varied with the level of viral transduction, we would be able to use an imagable marker gene to provide readout on the expression of the other genes (e.g., therapeutic transgenes) expressed from within the same viral vector. Therefore, we designed and produced a herpes amplicon vector that expressed three different transgenes (CYP2B1, ETR, and LacZ) under control of separate and individual promoters (see Figure 1). The results presented here are the first to demonstrate that expression of multiple gene products from a single gene therapy vector can be monitored using MRI. These data demonstrate that it is possible to: 1) use different constitutive promoters to drive the expression of several different transgenes from a single amplicon; 2) that the expression of each of the transgenes is closely correlated to the others at different levels of expression; 3) that individual cells express all the gene products simultaneously; and 4) that the function of the individual transgenes is maintained.
By combining the HSV amplicon system with the ETR/Tf-S-S-CLIO imaging system, we have taken advantage of the qualities of each to generate a gene therapy vector with the capacity to efficiently deliver a large number of genes into target tissues whose expression can be surrogately monitored noninvasively by MRI. The use of the HSV amplicon vectors for devising this system has several significant advantages over other viral transduction systems allowing for the largest versatility in applying this imaging system to biological questions. First, the amplicons have an enormous capacity to carry exogenous DNA (up to 150 kb of added DNA), making the delivery of several different therapeutic or imaging transgenes (and use of other imaging modalities) possible [33]. Second, generation of amplicons containing novel gene products is relatively straightforward [33], Third, HSV-encapsulated amplicons have a wide tropism (crossspecies), efficiently transducing both primary cells and cell lines in vitro, and transducing a variety of tissues in vivo [24,34]. Finally, packaged amplicons can be generated free of any contaminating replication-competent helper virus [25]. In addition to the attributes of the amplicon, imaging by MRI has several useful features, including little to no toxicity in vitro or in vivo for the parent iron oxide particles, relatively rapid in vivo clearance of the imaging agent, iron oxide nanoparticles [35], and high anatomical resolution of MRI. These features potentially would permit excellent detection of gene expression by MRI and are very convenient in applications in which repeated evaluation of gene expression over short periods of time is needed, potentially allowing for repeated MRI in vivo as well as in vitro.
When using ETR as a surrogate marker for expression of other transgenes, a potential limitation might be that selective pressures in vitro differentially modulate expression of the ETR reporter gene and the other transgenes. To assess such interactions, we performed semiquantitative Western Blots and qualitative immunohistochemical analysis to demonstrate that expression of all three transgene products appears to be linearly related to viral transduction and that all three are expressed in all observed cells, simultaneously. Therefore, the strategy to use different promoters to drive each transgene within the same vector is apparently not susceptible to the in vitro variations tested here (i.e., time after infection or MOI). These data also suggest that the use of IRES-containing bicistronic mRNA [36] is not necessary to achieve simultaneous and linear expression ratios between gene products, in agreement with other studies using separate adenoviral vectors [37]. Furthermore, the ability to employ strong promoters to drive expression of both the therapeutic transgene and the imaging transgene is advantageous compared to bicistronic mRNA, which often show several-fold reduction in expression from genes placed downstream of the IRES. This vector system may therefore prove to be advantageous when high expression levels of both therapeutic and imaging transgenes are required or when the expression of two or more transgenes is required.
We previously demonstrated that the imaging transgene, ETR, was effective both in vivo and in vitro for MRI of transgene expression when combined with targeted iron oxide nanoparticles. Here we demonstrate that using a viral transduction system, the ETR transgene can be successfully transferred into tissue culture cells, resulting in a ignificant change in T2-weighted MR signal intensity (Figure 3A). The change in signal was dependent on the time after infection as well as MOI (data not shown), suggesting that imaging the ETR transgene faithfully represents transduction and expression efficiency as monitored by Western blotting (Figure 2). These characteristics would make the ETR transgene useful for following gene transfer in vivo during gene therapy protocols. We also demonstrated that infection of cells with the EZ-ETR-P450 amplicon resulted in increased sensitivity to prodrug administration (Figure 3B). Taken together, these data demonstrate the utility of this amplicon for imaging therapeutic gene transfer and function.
Finally, because of the ability of this modality to generate high-resolution anatomical images, this system may supplement or replace systems, which use reporter genes in transgenic animals to analyze tissue-specific promoters sequences and/or effects of drugs or environment on gene expression. Currently, nuclear and optical modalities are limited to detecting changes in transgene expression without being able to ascribe the tissue, or individual cells responsible for signal generation. Using these (or other) MRI transgenes may make it possible to assign gene expression to small groups of cells within living animals and clearly define tissues or cells permissive for transgene expression. We are now performing studies aimed at moving these therapeutic imaging vectors into in vivo studies including pharmacokinetic analysis of contrast agents and HSV amplicons.
Acknowledgements
The authors would like to acknowledge the generosity of Cavanee and Huang (UCSD) for the gift of the ∂EGFR cDNA; Caroline A. Enns (Oregon Health Sciences University) for supplying antibodies against the TfR; David Louis (MGH/Harvard Medical School) for the parental Gli36 cell line; and Jun-ichi Miyazaki (Osaka University, Osaka, Japan) for providing CAG promoter sequences. Dagmar Högemann was supported by a fellowship from the Deutsche Forschungsgemeinschaft (Germany).
References
- 1.Rainov NG, Sena-Esteves M, Fraefel C, Dobberstein KU, Chiocca EA, Breakefield XO. A chimeric fusion protein of cytochrome CYP4B1 and green fluorescent protein for detection of pro-drug activating gene delivery and for gene therapy in malignant glioma. Adv Exp Med Biol. 1998;451:393–403. doi: 10.1007/978-1-4615-5357-1_61. [DOI] [PubMed] [Google Scholar]
- 2.Runnebaum IB. Basics of cancer gene therapy. Anticancer Res. 1997;17:2887–2890. [PubMed] [Google Scholar]
- 3.Zwacka RM, Dunlop MG. Gene therapy for colon cancer. Hematol Oncol Clin North Am. 1998;12:595–615. doi: 10.1016/s0889-8588(05)70010-1. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda K, Ichikawa T, Wakimoto H, Silver J, Deisboeck T, Finkelstein D, Harsh GI, Louis D, Bartus R, Hochberg F, Chiocca E. Oncolytic virus therapy of multiple tumors in the brain requires suppression of innate and elicited antiviral responses. Nat Med. 1999;5:881–887. doi: 10.1038/11320. [DOI] [PubMed] [Google Scholar]
- 5.Carter BS, Zervas NT, Chiocca EA. Neurogenetic surgery: current limitations and the promise of gene- and virus-based therapies. Clin Neurosurg. 1999;45:226–246. (in process citation) [PubMed] [Google Scholar]
- 6.Chung RY, Saeki Y, Chiocca EA. B-myb promoter retargeting of herpes simplex virus gamma34. gene-mediated virulence toward tumor and cycling cells. J Virol. 1999;73:7556–7564. doi: 10.1128/jvi.73.9.7556-7564.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Markert J. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase 1 trial. Gene Ther. 2000;7:867–874. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 8.Sandmair AM, Loimas S, Puranen P, Immonen A, Kossila M, Puranen M, Hurskainen H, Tyynela K, Turunen M, Vanninen R, Lehtolainen P, Paljarvi L, Johansson R, Vapalahti M, Yla-Herttuala S. Thymidine kinase gene therapy for human malignant glioma, using replication-deficient retroviruses or adenoviruses. Hum Gene Ther. 2000;11:2197–2205. doi: 10.1089/104303400750035726. [DOI] [PubMed] [Google Scholar]
- 9.Klatzmann D, Cherin P, Bensimon G, Boyer O, Coutellier A, Charlotte F, Boccaccio C, Salzmann JL, Herson S. A phase I/II dose-escalation study of herpes simplex virus type 1 thymidine kinase “suicide” gene therapy for metastatic melanoma. Study group on gene therapy of metastatic melanoma. Hum Gene Ther. 1998;9:2585–2594. doi: 10.1089/hum.1998.9.17-2585. (in process citation) [DOI] [PubMed] [Google Scholar]
- 10.Harsh GR, Deisboeck TS, Louis DN, Hilton J, Colvin M, Silver JS, Qureshi NH, Kracher J, Finkelstein D, Chiocca EA, Hochberg FH. Thymidine kinase activation of ganciclovir in recurrent malignant gliomas: a gene-marking and neuropathological study. J Neurosurg. 2000;92:804–811. doi: 10.3171/jns.2000.92.5.0804. [DOI] [PubMed] [Google Scholar]
- 11.Gambhir SS, Herschman HR, Cherry SR, Barrio JR, Satyamurthy N, Toyokuni T, Phelps ME, Larson SM, Balatoni J, Finn R, Sadelain M, Tjuvajev J, Blasberg R. Imaging transgene expression with radionuclide imaging technologies. Neoplasia (New York) 2000;2:118–138. doi: 10.1038/sj.neo.7900083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tjuvajev JG, Chen SH, Joshi A, Joshi R, Guo ZS, Balatoni J, Ballon D, Koutcher J, Finn R, Woo SL, Blasberg RG. Imaging adenoviral-mediated herpes virus thymidine kinase gene transfer and expression in vivo. Cancer Res. 1999;59:5186–5193. [PubMed] [Google Scholar]
- 13.Wunderbaldinger P, Bogdanov A, Weissleder R. New approaches for imaging in gene therapy. Eur J Radiol. 2000;34:156–165. doi: 10.1016/s0720-048x(00)00196-0. [DOI] [PubMed] [Google Scholar]
- 14.Bremer C, Weissleder R. In vivo imaging of gene expression. Acad Radiol. 2001;8:15–23. doi: 10.1016/s1076-6332(03)80739-0. (see comments) [DOI] [PubMed] [Google Scholar]
- 15.Jacobs A, Voges J, Reszka R, Lercher M, Gossmann A, Kracht L, Kaestle C, Wagner R, Wienhard K, Heiss W. Positron-emission tomography of vector-mediated gene expression in gene therapy for gliomas. Lancet. 2001;358:727–729. doi: 10.1016/s0140-6736(01)05904-9. [DOI] [PubMed] [Google Scholar]
- 16.Rehemtulla A, Hall DE, Stegman LD, Prasad U, Chen G, Bhojani MS, Chenevert TL, Ross BD. Molecular imaging of gene expression and efficacy following adenoviral-mediated tumor gene therapy. Mol Imaging. 2002;1:43–55. doi: 10.1162/15353500200200005. [DOI] [PubMed] [Google Scholar]
- 17.Kristensen CA, Askenasy N, Jain RK, Koretsky AP. Creatine and cyclocreatine treatment of human colon adenocarcinoma xenografts: 31P and 1H magnetic resonance spectroscopic studies. Br J Cancer. 1999;79:278–285. doi: 10.1038/sj.bjc.6690045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegman LD, Rehemtulla A, Beattie B, Kievit E, Lawrence TS, Blasberg R, Tjuvajev J, Ross BD. Noninvasive quantitation of cytosine deaminase transgene expression in human tumor xenographs with in vivo magnetic resonance spectroscopy. Proc Natl Acad Sci. 1999;96:9821–9826. doi: 10.1073/pnas.96.17.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6:351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 20.Moore A, Basilion J, Chiocca A, Weissleder R. Measuring transferrin receptor gene expression by NMR imaging. Biochim Biophys Acta. 1998;1402:239–249. doi: 10.1016/s0167-4889(98)00002-0. [DOI] [PubMed] [Google Scholar]
- 21.Högemann D, Josephson L, Weissleder R, Basilion JP. Improvement of MRI probes to allow efficient detection of gene expression. Bioconjug Chem. 2000;11:941–946. doi: 10.1021/bc000079x. (in process citation) [DOI] [PubMed] [Google Scholar]
- 22.Nagane M, Coufal F, Lin H, Bogler O, Cavenee WK, Huang HJ. A common mutant epidermal growth factor receptor confers enhanced tumorigenicity on human glioblastoma cells by increasing proliferation and reducing apoptosis. Cancer Res. 1996;56:5079–5086. [PubMed] [Google Scholar]
- 23.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 24.Saeki Y, Ichikawa T, Saeki A, Chiocca EA, Tobler K, Ackermann M, Breakefield XO, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 25.Saeki Y, Fraefel C, Ichikawa T, Breakefield XO, Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol Ther J Am Soc Gene Ther. 2001;3:591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- 26.Nishikawa R, Ji XD, Harmon RC, Lazar CS, Gill GN, Cavenee WK, Huang HJ. A mutant epidermal growth factor receptor common in human glioma confers enhanced tumorigenicity. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rutledge EA, Gaston I, Root BJ, McGraw TE, Enns CA. The transferrin receptor cytoplasmic domain determines its rate of transport through the biosynthetic pathway and its susceptibility to cleavage early in the pathway. J Biol Chem. 1998;273:12169–12175. doi: 10.1074/jbc.273.20.12169. [DOI] [PubMed] [Google Scholar]
- 28.Fraefel C, Breakefield XO, Jacoby D. In: HSV-1 Amplicon in Gene Therapy for Neurological Disorders and Brain Tumors. Chiocca E, Breakefield X, editors. Totowa, NJ: Humana Press; 1998. pp. 63–82. [Google Scholar]
- 29.Eriksson LC, Torndal UB, Andersson GN. The transferrin receptor in hepatocyte nodules: binding properties, subcellular distribution and endocytosis. Carcinogenesis. 1986;7:1467–1474. doi: 10.1093/carcin/7.9.1467. [DOI] [PubMed] [Google Scholar]
- 30.Lamb JE, Ray F, Ward JH, Kushner JP, Kaplan J. Internalization and subcellular localization of transferrin and transferrin receptors in HeLa cells. J Biol Chem. 1983;258:8751–8758. [PubMed] [Google Scholar]
- 31.Wei MX, Tamiya T, Chase M, Boviatsis EJ, Chang TK, Kowall NW, Hochberg FH, Waxman DJ, Breakefield XO, Chiocca EA. Experimental tumor therapy in mice using the cyclophosphamide-activating cytochrome P450 2B1 gene. Hum Gene Ther. 1994;5:969–978. doi: 10.1089/hum.1994.5.8-969. [DOI] [PubMed] [Google Scholar]
- 32.Monier S, Van Luc P, Kreibich G, Sabatini DD, Adesnik M. Signals for the incorporation and orientation of cytochrome P450 in the endoplasmic reticulum membrane. J Cell Biol. 1988;107:457–470. doi: 10.1083/jcb.107.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wade-Martins R, Smith ER, Tyminski E, Chiocca EA, Saeki Y. An infectious transfer and expression system for genomic DNA loci in human and mouse cells. Nat Biotechnol. 2001;19:1067–1070. doi: 10.1038/nbt1101-1067. [DOI] [PubMed] [Google Scholar]
- 34.Geller AI, Breakefield XO. A defective HSV-1 vector expresses Escherichia coli beta-galactosidase in cultured peripheral neurons. Science. 1988;241:1667–1669. doi: 10.1126/science.241.4873.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weissleder R, Stark DD, Engelstad BL, Bacon BR, Compton CC, White DL, Jacobs P, Lewis J. Superparamagnetic iron oxide: pharmacokinetics and toxicity. AJR, Am J Roentgenol. 1989;152:167–173. doi: 10.2214/ajr.152.1.167. [DOI] [PubMed] [Google Scholar]
- 36.Tjuvajev JG, Joshi A, Callegari J, Lindsley L, Joshi R, Balatoni J, Finn R, Larson SM, Sadelain M, Blasberg RG. A general approach to the non-invasive imaging of transgenes using cis-linked herpes simplex virus thymidine kinase. Neoplasia (New York) 1999;1:315–320. doi: 10.1038/sj.neo.7900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaghoubi SS, Wu L, Liang Q, Toyokuni T, Barrio JR, Namavari N, Satyamurthy N, Phelps ME, Herschman HR, Gambhir SS. Direct correlation between positron emission tomographic images of two reporter genes delivered by distinct adenoviral vectors. Gene Ther. 2001;8:1072–1080. doi: 10.1038/sj.gt.3301490. [DOI] [PubMed] [Google Scholar]