Abstract
Human chromosome 4 was previously shown to elicit features of senescence when introduced into cell lines that map to complementation group B for, senescence including HeLa cells. Subsequently, a DNA segment encoding the pseudogene Mortality Factor 4 (MORF4) was shown to reproduce some of the effects of the intact chromosome 4 and was suggested to be a candidate mortality gene. We have identified multiple MORF4 alleles in several cell lines and tissues by sequencing and have failed to detect any cancer-specific mutations in three of the complementation group B lines (HeLa, T98G, and J82). Furthermore, MORF4 was heterozygous in these lines. These results question whether MORF4 is the chromosome 4 mortality gene. To map other candidate mortality gene(s) on this chromosome, we employed microcell-mediated monochromosome transfer to introduce either a complete copy, or defined fragments of the chromosome into HeLa cells. The introduced chromosome 4 fragments mapped the mortality gene to a region between the centromere and the marker D4S2975 (4q27), thus excluding MORF4, which maps to 4q33-q34.1. Analysis of microsatellite markers on the introduced chromosome in 59 immortal segregants identified a frequently deleted region, spanning the markers BIR0110 and D4S1557. This defines a new candidate interval of 130 kb at 4q22-q23.
Keywords: gene mapping, microcell-mediated monochromosome transfer, chromosome 4, MORF4, complementation group B
Abbreviations: MORF4, Mortality Factor 4; MMCT, microcell-mediated monochromosome transfer; MPD, mean population doublings
Introduction
Microcell-mediated monochromosome transfer (MMCT) experiments suggest that several cancer mortality genes may exist [6,11,13,15,19,21,22,26–28,31,33–35,38,41,44], but in most cases, the genes responsible have not been identified, and with the exception of those that downregulate telomerase [13,28,38], their function is unknown. Chromosome 4 has been shown to carry a mortality gene(s) for cell lines mapping to mortality complementation group B [26] and a candidate for one of these genes, Mortality Factor 4 (MORF4), was described recently [6].
We investigated the candidacy of the MORF4 mortality gene and found no evidence for cancer-specific alterations in its coding sequence [9] and have extended this observation to include the complementation group B cell lines (HeLa, T98G, and J82). We, therefore, searched for other candidate regions of chromosome 4 by a combination of MMCT and deletion analysis. Our investigations revealed a candidate interval of approximately 130 kb for the chromosome 4 mortality gene.
Materials and Methods
Cell Culture
HeLa, T98G, and 143B cells were cultured in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS). Mouse A92 cells containing a single copy of human chromosome 4 or chromosome 4 fragments carrying the selectable markers for hygromycin B and G418 resistance (HyTK4/4i24/4i30/4i33 [14,17] was grown in DMEM plus 10% FBS and 800 U/ml hygromycin B; Calbiochem Novachem, UK).
DNA Extraction
DNA was extracted from typically 106 to 107 cultured cells using the QIAamp tissue kit (QIAgen, Crawley, West Sussex, UK) according to the manufacturer's “blood and body fluid” protocol.
MORF4 Sequencing
A 1.5-kb genomic fragment containing the MORF4 sequence was amplified, purified, and sequenced as previously described [9].
MMCT
Microcells were prepared as described previously [14] and the purified microcell pellet was then resuspended in 50 µg/ml phytohemagglutinin in serum-free medium and applied to 70% to 80% confluent recipient cells. Plates were left for 2 hours to allow attachment of microcells before fusion to the recipient cells in the presence of 42.5% polyethylene glycol (PEG 1000; Sigma, Poole, Dorset, UK) and 8.5% DMSO in serum-free medium. Following a 24-hour recovery period in complete medium, fused cells were replated in standard medium and the following day placed under selection in medium containing 300 (HeLa) and 500 (143B) U/ml hygromycin B. Doses represented the minimum required for complete killing of mock-fused cultures of each line and hence the minimum dose required. The careful selection of drug dose avoids artifacts due to incomplete hygromycin resistance. Those colonies, which overcame selection in hygromycin B, were picked and cultured for a further 5 to 6 weeks before harvesting and extracting DNA. An immortal segregant chromosome 4 hybrid culture was classified as one that proliferated through at least 25 population doublings and the control hybrids were cultured for the same number of doublings prior to molecular genetic analysis. The immortal segregants were all maintained in the appropriate dose of hygromycin.
Chromosome Painting
Metaphase chromosome spreads were prepared essentially as described by Ref. [14]. Metaphase spreads were hybridized with a chromosome 4 probe prepared from flow-sorted chromosome 4 DNA and chromosome painting carried out as described previously [12].
Marker Alleles Analysis
Total reaction volumes were 10 µl containing 40 ng of genomic DNA, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, 10% dimethylsulphoxide, 1 µM of each oligonucleotide primer, 200 µM concentrations of each deoxynucleotide triphosphate, 0.4 µl of [α32P]dCTP (24.7 kBq/µl), and 0.5 U of Taq polymerase (Advanced Biotechnologies, Epsom, Surrey, UK). Reactions were subjected to an initial denaturation phase of 5 minutes at 94°C, followed by 30 cycles of 30 seconds at 94°C, 30 seconds at 55 to 65°C, and 30 seconds at 72°C, and a final extension phase of 7 minutes at 72°C. Radiolabeled samples were electrophoresed on 4% to 10% polyacrylamide gels under denaturing conditions. Following a drying step, gels were then exposed to X-ray film to visualize the resolved reaction products.
Statistics
The Fisher's exact test was used for the comparison of allele deletion frequencies in different groups of cells. In the case of the immortal segregant colonies resulting from the MMCT of chromosome 4, the allele deletion frequencies of the transferred, wild-type chromosome 4 were compared with the deletion frequencies of the same allele in a panel of control chromosome 4 hybrids. The control panel of hybrids comprised one HeLa-chromosome 4 hybrid that displayed a mortal phenotype and eleven 143B-chromosome 4 hybrids that all retained an immortal phenotype. All of these control fusions therefore constitute a measurement of the spontaneous loss of each allele of the transferred chromosome that may not be consistent throughout the length of the chromosome.
Results
MORF4 is Normal and Heterozygous in Complementation Group B Cell Lines
We have sequenced MORF4 in three complementation group B lines — HeLa, J82, and T98G. We found no mutations or deletions. Any sequence differences from the reported MORF4 sequence [6] were at polymorphic loci previously identified in a study of squamous cell carcinoma lines [9] and are not tumor line-specific as they were also all found in normal fibroblasts. Table 1 lists the base present at each of the polymorphic loci of the MORF4 sequence in the different lines. At base positions 108 and 115 in HeLa, and at position 49 in J82 and T98G, two different alleles are present in the sequence. Therefore, each of the complementation group B lines we have examined is heterozygous at the MORF4 locus.
Table 1.
Polymorphic Loci in the MORF4 Gene.
| Allele at Polymorphic Loci | |||||||
| 49 | 108 | 115 | 428 | 615 | 730 | 865 | |
| Tumor Lines | |||||||
| HeLa | G | T | A/G | A/G | T | T | G |
| J82 | A/G | T | A | G | C | A | A |
| T98G | A/G | T | A | G | C | A | A |
| Fibroblasts | |||||||
| 31F | A/G | C/T | A | G | C/T | A/T | A/G |
| 63F | G | T | A/G | A/G | T | T | G |
Nucleotide(s) at the seven polymorphic loci in the MORF4 gene (base positions as previously described in Ref. [9]) in three complementation group B lines. Each of the lines is heterozygous in at least one position. Allelotypes from normal fibroblast controls containing the same polymorphisms are shown for comparison.
Introduction of Chromosome 4/Chromosome 4cen-q27 Containing Fragments Elicits a Mortal Phenotype in HeLa Cells
We introduced, using MMCT, an intact copy of chromosome 4 into either HeLa cells or, as a control, the complementation group C line 143B. Following selection, 44% (78/178) of the HeLa-chromosome 4 hybrid clones exhibited a mortal phenotype following a variable delay of approximately 4 to 25 mean population doublings (MPD). The remaining 56% of the hybrid clones continued to grow vigorously beyond at least 25 MPD and were considered to have retained an immortal phenotype. We performed chromosome painting experiments to confirm that hybrid clones displaying the immortal phenotype had retained an extra introduced copy of chromosome 4 (Figure 1). Analysis of metaphase spreads from six different hybrid clones revealed an average of four copies of chromosome 4 per cell compared to an average of three for the control HeLa metaphases. Cells displaying the mortal phenotype took on a flattened morphology with an enlarged cytoplasm, reminiscent of senescent epithelial cells (Figure 2). These cells were often multinucleate and stained positive for the senescence-associated β-galactosidase activity (data not shown). All (11/11) of the 143B-chromosome 4 hybrids retained vigorous growth beyond 25 MPD and were judged immortal by this criterion. To identify regions of chromosome 4 responsible for the restoration of mortality effect, we introduced defined fragments of chromosome 4 (described previously in Ref. [17]) into HeLa cells. All three fragments tested induced a mortal phenotype at around the same frequency as the whole chromosome (Table 2). The introduced chromosome 4 fragments share the region between the markers D4S2978 and D4S2975 in common (approximately 4cen-q27), therefore mapping the activity to this interval.
Figure 1.
Chromosome 4-specific painting in HeLa and HeLa-chromosome 4 hybrid cells. Metaphase spreads from (A) HeLa-chromosome 4 clone and (B) untransferred HeLa cells hybridized with a chromosome 4-specific paint, demonstrating the gain of an extra copy of chromosome 4 in hybrid cells. Arrows indicate painted chromosomes.
Figure 2.
Mortal clones develop a phenotype reminiscent of senescence. Phase contrast images of cells exhibiting (A) the characteristic flattened morphology and enlarged cytoplasm of mortal hybrid clones and (B) cells from an immortal hybrid clone. Scale bar indicates 100 µm.
Table 2.
Effect of Introduction of Chromosome 4/4 Fragments into HeLa.
| MMCT Hybrid Clones | ||||
| Mortal | Immortal | |||
| HeLa-4 | 78 | 44% | 100 | 56% |
| HeLa-4i24 | 15 | 44% | 19 | 56% |
| HeLa-4i32 | 5 | 29% | 12 | 71% |
| HeLa-4i33 | 1 | 33% | 2 | 66% |
Proportions of clones exhibiting mortal or immortal phenotypes following the introduction of chromosome 4 or three different chromosome 4 fragments into HeLa cells.
Deletions on the Introduced Chromosome Identify a Candidate Interval Between the Markers BIR0110 and D4S1557
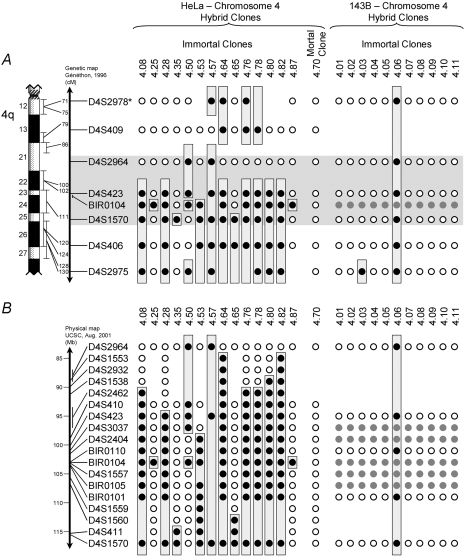
We examined hybrid clones that retain an immortal phenotype for the loss of material from the introduced chromosome 4, as a common region of loss might suggest selection against a suppressor of the immortal phenotype at that locus. Initially, we screened 59 HeLa-chromosome 4 immortal hybrids, the eleven 143B-chromosome 4 immortal control hybrids, and as an additional control a HeLa-chromosome 4 clone (4.70), which displayed a mortal phenotype. Using 23 markers spaced at approximately 10 cM intervals over the length of chromosome 4, we found allele loss from the introduced chromosome at one or more loci with 30 (51%) of the immortal HeLa clones, with loss of alleles at all loci examined in 12 of these clones. Figure 3A shows the losses from HeLa clones with discrete deletions from the D4S2978-D4S2975 marker interval compared to the control hybrids. Using the Fisher's exact test, we calculated the probability that the losses at the microsatellite loci have occurred by chance, as assessed by comparing the frequency of loss at each locus in the immortal HeLa hybrids and the control clones. The significant losses with the markers D4S423 (P<.04), D4S1570 (P<.03), and D4S406 (P<.04) and the frequent breakage of the introduced chromosome between the markers D4S2964 and D4S423 directed us to examine the interval from D4S2964 to D4S1570 in more detail. We used all available polymorphic markers from the public domain databases and a number of novel markers that we developed [17] to generate a detailed map of the losses in this interval. The results for key markers are shown in Figure 3B. There is no region that is lost in all the clones tested; however, three clones lose the marker BIR0104 while retaining the nearest surrounding markers BIR0110 and D4S1557. This region is overlapped by larger deletions in 23 other clones, and as such comprises the most frequently deleted locus in the 4cen-q27 interval. We did not find discrete deletions involving a single marker at any other locus, and therefore consider the interval from BIR0110 to D4S1557 to be the minimal candidate interval for the immortality-suppressing activity. Although deletion analysis can be confounded by the presence of fragile sites and sites of high recombination, the analysis of our control 143B hybrids enabled us to eliminate regions that might be lost spontaneously. For example, D4S2975 has high rates of allele loss in the HeLa-chromosome 4 immortal segregants, but similar high rates of loss in the control hybrids reduce the impact of its candidacy (P=.20). We found no allele losses at any marker loci, including BIR0104, with the mortal HeLa-chromosome 4 hybrid clone 4.70.
Figure 3.
Deletion analysis in immortal HeLa-chromosome 4 hybrid clones and 143B-chromosome 4 hybrid controls. Maps of allele loss from the exogenous chromosome 4 in selected HeLa-chromosome 4 hybrid clones and all 143B-chromosome 4 hybrid clones. Open circles (◯) indicate retained alleles; dark filled circles (●) indicate allele loss; and light filled circles (●) indicate marker not informative. Boxes indicate contiguous regions of allele loss. Map (A) shows markers from the 4cen-q27 region common to the chromosome 4 fragments (with 143B-chromosome 4 hybrids, the marker D4S1583 was used instead of D4S2978). Marker positions were derived from the Généthon genetic map (available at http://www.gdb.org). A shaded box indicates the common region of loss between the markers D4S2964 and D4S1570. A more detailed map of this region is shown in (B). Marker order on this finer scale is derived from the University of California at Santa Cruz draft human genome assembly (August 2001 data freeze; http://genome.ucsc.edu).
Discussion
Heterozygosity of MORF4 in Immortality Complementation Group B Lines and an Absence of Cancer-Specific Alleles
MORF4 has been suggested as a candidate for the complementation group B mortality gene [6]. We have examined the sequence of MORF4 in a number of cell lines and tissues, including the three complementation group B lines (HeLa, J82, and T98G). We found no evidence of cancer-specific alterations of MORF4 in these lines and the MORF4 gene was heterozygous in all three lines. Previous work had already shown that MORF4 showed no differences in expression between cell lines from the different complementation groups [6]. These results, coupled with the mapping of the chromosome 4 activity using fragment MMCT to a region of the chromosome that does not include MORF4, strongly suggested that MORF4 is not the complementation group B gene. Therefore, we continued to search for other candidate loci on chromosome 4 that might harbor a mortality gene.
A Novel Chromosome 4 Locus is Associated with HeLa Cell Immortality
First, we introduced chromosome 4 into HeLa cells and the complementation group C line 143B as a control and showed specific induction of the mortal phenotype in HeLa cells, as reported previously and only when chromosome 4 was transferred [26]. The frequency of MMCT hybrids displaying a mortal phenotype was only around 40%. This is most likely explained by the relatively large fraction of the transferred chromosomes that harbor deletions of the candidate region (see below). However, it is also possible that the effectiveness of the reintroduced chromosome may be subject to clonal variation of the target cell population.
Chromosome 4 fragment analysis mapped the activity crudely to a region bounded by the markers D4S2978-D4S2975. Subsequent deletion analysis of immortal clones resulting from the MMCT revealed a minimal 130-kb region of loss of exogenous chromosome 4 alleles mapping to between the markers BIR0110 and D4S1557, within the D4S2978-D4S2975 interval. The region between D4S1570 and D4S2975 did show a significant level of loss at D4S406 and cannot, at this time, be excluded. However, we have recently shown that a chromosomal fragment lacking the region telomeric to BIR0101 can still induce mortality in human squamous cell carcinoma cells [17]. Thus, we favor the BIR0110-D4S1557 interval as the likely position of the HeLa mortality gene also.
Mechanism of Action of the Mortality Gene
The HeLa cell line derives from a HPV18-positive cervical adenocarcinoma. As a result of this, the viral oncoproteins E6 and E7 disrupt, respectively, the p53 and pRB molecular pathways and, as such, it is unlikely that the introduction of chromosome 4 complements these defects in the control of proliferative lifespan. It is also unlikely that cells with compromised p53 and pRB can effect a genuine senescence checkpoint; it seems probable therefore that chromosome 4 generates a phenotype more akin to crisis. It was recently reported that the introduction of an exogenous copy of chromosome 4 into HeLa cells resulted in the suppression of the telomerase enzyme in a subset of hybrid clones and that this correlated with the acquisition of a senescence-like phenotype [3]. However, the average telomere restriction fragment (TRF) length in a population of HeLa cells is 6.7 kb [16,17]. This relatively long telomere length is inconsistent with maximum number of divisions observed in the HeLa clones displaying a phenotype following the introduction of chromosome 4. Furthermore, a proportion of subclones of HeLa cells has undetectable levels of telomerase activity [8]; therefore, it remains unclear whether the hybrid clones with low telomerase activity result from this subpopulation of cells. In a recent study, we have also shown that telomerase activity is reduced in mortal clones of squamous cell carcinoma cells harboring normal copies of chromosome 4 [17]. However, the phenotype is not reversed by ectopic telomerase and is TRF length-independent [17]. This, nonetheless, does not rule out the possibility that chromosome 4 acts to restrict the action of the telomerase enzyme complex, or acts on the stability of the telomere structure itself.
The Candidate Region may Contain a Tumor Suppressor In Vivo
A number of tumor suppressor genes appear to have a role in the control of human cancer immortality [24]. The mortality gene at the locus, which we have identified, may be a target for deletion in human cancer. Analysis of markers in human tumors has detected loss of heterozygosity (LOH) at loci that coincide with our candidate interval. Reports include LOH in squamous cell carcinoma of the head and neck [29,36,43], and also esophageal [20,32], cervical [18,25], and hepatic carcinoma [10]. The candidate intervals identified for hepatocellular carcinoma [7] include an 18-Mb interval centered on our locus. Moreover, this overlaps with a hemizygous 8-cM germline deletion recently reported in a case of hepatoblastoma [40]. The LOH studies are supported by comparative genomic hybridization investigations that detect a reduced dosage of 4q in breast [39], colon [2], and prostate [42] carcinomas and a high incidence of loss (86%) at 4q11-q23 in cases of small cell cancer of the lung [29]. However, it should be stressed that there is no evidence that any of the complementation group B lines, including HeLa, shows monoallelic regions suggestive of LOH. It is therefore possible that the chromosome 4 mortality gene may be inactivated by mechanisms, such as biallelic methylation in many cases, and the list of candidate tumor suppressor genes that can be inactivated in this way is expanding [4].
Further evidence to link our candidate region to tumor development stems from a rare heritable skin disorder, Huriez syndrome (MIM 181600), which can predispose to squamous cell carcinoma of the skin and perhaps other tissues. Linkage analysis and recombination events in affected individuals have mapped this disorder to a 24-Mb region [23], which spans our candidate interval. On a related issue, a quantitative trait loci scan in individuals with exceptional longevity found evidence for linkage to a region on 4q [30]. This includes the region identified by our study suggesting that the locus could also be associated with aging in vivo.
Candidate Genes in the Region
No previously identified genes, spliced ESTs, or in silico predicted genes are contained within, or overlap with the BIR0110-D4S1557 candidate interval. The nearest gene centromeric to this interval (90 kb from BIR0110) is ATOH1. This encodes a basic helix-loop-helix domain-containing molecule, which is the human homologue of the Drosophila “atonal” gene product [5]. It is proposed to play an important role in the development of the sensory epithelia of the inner ear, but having a non-skin epithelial expression pattern suggests that it is not a good candidate for the mortality gene.
On the telomeric side, the ETL1 gene (also known as SMARCAD1 [1] and KIAA1122) shows a relatively ubiquitous expression and presents a more plausible candidate. It is the human homologue of murine Etl1 [37] and may have a role in ATP-driven chromatin remodeling through a SNF2-related domain. The 5′ end of the gene maps 160 kb distal to D4S1557 and is only targeted by 23/30 of the deletions in the most informative MMCT hybrid clones and by none of the three small, discrete deletions (Figure 3B).
To date, we have found no somatic mutations in ETL1 in any cell lines, including HeLa and Northern blot analysis, which showed that HeLa expressed equivalent levels of ETL1 to normal keratinocytes (N. Barr, unpublished data). This eliminates the possibility that a small reduction in the dosage of the gene could contribute to the immortality of HeLa cells.
A further candidate emerges from the sequence of a single cDNA clone (IMAGE 3935474), which maps onto eight exons between 70 and 120 kb telomeric to D4S1557. No open reading frame is evident within this expressed sequence, although it has not been ruled out that more of this gene remains to be identified. Preliminary experiments suggest that this transcript is not expressed in primary keratinocyte (epithelial) cultures (data not shown), making it an unlikely candidate.
Although it is possible that there may be a defect affecting ETL1 protein expression that may have been undetected by the methods we have used, it is more likely that another, as yet unidentified, gene spanning the marker BIR0104 is responsible for the mortality effect of chromosome 4. The genome sequence for this region has now been completed and presents an excellent resource to aid in the identification of novel candidate genes in this interval.
In summary, the identification of the chromosome 4 mortality gene at the locus described here may well identify additional pathways that must be disrupted in the immortalization of human cancer cells and that may also contribute to the aging of human cells in a more general way.
Acknowledgements
The authors thank the Cancer Research UK and the Association for International Cancer Research for financial support and John Wyke and Paul Harrison for critical review of the manuscript. We also thank Olivia Pereira-Smith for access to MORF4 sequences prior to original publication.
References
- 1.Adra CN, Donato JL, Badovinac R, Syed F, Kheraj R, Cai H, Moran C, Kolker MT, Turner H, Weremowicz S, Shirakawa T, Morton CC, Schnipper LE, Drews R. SMARCAD1, a novel human helicase family-defining member associated with genetic instability: cloning, expression, and mapping to 4q22-q23, a band rich in breakpoints and deletion mutants involved in several human diseases. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- 2.Arribas R, Risques RA, Gonzalez-Garcia I, Masramon L, Aiza G, Ribas M, Capella G, Peinado MA. Tracking recurrent quantitative genomic alterations in colorectal cancer: allelic losses in chromosome 4 correlate with tumor aggressiveness. Lab Invest. 1999;79:111–122. [PubMed] [Google Scholar]
- 3.Backsch C, Wagenbach N, Nonn M, Leistritz S, Stanbridge E, Schneider A, Durst M. Microcell-mediated transfer of chromosome 4 into HeLa cells suppresses telomerase activity. Genes Chromosomes Cancer. 2001;31:196–198. doi: 10.1002/gcc.1134. [DOI] [PubMed] [Google Scholar]
- 4.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1:299–305. doi: 10.1016/s1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5:1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- 6.Bertram MJ, Berube NG, Hang-Swanson X, Ran Q, Leung JK, Bryce S, Spurgers K, Bick RJ, Baldini A, Ning Y, Clark LJ, Parkinson EK, Barrett JC, Smith JR, Pereira-Smith OM. Identification of a gene that reverses the immortal phenotype of a subset of cells and is a member of a novel family of transcription factor-like genes. Mol Cell Biol. 1999;19:1479–1485. doi: 10.1128/mcb.19.2.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bluteau O, Beaudoin JC, Pasturaud P, Belghiti J, Franco D, Bioulac-Sage P, Laurent-Puig P, Zucman-Rossi J. Specific association between alcohol intake, high grade of differentiation and 4q34-q35 deletions in hepatocellular carcinomas identified by high resolution allelotyping. Oncogene. 2002;21:1225–1232. doi: 10.1038/sj.onc.1205197. [DOI] [PubMed] [Google Scholar]
- 8.Bryan TM, Englezou A, Dunham MA, Reddel RR. Telomere length dynamics in telomerase-positive immortal human cell populations. Exp Cell Res. 1998;239:370–378. doi: 10.1006/excr.1997.3907. [DOI] [PubMed] [Google Scholar]
- 9.Bryce SD, Forsyth NR, Fitzsimmons SA, Clark LJ, Bertram MJ, Cuthbert AP, Newbold RF, Pereira-Smith OM, Parkinson EK. Genetic and functional analyses exclude Mortality Factor 4 (MORF4) as a keratinocyte senescence gene. Cancer Res. 1999;59:2038–2040. [PubMed] [Google Scholar]
- 10.Buetow KH, Murray JC, Israel JL, London WT, Smith M, Kew M, Blanquet V, Brechot C, Redeker A, Govindarajah S. Loss of heterozygosity suggests tumor suppressor gene responsible for primary hepatocellular carcinoma. Proc Natl Acad Sci USA. 1989;86:8852–8856. doi: 10.1073/pnas.86.22.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casey G, Plummer S, Hoeltge G, Scanlon D, Fasching C, Stanbridge EJ. Functional evidence for a breast cancer growth suppressor gene on chromosome 17. Hum Mol Genet. 1993;2:1921–1927. doi: 10.1093/hmg/2.11.1921. [DOI] [PubMed] [Google Scholar]
- 12.Cottage A, Dowen S, Roberts I, Pett M, Coleman N, Stanley M. Early genetic events in HPV immortalised keratinocytes. Genes Chromosomes Cancer. 2001;30:72–79. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1060>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Cuthbert AP, Bond J, Trott DA, Gill S, Broni J, Marriott A, Khoudoli G, Parkinson EK, Cooper CS, Newbold RF. Telomerase repressor sequences on chromosome 3 and induction of permanent growth arrest in human breast cancer cells. J Natl Cancer Inst. 1999;91:37–45. doi: 10.1093/jnci/91.1.37. [DOI] [PubMed] [Google Scholar]
- 14.Cuthbert AP, Trott DA, Ekong RM, Jezzard S, England NL, Themis M, Todd CM, Newbold RF. Construction and characterization of a highly stable human: rodent monochromosomal hybrid panel for genetic complementation and genome mapping studies. Cytogenet Cell Genet. 1995;71:68–76. doi: 10.1159/000134066. [DOI] [PubMed] [Google Scholar]
- 15.England NL, Cuthbert AP, Trott DA, Jezzard S, Nobori T, Carson DA, Newbold RF. Identification of human tumour suppressor genes by monochromosome transfer: rapid growth-arrest response mapped to 9p21 is mediated solely by the cyclin-D-dependent kinase inhibitor gene, CDKN2A (p16INK4A) Carcinogenesis. 1996;17:1567–1575. doi: 10.1093/carcin/17.8.1567. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu CP, Adams RR, Chang E, Allsopp RC, Yu J, Le S, West MD, Harley CB, Andrews WH, Greider CW, Villeponteau B. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 17.Forsyth N, Morrison V, Craig NJ, Fitzsimmons SA, Barr NI, Ireland H, Gordon KE, Dowen S, Cuthbert AP, Newbold RF, Bryce SD, Parkinson EK. Functional evidence for a squamous cell carcinoma mortality gene(s) on human chromosome 4. Oncogene. 2002;21:5135–5147. doi: 10.1038/sj.onc.1205688. [DOI] [PubMed] [Google Scholar]
- 18.Hampton GM, Larson AA, Baergen RN, Sommers RL, Kern S, Cavenee WK. Simultaneous assessment of loss of heterozygosity at multiple microsatellite loci using semi-automated fluorescence -based detection: subregional mapping of chromosome 4 in cervical carcinoma. Proc Natl Acad Sci USA. 1996;93:6704–6709. doi: 10.1073/pnas.93.13.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hensler PJ, Annab LA, Barrett JC, Pereira-Smith OM. A gene involved in control of human cellular senescence on human chromosome 1q. Mol Cell Biol. 1994;14:2291–2297. doi: 10.1128/mcb.14.4.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu N, Roth MJ, Polymeropolous M, Tang ZZ, Emmert-Buck MR, Wang QH, Goldstein AM, Feng SS, Dawsey SM, Ding T, Zhuang ZP, Han XY, Reid T, Giffen C, Taylor PR. Identification of novel regions of allelic loss from a genomewide scan of esophageal squamous-cell carcinoma in a high-risk Chinese population. Genes Chromosomes Cancer. 2000;27:217–228. doi: 10.1002/(sici)1098-2264(200003)27:3<217::aid-gcc1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson C, Stenman G, Vojta PJ, Bongcam-Rudloff E, Barrett JC, Westermark B, Paulsson Y. Escape from senescence in hybrid cell clones involves deletions of two regions located on human chromosome 1q. Cancer Res. 1996;56:241–245. [PubMed] [Google Scholar]
- 22.Koi M, Johnson LA, Kalikin LM, Little PF, Nakamura Y, Feinberg AP. Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science. 1993;260:361–364. doi: 10.1126/science.8469989. [DOI] [PubMed] [Google Scholar]
- 23.Lee YA, Stevens HP, Delaporte E, Wahn U, Reis A. A gene for an autosomal dominant scleroatrophic syndrome predisposing to skin cancer (Huriez syndrome) maps to chromosome 4q23. Am J Hum Genet. 2000;66:326–330. doi: 10.1086/302718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loughran O, Clark LJ, Bond J, Baker A, Berry IJ, Edington KG, Ly IS, Simmons R, Haw R, Black DM, Newbold RF, Parkinson EK. Evidence for the inactivation of multiple replicative lifespan genes in immortal human squamous cell carcinoma keratinocytes. Oncogene. 1997;14:1955–1964. doi: 10.1038/sj.onc.1201028. [DOI] [PubMed] [Google Scholar]
- 25.Mitra AB, Murty VV, Li RG, Pratap M, Luthra UK, Chaganti RS. Allelotype analysis of cervical carcinoma. Cancer Res. 1994;54:4481–4487. [PubMed] [Google Scholar]
- 26.Ning Y, Weber JL, Killary AM, Ledbetter DH, Smith JR, Pereira-Smith OM. Genetic analysis of indefinite division in human cells: evidence for a cell senescence-related gene(s) on human chromosome 4. Proc Natl Acad Sci USA. 1991;88:5635–5639. doi: 10.1073/pnas.88.13.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogata T, Ayusawa D, Namba M, Takahashi E, Oshimura M, Oishi M. Chromosome 7 suppresses indefinite division of nontumorigenic immortalized human fibroblast cell lines KMST-6 and SUSM-1. Mol Cell Biol. 1993;13:6036–6043. doi: 10.1128/mcb.13.10.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohmura H, Tahara H, Suzuki M, Ide T, Shimizu M, Yoshida MA, Tahara E, Shay JW, Barrett JC, Oshimura M. Restoration of the cellular senescence program and repression of telomerase by human chromosome 3. Jpn J Cancer Res. 1995;86:899–904. doi: 10.1111/j.1349-7006.1995.tb02998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pershouse MA, El-Naggar AK, Hurr K, Lin H, Yung WK, Steck PA. Deletion mapping of chromosome 4 in head and neck squamous cell carcinoma. Oncogene. 1997;14:369–373. doi: 10.1038/sj.onc.1200836. [DOI] [PubMed] [Google Scholar]
- 30.Puca AA, Daly MJ, Brewster SJ, Matise TC, Barrett J, Shea-Drinkwater M, Kang S, Joyce E, Nicoli J, Benson E, Kunkel LM, Perls T. A genome-wide scan for linkage to human exceptional longevity identifies a locus on chromosome 4. Proc Natl Acad Sci USA. 2001;98:10505–10508. doi: 10.1073/pnas.181337598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rimessi P, Gualandi F, Morelli C, Trabanelli C, Wu Q, Possati L, Montesi M, Barrett JC, Barbanti-Brodano G. Transfer of human chromosome 3 to an ovarian carcinoma cell line identifies three regions on 3p involved in ovarian cancer. Oncogene. 1994;9:3467–3474. [PubMed] [Google Scholar]
- 32.Rumpel CA, Powell SM, Moskaluk CA. Mapping of genetic deletions on the long arm of chromosome 4 in human esophageal adenocarcinomas. Am J Pathol. 1999;154:1329–1334. doi: 10.1016/S0002-9440(10)65386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandhu AK, Hubbard K, Kaur GP, Jha KK, Ozer HL, Athwal RS. Senescence of immortal human fibroblasts by the introduction of normal human chromosome 6. Proc Natl Acad Sci USA. 1994;91:5498–5502. doi: 10.1073/pnas.91.12.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandhu AK, Kaur GP, Reddy DE, Rane NS, Athwal RS. A gene on 6q 14-21 restores senescence to immortal ovarian tumor cells. Oncogene. 1996;12:247–252. [PubMed] [Google Scholar]
- 35.Sasaki M, Honda T, Yamada H, Wake N, Barrett JC, Oshimura M. Evidence for multiple pathways to cellular senescence. Cancer Res. 1994;54:6090–6093. [PubMed] [Google Scholar]
- 36.Shah SI, Yip L, Greenberg B, Califano JA, Chow J, Eisenberger CF, Lee DJ, Sewell DA, Reed AL, Lango M, Jen J, Koch WM, Sidransky D. Two distinct regions of loss on chromosome arm 4q in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2000;126:1073–1076. doi: 10.1001/archotol.126.9.1073. [DOI] [PubMed] [Google Scholar]
- 37.Soininen R, Schoor M, Henseling U, Tepe C, Kisters-Woike B, Rossant J, Gossler A. The mouse Enhancer trap locus 1 (Etl-1): a novel mammalian gene related to Drosophila and yeast transcriptional regulator genes. Mech Dev. 1992;39:111–123. doi: 10.1016/0925-4773(92)90030-n. [DOI] [PubMed] [Google Scholar]
- 38.Steenbergen RD, Kramer D, Meijer CJ, Walboomers JM, Trott DA, Cuthbert AP, Newbold RF, Overkamp WJ, Zdzienicka MZ, Snijders PJ. Telomerase suppression by chromosome 6 in a human papillomavirus type 16-immortalized keratinocyte cell line and in a cervical cancer cell line. J Natl Cancer Inst. 2001;93:865–872. doi: 10.1093/jnci/93.11.865. [DOI] [PubMed] [Google Scholar]
- 39.Tanner MM, Karhu RA, Nupponen NN, Borg A, Baldetorp B, Pejovic T, Ferno M, Killander D, Isola JJ. Genetic aberrations in hypodiploid breast cancer: frequent loss of chromosome 4 and amplification of cyclin D1 oncogene. Am J Pathol. 1998;153:191–199. doi: 10.1016/S0002-9440(10)65560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terada Y, Imoto I, Nagai H, Suwa K, Momoi M, Tajiri T, Onda M, Inazawa J, Emi M. An 8-cM interstitial deletion on 4q21-q22 in DNA from an infant with hepatoblastoma overlaps with a commonly deleted region in adult liver cancers. Am J Med Genet. 2001;103:176–180. doi: 10.1002/ajmg.1521. [DOI] [PubMed] [Google Scholar]
- 41.Uejima H, Mitsuya K, Kugoh H, Horikawa I, Oshimura M. Normal human chromosome 2 induces cellular senescence in the human cervical carcinoma cell line SiHa. Genes Chromosomes Cancer. 1995;14:120–127. doi: 10.1002/gcc.2870140206. [DOI] [PubMed] [Google Scholar]
- 42.Virgin JB, Hurley PM, Nahhas FA, Bebchuk KG, Mohamed AN, Sakr WA, Bright RK, Cher ML. Isochromosome 8q formation is associated with 8p loss of heterozygosity in a prostate cancer cell line. Prostate. 1999;41:49–57. doi: 10.1002/(sici)1097-0045(19990915)41:1<49::aid-pros7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 43.Wang XL, Uzawa K, Imai FL, Tanzawa H. Localization of a novel tumor suppressor gene associated with human oral cancer on chromosome 4q25. Oncogene. 1999;18:823–825. doi: 10.1038/sj.onc.1202318. [DOI] [PubMed] [Google Scholar]
- 44.Wang XW, Lin X, Klein CB, Bhamra RK, Lee YW, Costa M. A conserved region in human and Chinese hamster X chromosomes can induce cellular senescence of nickel-transformed Chinese hamster cell lines. Carcinogenesis. 1992;13:555–561. doi: 10.1093/carcin/13.4.555. [DOI] [PubMed] [Google Scholar]