Abstract
The crystal structure of the synaptic SNARE complex reveals a parallel four-helix coiled-coil arrangement; buried in the hydrophobic core of the complex is an unusual ionic layer composed of three glutamines and one arginine, each provided by a separate α-helix. The presence of glutamine or arginine residues in this position is highly conserved across the t- and v-SNARE families, and it was recently suggested that a 3Q:1R ratio is likely to be a general feature common to all SNARE complexes. In this study, we have used genetic and biochemical assays to test this prediction with the yeast exocytic SNARE complex. We have determined that the relative position of Qs and Rs within the layer is not critical for biological activity and that Q-to-R substitutions in the layer reduce complex stability and result in lethal or conditional lethal growth defects. Surprisingly, SNARE complexes composed of four glutamines are fully functional for assembly in vitro and exocytic function in vivo. We conclude that the 3Q:1R layer composition is not required within the yeast exocytic SNARE complex because complexes containing four Q residues in the ionic layer appear by all criteria to be functionally equivalent. The unexpected flexibility of this layer suggests that there is no strict requirement for the 3Q:1R combination and that the SNARE complexes at other stages of transport may be composed entirely of Q-SNAREs or other noncanonical combinations.
INTRODUCTION
Vesicle trafficking in eukaryotic cells requires the participation of a large family of proteins called SNAREs (Bennett, 1995; Pelham, 1999). Specific interactions between SNARE proteins present on target membranes (t-SNAREs) and proteins on vesicle membranes (v-SNAREs) are thought to play a critical role in the fidelity of vesicle docking and fusion events (Sollner et al., 1993b; Rothman, 1994). The initial association of v-SNAREs to t-SNAREs results in the formation of a protein complex bridging the two opposed bilayers, sometimes referred to as a trans-SNARE complex. The subsequent formation of a highly stable cis-SNARE complex is thought to play a key role in vesicle-target membrane fusion (Fasshauer et al., 1997; Hanson et al., 1997; Sutton et al., 1998). After SNARE complex formation, the ATPase NSF/sec18 is recruited to the membrane fusion site in conjunction with its soluble cofactor, α-SNAP/sec17; NSF/sec18 dissociates the SNARE complex, allowing recycling and reactivation of the component SNAREs for participation in additional rounds of fusion (Sollner et al., 1993a).
Despite relatively low sequence homology, SNARE proteins from such evolutionarily distant organisms as yeast and human form protein complexes remarkably similar in overall topology. Electron microscopy studies have shown that the yeast post-Golgi SNARE complex, for example, is a parallel four-helix bundle, roughly 140 Å long, consisting of two t-SNAREs (one of which contributes two helices) and one v-SNARE (Katz et al., 1998). Atomic-level structural data from the neuronal SNARE complex containing SNAP-25b, synaptobrevin-II, and syntaxin-1A demonstrated that the neuronal SNARE complex looks remarkably similar; the core of the four-helix bundle contains 16 distinct “layers” of packing interactions. Each layer is formed by the interdigitation of four side chains, one provided by each SNARE α-helix, with geometry approximating a canonical coiled-coil structure (Sutton et al., 1998). Most of the layers are composed entirely of hydrophobic residues, but roughly halfway along the axis of the helical bundle is a unique ionic layer (also known as the “zero” layer) consisting of an arginine and three glutamines. Within this layer, the arginine is contributed by the v-SNARE synaptobrevin and the glutamines are contributed by the t-SNAREs SNAP-25 and syntaxin. The glutamine and arginine residues form an extensive network of hydrogen bonds with each other (Figure 1). The flanking hydrophobic layers stabilize the ionic layer by protecting the polar interactions from exposure to the surrounding environment (Sutton et al., 1998).
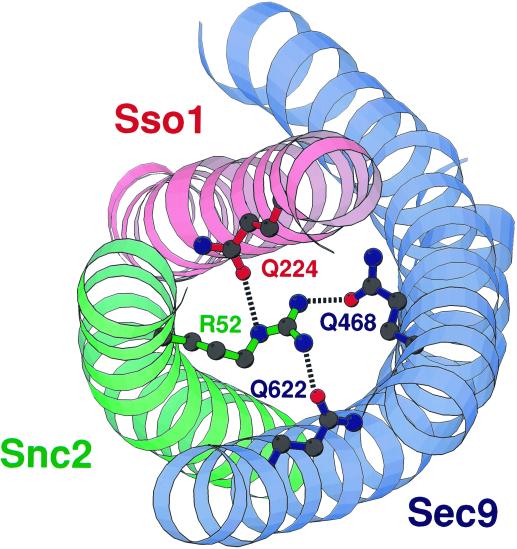
Figure 1.
Model of the ionic layer of the yeast post-Golgi SNARE complex. Names of the residues proposed to constitute an ionic layer in the yeast Sec9/Sso1/Snc2 SNARE complex have been imposed onto the molecular structure of the neuronal SNARE complex ionic layer, represented as a Molscript-generated ribbon diagram. According to a sequence alignment–based prediction, the t-SNARE Sec9 contributes Q468 and Q622 from its first and second helices (respectively), the t-SNARE Sso1 contributes Q224, and the v-SNARE contributes R52 to the putative ionic layer.
A primary sequence alignment of all known SNARE proteins suggested that the ionic layer observed in the neuronal system is a general property of SNARE complexes and that residues in this layer are among the most highly conserved across species. With a few exceptions, SNARE proteins contain either a glutamine or an arginine in the apparent 0 position (Fasshauer et al., 1998b; Weimbs et al., 1998), and mutation of these residues often results in complete inactivation of the corresponding genes (Novick et al., 1980; Ossig et al., 1991). In instances in which it has been possible to determine how trafficking proteins interact to form active complexes, it seems that Q- and R-SNAREs generally combine to produce an ionic layer with one arginine and three glutamines, as in the neuronal complex (Fasshauer et al., 1998b).
We used genetic analyses to examine the potential of the crystal structure–based alignment to predict the residues contributing to this layer in the yeast post-Golgi SNARE complex. We established that the ionic layer in yeast is composed of glutamines 468 and 622 of Sec9, glutamine 224 of Sso1, and arginine 52 of Snc2, as predicted by the amino acid alignment. We then tested the functional consequences of alterations in the ionic layer and demonstrated that Q-to-R mutations in the layer result in lethal or conditional lethal phenotypes and affect SNARE complex stability in vitro. Introduction of a complementary amino acid restoring the 3Q:1R ratio can restore biological function, demonstrating that the precise distribution of Qs and Rs within the ionic layer of the Sec9/Sso1/Snc2 complex is not critical for protein trafficking in yeast. Notably, four-helix bundles containing only glutamines in the ionic layer were completely functional in vivo and in vitro, suggesting the inherent flexibility in the formation of biologically active SNARE complexes.
MATERIALS AND METHODS
Plasmid Construction and Protein Purification
sec9-Q468R, sec9-Q622R, SNC2-R52Q, and sso1-Q224R mutants were generated by fusion PCR with the use of oligonucleotides containing mutant sequences. All constructs contained the entire ORF plus the upstream promoter (∼800 bases) and downstream flanking sequences (∼300 bases) to ensure proper gene expression. SEC9 fragments were subcloned into the pRS315 vector with the use of PstI–SacI restriction sites, and SNC2 fragments were subcloned into the pRS316 vector with the use of BamHI–SalI sites. All constructs were verified by sequence analysis to ensure that they carried the desired nucleotide changes.
Sec9(402–651), Sec9-Q468R(402–651), Sec9-Q622R(402–651), Sso1(1–265), Sso1-Q224R(1–265), Snc2(1–94), and Snc2-R52Q(1–94) were subcloned into the pGEX-4T1 expression vector (Pharmacia Biotech, Piscataway, NJ) with the use of PCR-amplified fragments with BamHI–SalI restriction sites incorporated into the PCR primers. Recombinant proteins were expressed and purified as described by Rossi et al. (1997) with the exception of Sso1(188–265), which was generously provided by Luke Rice (Yale University, New Haven, CT). Protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, IL) and by comparison to purified standards with the use of SDS-PAGE followed by Coomassie blue staining.
Genetic Analysis of Mutants
Wild-type and mutant constructs were introduced into BY68 (a; sec9-4; ura3-52; leu2-3,112; his3-200) and BY386 (a; sec9-7; ura3-52; leu2-3,112) strains by yeast transformation. Transformants were picked and tested for their ability to grow at a restrictive temperature of 37°C.
To study the phenotypes of mutants carrying sec9-Q468R and sec9-Q622R alleles as the only copy of SEC9, the constructs were introduced into the diploid strain BY153 (a/α; SEC9/s9Δ::HIS3; ura3-52/ura3-52; leu2-3,112/leu2-3,112; his3Δ200/his3Δ200). The transformants were sporulated, and tetrads were dissected with the use of a micromanipulator on YPD plates. The plates were grown at 25°C, and the haploid progeny were analyzed for the presence of the mutants (scored as leu+), the absence of the wild-type SEC9 (scored as his+), viability, and conditional growth defects.
To examine the effect of mutations in SNC2 and SSO1, strain BY101 (a; snc1Δ::URA3; snc2Δ::ADE8; his3-Δ200; leu2-3,112; pGAL1-SNC1) was transformed with SNC2 or SNC2-R52Q alleles on a CEN-LEU2 plasmid; strain BY103 (a; sso1Δ::LEU2; sso2Δ::HIS3; ade2-1; his3-11,15; ura3-1; trp1-1, pGAL-SSO1) was transformed with SSO1 or sso1-Q224R genes on a CEN-URA3 plasmid. Transformants were selected on medium containing galactose and tested for phenotypes in the presence of galactose or glucose as a carbon source.
To assess the effect of overexpression of the mutant alleles, DNA fragments were subcloned behind the GAL1 promoter in LEU2 (pNB527) or URA3 (pNB529) integrating vectors, linearized with ClaI (pNB527) or PstI (pNB529), transformed into strain BY17 (a; GAL+; leu2-3,112; ura3-52), and monitored for growth at 25 and 37°C on YPD or YP-Gal plates.
Binding Assays
Assays were performed as described in detail by Rossi et al. (1997). Briefly, for the ternary assays, GST–Snc2 beads were incubated with soluble Sso1 and Sec9 in a 100-μl reaction volume. Immobilized proteins were added to 0.5 μM soluble Sso1 to 1 μM final concentration in a binding buffer containing 10 mM HEPES/KOH, pH 7.4, 140 mM KCl, 2 mM MgCl2, and 0.5 mM Triton X-100. Sec9 protein concentrations were varied from 0.01 to 10 μM. The reactions were incubated for 20 h at 4°C; bound proteins were separated by centrifugation. Beads were washed three times with binding buffer and then boiled in 900 μl of sample buffer. Equal volumes of samples were subjected to SDS-PAGE, with subsequent Western blotting. Membranes were incubated with affinity-purified α-Sec9 antibody, detected with 125I-protein A, and quantified on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). Fifty percent effective concentrations (EC50s) were determined with the use of the GraphPad Prism program (GraphPad Software, San Diego, CA). Binary assays were performed as described above with GST–Sso1 at 0.5 μM and soluble Sec9 at 0.01 to 10 μM. Titration curves for Sec9 proteins were determined to ensure that concentrations used for saturation binding curves fell within the range of linear detection by the α-Sec9 antibody.
Invertase Assays
Invertase assays were performed on SNC1/2 disruption strains (BY101) containing CEN-SNC2 or CEN-SNC2-R52Q plasmids. Cultures were grown overnight to midlog phase in YP medium supplemented with 2% glucose. To stimulate production of invertase, strains were shifted to 0.1% glucose for 90 min at 30°C. Internal and external invertase activities were measured as described by Nair et al. (1990). The percentage of total invertase secretion was calculated as Δexternal/(Δexternal + Δinternal).
RESULTS
The Ionic Layer in Yeast Is Composed of Three Glutamines and One Arginine
Several different alignments of the yeast and neuronal SNAREs have been proposed; alignment of the neuronal SNAP-25 and the yeast Sec9 sequences has been most challenging because of the low degree of amino acid similarity (Rossi et al., 1997; Weimbs et al., 1997; Fasshauer et al., 1998b). The most recent crystal structure–based sequence comparison predicted that, like the synaptic SNARE complex, the post-Golgi SNARE complex of the yeast Saccharomyces cerevisiae would contain an ionic layer formed by the side chains of R52 from Snc2, Q468 and Q622 from the H1 and H2 helices (respectively) of Sec9, and Q224 from Sso1 (Figure 1). To verify the prediction of an ionic layer and to assess its physiological significance in yeast, we carried out a structure–function analysis of these four residues. We used two different mutant strains, sec9-4 and sec9-7, both of which contain temperature-sensitive mutations in the t-SNARE Sec9. sec9-4 (Gly-458 to Asp) and sec9-7 (Leu-627 to His) mutations map to different sites of the protein and are distinct in the structural nature of the defects. The Sec9-4 mutant protein is profoundly impaired in its ability to interact with Sso1 and Snc2. In contrast, the Sec9-7 protein can be assembled into the ternary complex but appears to be defective for a step after the initial assembly of the SNARE complex (Rossi et al., 1997; Katz et al., 1998). Therefore, suppression of both sec9-4 and sec9-7 mutants is likely to represent a general restoration of Sec9 function rather than a specific restoration of a defect associated with either one of these alleles.
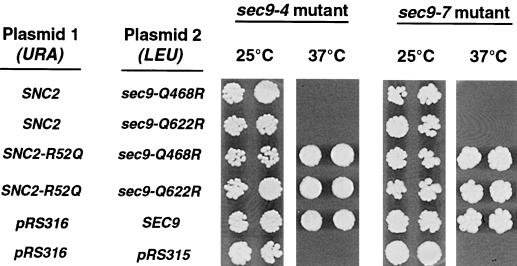
When CEN plasmids containing the sec9-Q468R or sec9-Q622R mutant were transformed into these strains together with a wild-type form of the v-SNARE SNC2, the ratio of Qs to Rs at the putative ionic layer of the resulting complexes with endogenous Sso1 was 2:2. Under these conditions, we observed no rescue of the temperature-sensitive phenotype in either the sec9-4 or sec9-7 strain. In contrast, the expression of Sec9-Q468R or Sec9-Q622R in combination with the Snc2-R52Q mutant is predicted to restore the 3Q:1R ratio in the ionic layer and should result in the formation of active SNARE complexes. We observed that in the corresponding transformed sec9-4 and sec9-7 strains, lethality at the restrictive temperature was in fact suppressed (Figure 2). When sec9-Q468R or sec9-Q622R was transformed together with an R55Q substitution in Snc2 (three residues away from the predicted ionic-layer arginine), no rescue of temperature sensitivity was observed. The fact that an R-to-Q mutation at SNC2-R52 (but not R55) can complement sec9-Q468R and sec9-Q622R suggests that these amino acids do actually contribute to a layer of residues that interact in functional SNARE complexes. Furthermore, it appears that the ratio, but not the precise position, of Q and R residues at the ionic layer is important for biological function, perhaps because the hydrogen bonding structure of the layer is invariant to “rotation” of the side chain positions.
Figure 2.
The sec9-Q468R and sec9-Q622R mutants are functional only in combination with an SNC2 allele containing a compensatory R-to-Q mutation. Each of the alleles shown was subcloned into a single-copy CEN vector and introduced into the sec9-4 or sec9-7 strain by transformation. Transformants were picked into microtiter wells, transferred onto YPD plates, and allowed to grow at the indicated temperatures for 2–3 d. In a separate experiment, we have shown that the SNC2-R52Q mutant has no ability to suppress the sec9-4 and sec9-7 mutants on its own (our unpublished data)
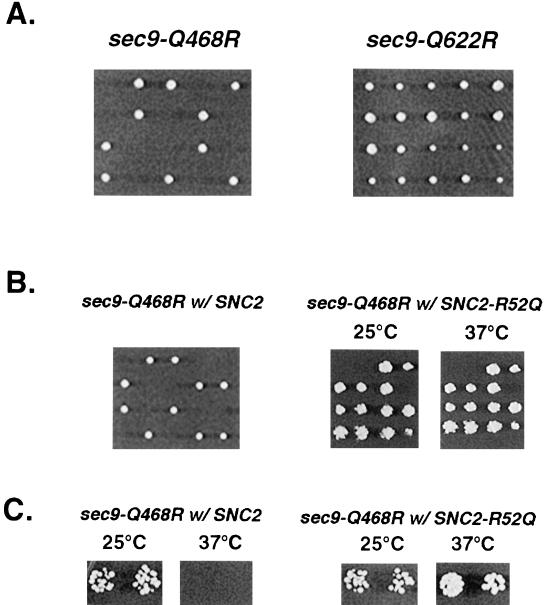
To assess the biological significance of the ionic layer, we constructed yeast strains that contained Q-to-R mutant forms as the only source of Sec9. The sec9-Q468R and sec9-Q622R mutants were subcloned into a CEN-LEU2 plasmid and introduced into a heterozygous diploid strain that had one allele of SEC9 replaced by a HIS3 marker. After allowing the strain to sporulate, we analyzed haploid progeny for viability and/or conditional growth defects. All tetrads derived from a strain carrying a Q468R mutant contained two viable and two inviable spores, indicating that the sec9-Q468R mutation is lethal (two viable spores carried a wild-type SEC9 gene and were his−) (Figure 3A). The Q622R mutant haploid progeny (scored as his+ and ura−) was viable, but two of the four spores were slow growing at 25°C and inviable at 37°C (Figure 3A). These results demonstrated that disruption of the ionic layer has profound consequences in vivo. We reasoned that restoration of the 3Q:1R ratio in the layer by introducing a complementing glutamine in Snc2, as in the sec9-4 and sec9-7 studies, should suppress the growth defects of the SEC9 mutant strains. For the sec9-Q468R mutant, we again used the heterozygous sec9Δ::HIS3 strain but introduced genes for wild-type SNC2 or SNC2-R52Q on a URA3 plasmid together with the LEU2 plasmid encoding Sec9-Q468R. All haploid progeny of this strain were viable, indicating that the SNC2-R52Q mutation complemented the Q468R mutation in Sec9 to produce functional SNARE complexes. Supplying a wild-type form of Snc2 (maintaining the ionic layer residue stoichiometry at 2Q:2R) had no positive effect on the viability of the sec9-Q468R–containing strain (Figure 3B). Similarly, the sec9-Q622R–related defect was completely suppressed by SNC2-R52Q but not by the wild-type form of SNC2 (Figure 3C), as in our studies with the temperature-sensitive sec9 mutant strains.
Figure 3.
Mutations in either the Sec9 H1 or H2 helix result in loss of function that is suppressed by the expression of SNC2 with an R-to-Q substitution. (A) The sec9-Q468R mutant is lethal and the sec9-Q622R mutant is temperature-sensitive as the only source of SEC9. A diploid strain heterozygous for a deletion of SEC9 (a/α; sec9Δ::HIS3/sec9; ura3-52/ura3-52; leu2-3,112/leu2-3,112; his3-Δ200/his3-Δ200) was transformed with sec9-Q468R or sec9-Q622R CEN-LEU2 plasmids. Transformants were sporulated, and tetrads were picked for each mutant and allowed to grow for 3 d on YPD plates at 25°C. (B) Rescue of sec9-Q468R lethality by SNC2-R52Q. Diploids were cotransformed with either SNC2 (left panel) or SNC2-R52Q (right panel) carrying plasmids along with sec9-Q468R and analyzed as described in A. The left panel shows the resulting tetrads after cotransformation with SNC2 and sec9-Q468R plasmids. Subsequent analysis of the viable spores indicated that both plasmids were present but were unable to support growth in the spores containing the deleted sec9Δ::HIS3 allele. The right panel shows replica-plate analysis of four tetrads resulting from cotransformation with the SNC2-R52Q and sec9-Q468R plasmids. Rescue of the deleted SEC9 by the combination of SNC2-R52Q and sec9-Q468R plasmids was clearly evident by the presence of more than two viable spores and by the analysis of auxotrophic markers, which demonstrated that each of the viable spores contained either the wild-type SEC9 allele (his−) or sec9Δ::HIS3 disruption (his+) and both CEN-SNC2-R52Q (ura+) and CEN/sec9-Q468R (leu+). Rescued segregants show wild-type growth on YPD medium at both 25 and 37°C. (C) The SNC2-R52Q allele dominantly suppresses the temperature sensitivity of the sec9-Q622R mutant. The sec9-Q622R mutant sec9Δ::HIS3; CEN/sec9-Q622R (ts-) strain constructed in A was transformed with the SNC2 or SNC2-R52Q alleles on CEN/URA3 plasmids. Transformants were replica-plated onto YPD plates at 25 and 37°C.
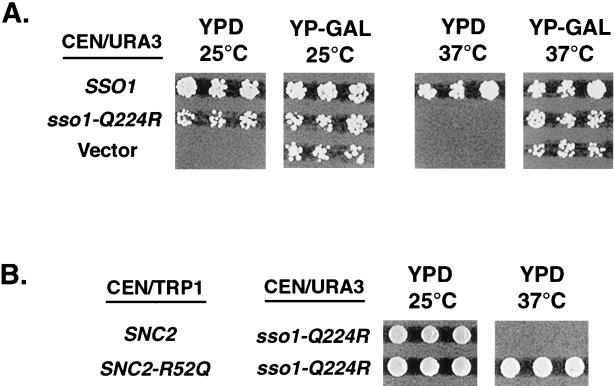
If 3Q:1R stoichiometry is required for proper biological function, then the substitution of the arginine at position 224 of Sso1 should produce the phenotype similar to the Q-to-R mutations in Sec9. We used the yeast strain containing the disrupted SSO1/SSO2 loci and with Sso1 being expressed from the GAL1 promoter (inducible by galactose and repressible by glucose). When such strains are grown in medium containing glucose, they are inviable at any temperature because of the lack of the essential proteins Sso1/2. Wild-type and mutant alleles of SSO1 were introduced into this strain on a CEN-URA3 plasmid, and the effect of the Q-to-R substitution was examined by growing transformants on glucose- or galactose-containing medium. We observed that the introduction of the sso1-Q224R allele produced a partial defect similar to that of sec9-Q622R; functional SNARE complexes with the 2Q:2R composition were viable at the ambient temperature of 25°C (growth in the presence of glucose) but not at 37°C (Figure 4A). Supplying the mutant allele of SNC2 (SNC2-R52Q) produced the normal ratio of Q to R residues in this layer and resulted in complete restoration of cell viability and growth at the restrictive temperature (Figure 4B).
Figure 4.
The SSO1 Q-to-R mutant displays a conditional growth defect that is suppressed by the expression of SNC2 with an R-to-Q substitution. (A) Each of the plasmids shown was introduced into SSO1/2 disruption strain (a; SSO1Δ::LEU2; SSO2Δ::HIS3; ade2-1; his3-11,15; ura3-1; trp1-1, pGAL-SSO1); transformants were selected on medium containing galactose and tested for viability on 2% glucose (YPD) and 3% galactose (YP-Gal) at the indicated temperatures. (B) SNC2 or SNC2-R52Q plasmids (CEN-TRP1) were introduced into the yeast strains containing sso1-Q224R (CEN-URA3) as the only source of Sso1 protein (generated as described in A). Transformants were selected on glucose medium lacking tryptophan and uracil and tested for viability at the permissive and restrictive temperatures.
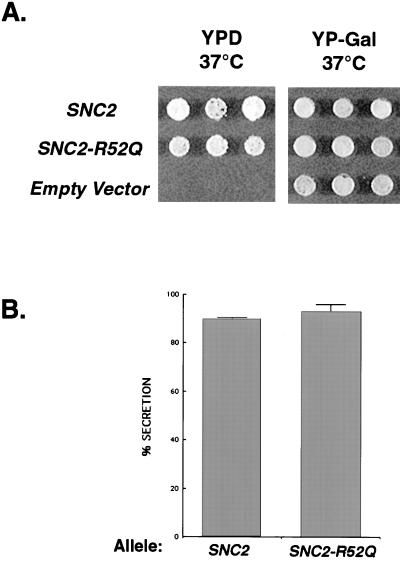
Finally, using the same “GAL shut-off” strategy described for Sso, we examined how the R-to-Q substitution in Snc2 would affect the phenotype of the recipient yeast strain. Surprisingly, we found that the SNC2-R52Q mutation resulted in completely normal growth when it was the only source of Snc in the cell under all conditions tested (Figure 5A). Moreover, when we compared the secretory capacity of SNC2-R52Q– and SNC2-containing strains as the only source of Snc, we found that the SNC2-R52Q mutant secreted normal levels of invertase and was again indistinguishable from strains containing wild-type SNC2 (Figure 5B). Therefore, yeast post-Golgi SNARE complexes containing four glutamines—i.e., four Q-SNARE helices—at the core layer are fully functional for growth and secretion.
Figure 5.
R-to-Q mutation in SNC2 has no effect on growth or secretion. (A) Each of the alleles shown was transformed into a strain containing the sole source of Snc1/2 protein under the control of the regulatable GAL1 promoter (a; snc1Δ::URA3; snc2Δ::ADE8; his3-Δ200; leu2-3,112; pGAL1-SNC1; from J. Gerst) (Weizman Institute, Rehovot, Israel), selected on galactose-containing medium, and tested for viability and growth on 2% glucose (YPD) and 3% galactose (YP-Gal). Similar results were obtained at 25 and 37°C. (B) Invertase assays were performed on the yeast strains containing CEN-SNC2 or CEN-SNC2-R52Q plasmids as the only source of Snc2 (generated as described in A). Comparison of the percentage of invertase secreted into the periplasm is shown. All samples were measured in duplicate, and the SD was determined from two separate experiments.
SEC9 Mutants Act as Dominant Negative Alleles When Overexpressed behind the GAL1 Promoter
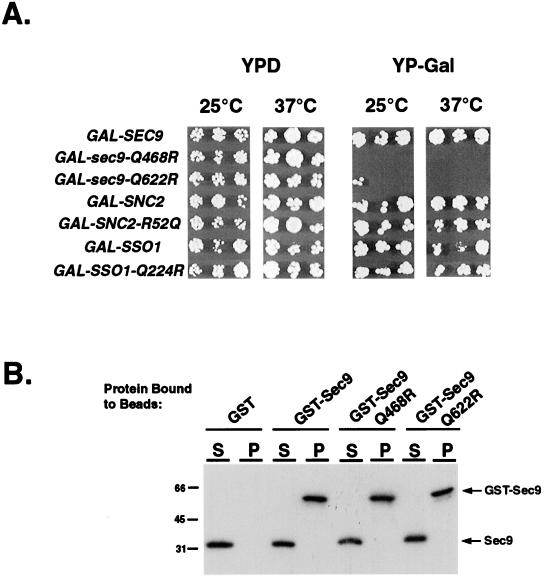
To test whether the expression level of wild-type and ionic-layer mutant SNAREs would influence complex formation and biological activity in vivo, we also studied wild-type yeast strains overexpressing wild-type and mutant Sec9, Snc2, and Sso1 proteins under the direction of the GAL1 promoter. Constructs were introduced into S. cerevisiae by integrative transformation and tested on different carbon sources at different temperatures. As might be expected, overexpression of the wild-type form of Sec9 or the biologically active Snc2-R52Q mutant had no effect under any conditions. In contrast, overexpression of either the Q468R or the Q622R mutant Sec9 proteins resulted, respectively, in an absolute or an almost complete failure to grow, even at 25°C. Surprisingly, overexpression of the sso1-Q224R temperature-sensitive mutant did not result in a dominant negative phenotype at any temperature (Figure 6).
Figure 6.
Mutations in SEC9 H1 and H2 but not in SNC2 or SSO1 act as dominant negative alleles when overexpressed, but heterodimerization with wild-type Sec9 is not a likely mechanism for dominant negativity. (A) Each of the alleles was subcloned into a GAL1 expression plasmid and introduced into a wild-type yeast strain by integrative transformation. Transformants were replica-plated on YPD or YP-Gal plates and incubated at the indicated temperatures for 2–3 d. (B) GST–Sec9 fusion proteins were tested for their ability to interact with excess-soluble Sec9. Binding reactions were performed as described in MATERIALS AND METHODS. Pellets containing bound proteins (P) were separated from unbound proteins (S) by centrifugation and subjected to the Western blotting with α-Sec9 antibody.
We considered the possibility that the dominant negativity of overexpressed (two helices) Sec9 mutants, but not of (single helix) Snc2 or Sso1 mutants, could result from a repression of wild-type Sec9 activity attributable to complex formation with mutant Sec9. Such unphysiological heterodimerization could conceivably occur at high intracellular protein concentrations and would result in the formation of four-helix bundles with the correct 3Q:1R ionic layer composition. We examined this possibility directly by testing the affinity of soluble wild-type Sec9 for wild-type and mutant GST–Sec9 fusion proteins conjugated to glutathione beads. After incubation and centrifugation to pellet the beads, bound (P) and unbound (S) protein fractions were analyzed by immunoblotting with the α-Sec9 antibody. Even at micromolar concentrations, we were unable to detect binding between wild-type and Q-to-R mutant Sec9 molecules, indicating that heterodimerization was not a likely mechanism for the dominant negativity (Figure 6).
Biochemical Characterization of the Ionic Layer
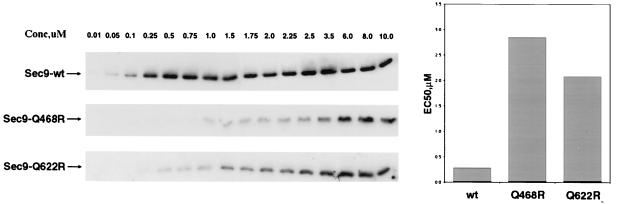
To determine whether we could observe a biochemical correlate for the ionic-layer complementarity effects we had detected in vivo, we examined the affinity of each of the mutant SNARE proteins for the other complex components in a simple binding assay. The SNAP-25 domains of Sec9 wild-type and Q468R or Q622R mutant proteins were added in a series of concentrations (0.01–10 μM) to a fixed mixture of the cytoplasmic domain of Sso1 (1.0 μM) and glutathione bead–conjugated GST–Snc2 (0.5 μM). The amounts of complexed (GST–Snc2-bound) Sec9 proteins were determined by Western blotting. Sec9 titration curves were obtained, and EC50 values were derived for each combination of proteins (Figure 7). Sec9 mutations, which introduced an additional arginine into the ionic layer, were clearly impaired in their ability to form complexes with the other SNARE proteins. With respect to wild-type Sec9 (EC50 = 0.28 μM), the Q468R and Q622R mutations produced a 10-fold and a 7-fold decrease, respectively, in the EC50 for assembly of the ternary complex (Table 1). The Q224R ionic-layer mutation in Sso, however, had little effect on SNARE complex formation, according to the GST-pulldown assay; an observed increase in EC50 (0.28–0.61 μM) was within the measurement error (Table 1).
Figure 7.
Q-to-R mutations in Sec9 produce a substantial decrease in binding affinity. Sec9 proteins were added in increasing concentrations (0.01–10 μM) to the assembly reactions, incubated overnight, and immunoblotted with α-Sec9 antibody (left panel). The amount of Sec9 incorporated into the SNARE complex was quantitated and used to calculate EC50 values (right panel).
Table 1.
Relative affinities of Sec9 monomers for ternary and binary SNARE complexes
| Sec9-H1 (Q468) | Sec9-H2 (Q622) | Sso1 (Q224) | Snc2 (R52) | log(EC50) Sso1 (1–265) | EC50, μM Sso1 (1–265) | log(EC50) Sso1 (188–265) | EC50, μM Sso1 (188–265) |
|---|---|---|---|---|---|---|---|
| Q | Q | Q | R | −6.55 ± 0.21 | 0.283 | −6.46 ± 0.08 | 0.345 |
| R | Q | Q | R | −5.55 ± 0.07 | 2.853 | −5.74 ± 0.12 | 1.811 |
| Q | R | Q | R | −5.68 ± 0.04 | 2.080 | −5.92 ± 0.08 | 1.209 |
| Q | Q | R | R | −6.22 ± 0.29 | 0.607 | n.d. | n.d. |
| Q | Q | Q | Q | −6.42 ± 0.15 | 0.383 | −6.34 ± 0.12 | 0.316 |
| R | Q | Q | Q | −5.74 ± 0.15 | 1.807 | −6.37 ± 0.16 | 0.426 |
| Q | R | Q | Q | −5.68 ± 0.10 | 2.098 | −6.41 ± 0.20 | 0.383 |
| Q | Q | Q | — | −6.52 ± 0.08 | 0.302 | n.d. | n.d. |
| R | Q | Q | — | −5.81 ± 0.26 | 1.561 | n.d. | n.d. |
| Q | R | Q | — | −5.78 ± 0.09 | 1.675 | n.d. | n.d. |
EC50 values were derived from saturation binding curves as shown in Figure 7 and described in MATERIALS AND METHODS. The last two columns represent assays done with an N-terminally truncated form of Sso1 that lacks the region known to drastically reduce the kinetics of SNARE assembly (Nicholson et al., 1998). The top seven rows represent ternary assays in which all three SNARE proteins are present, and the bottom three rows represent binary assays of association between Sec9 and Sso1. Mutant residues are indicated in bold italic type. n.d., not determined.
EC50 values were then determined from wild-type and mutant Sec9 titrations with Sso1 and glutathione beads conjugated instead to a GST–Snc2-R52Q mutant protein. We observed that the R52Q mutation in Snc2 did not affect SNARE complex stability; the EC50 for this combination of proteins was very similar to the wild-type value (Table 1). Surprisingly, we failed to detect a “biochemical suppression”; providing complementary glutamine in Snc2 together with Sec9-Q468R or Sec9-Q622R mutants did not restore Sec9-binding affinities under these conditions (Table 1).
The lack of biochemical suppression suggested that the defect in the SNARE protein association is likely to be due to the binary complex formation rather than ternary. To test this possibility directly, we examined the effect of mutations in Sec9 on the interaction with GST–Sso1. We observed a roughly fivefold lower affinity of GST–Sso1 for Sec9-Q468R and Sec9-Q622R than for wild-type Sec9 (EC50 = 0.30 μM), indicating that the defect in Sec9 mutants was in the formation of t-SNARE heterodimers (Table 1).
Because the rate-limiting step in the assembly of the ternary SNARE complexes is the formation of the Sec9/Sso1 heterodimer, we reexamined the binding affinities of wild-type and mutant Sec9 molecules for the SNARE complex containing the C-terminal (H3) portion of Sso1. The N terminus of Sso1 has been shown to inhibit SNARE complex assembly (Nicholson et al., 1998). Therefore, complexes of Sec9 and Snc2 with truncated Sso1 may produce binding affinities that more accurately capture the thermodynamic properties of the complex but not the kinetics of the SNARE association. We observed that, similar to our findings with full-length Sso1, both Sec9-Q468R and Sec9-Q622R proteins had a 3.5- to 5-fold increase in EC50 compared with the wild-type Sec9; importantly, in this assay, the binding defect now could be restored by Snc2-R52Q (Table 1).
The biochemical analysis of SNARE proteins suggests that in the absence of the N-terminal inhibitory domain of Sso1, the reduced stability of the Sso1/sec9-R t-SNARE heterodimer in vitro can be restored by providing the suppressing form of Snc2, in accordance with our in vivo observations. Most importantly, we find that under all conditions tested, the four-helix bundles containing only glutamines in the ionic layer are biochemically indistinguishable from the wild-type complexes.
DISCUSSION
In this study, we have determined that the ionic layer in the yeast post-Golgi SNARE complex is composed of three glutamines and one arginine, as predicted by a recent structure-based alignment. Q-to-R mutations in Sec9 or Sso1 helices, which together with the wild-type Snc2 would produce a 2Q:2R ratio in the ionic layer, resulted in the loss of biological function, presumably attributable to steric and electrostatic destabilization resulting from the presence of two bulky, positively charged side chains within the same layer. In vivo, these defects can be completely suppressed by the introduction of a compensatory R-to-Q mutation in the Snc2 protein. As predicted by the symmetry of the interactions within the ionic layer, there is no detectable difference in in vivo function regardless of which helix contributes the arginine, as long as the total number of arginines does not exceed one. Therefore, there is no functional or structural distinction between R-SNAREs and Q-SNAREs in this complex, because any one of the helices can contribute the arginine to this layer with equivalent results. This result, combined with our observation that four Q-SNARE helices are fully functional, demonstrates that the presence of glutamine or arginine in this layer is unlikely to contribute significantly to the specificity of SNARE interactions.
We observed that although sec9-Q468R and sec9-Q622R behave as recessive loss-of-function mutants when introduced behind their own promoter, they behave as dominant negative alleles when overexpressed behind the GAL1 promoter. The precise molecular mechanism for this dominant negative phenotype is not clear at present, but it has many aspects that appear similar to our previous analysis of the sec9-7 mutant (Rossi et al., 1997). In all three cases (sec9-7, sec9-Q468R, sec9-Q622R), the mutations appear as recessive loss-of-function mutants that have little or no defect in SNARE assembly in vitro. Such mutant proteins might be expected to assemble into complexes in vivo that would be impaired in their ability to promote fusion.
In contrast to the Sec9 Q-to-R mutants, cells overexpressing Sso1 with a Q-to-R substitution were viable on galactose-containing medium, and this mutant could form ternary complexes with wild-type affinity in vitro. This result suggests the possibility that a proofreading machinery capable of distinguishing arginines from glutamines may exist for syntaxins but not for SNAP-25 proteins. Such chaperone-like proteins would inhibit Sec9/Sso1-Q224R interaction in vivo. A good candidate for such a factor is Sec1. Sec1 binds to syntaxins and could fulfill this “editing” function by either preventing mutant SNARE complex assembly or by “discarding” such complexes as nonproductive after SNARE complex formation (Aalto et al., 1997; Carr et al., 1999; Dulubova et al., 1999). Alternatively, tomosyn/sro7/lethal giant larvae family members appear to have a role in SNARE assembly and thus could also provide such a proofreading function (Mechler et al., 1985; Fujita et al., 1998; Lehman et al., 1999). Finally, it is possible that the lack of the dominant negative phenotype with the sso1-Q224R is due to the fact that Sso1/2 are present at 5- to 10-fold higher concentrations than Sec9 (Brennwald et al., 1994) and thus may be more resistant to GAL1 expression of the Q-to-R allele.
Conservation of the ionic layer and the sensitivity of its constituent amino acids to mutations suggest that it is critical for some aspect of SNARE complex function, but its precise role remains unclear. Buried polar residues were observed in the dimeric and trimeric coiled coils of Myc/Max and GCN4; these interactions are thought to direct the relative orientation of helices and confer specificity of the oligomerization state (O'Shea et al., 1992; Lavigne et al., 1995; Oakley and Kim, 1997; Eckert et al., 1998). Similarly, residues of the ionic layer in the SNARE complex could be important to the kinetics of SNARE assembly if they promote correct “registration” of helices in the four-helix bundle (Harbury, 1998; Sutton et al., 1998; Weis and Scheller, 1998; Misura et al., 2000). Conformational properties of SNARE proteins may be especially significant if SNAREs must present specific epitopes for recognition by accessory or adapter proteins such as NSF/sec18 and SNAP/sec17. Substitutions in the SNARE core that preserve the stoichiometry of the ionic layer or do not affect the surface of the complex dramatically (such as an R-to-Q change) will not interfere with the SNARE function, whereas mutations that introduce a second arginine into the ionic layer could change the overall shape of the complex, disrupting interactions with the other factors such as disassembly by SNAP/sec17.
Recent evidence from an in vitro system to analyze SNAP-25 function has supported this possibility. Scales et al. (2000) found that Q-to-R or Q-to-A mutations in the ionic layer of either the H1 or H2 helix of SNAP-25 have no effect on the ability of the proteins to promote fusion and have only modest effects on the thermal stability of the resulting complexes. These authors suggest that this layer is not necessary for the fusion function but rather affect an event downstream, such as disassembly. Our results are entirely consistent with this notion; however, it is equally possible that the lack of an effect on fusion in this system may simply suggest that it is not as sensitive to perturbation of this layer as we observed in vivo. For example, a significant defect in the rate of fusion would not necessarily be apparent in the in vitro assay used by Scales et al. (2000). However, a similar defect in the rate of fusion could have a much more dramatic effect on our in vivo analysis, in which continual rounds of exocytosis are required for normal cell growth.
Biophysical analyses of SNARE complexes have previously suggested that assemblies containing only glutamines in the ionic layer are structurally permissible. For example, syntaxin trimers and SNAP-25–syntaxin heterodimers would be of a 3Q:0R composition, and such complexes were observed in vitro (Fasshauer et al., 1997, 1998a). Similarly, 4Q helical bundles with one SNAP-25 and two syntaxin molecules have been isolated biochemically, but these complexes are unlikely to be functional in vivo. (Fasshauer et al., 1997, 1998b; Fiebig et al., 1999). In this study, we report that mutant complexes of the 4Q:0R stoichiometry are indistinguishable from wild-type complexes for both in vitro assembly assays and in vivo functional assays. Therefore, the 3:1 ratio of glutamine to arginine side chains does not delineate a general rule for the formation of active SNARE complexes. Instead, we predict that as the composition of other SNARE complexes are determined, many will be composed of SNAREs that contain four Q-SNAREs (as well as perhaps other non-arginine-containing combinations) in the core of the four-helix bundle.
ACKNOWLEDGMENTS
We thank Dr. L. Rice for providing the Sso1(188–265) used in Table 1, Drs. J. Gerst and S. Keranen for strains, Dr. A. Jasanoff for help with Figure 1, computer modeling, and editing of the text, and J. Adamo and Dr. G. Rossi for critical comments on the manuscript. This work was supported by grants from the Mathers Charitable Foundation, the Pew Scholars in Biomedical Sciences program, and the National Institutes of Health (GM54712).
REFERENCES
- Aalto MK, Jantti J, Ostling J, Keranen S, Ronne H. Mso1p: a yeast protein that functions in secretion and interacts physically and genetically with Sec1p. Proc Natl Acad Sci USA. 1997;94:7331–7336. doi: 10.1073/pnas.94.14.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK. SNAREs and the specificity of transport vesicle targeting. Curr Opin Cell Biol. 1995;7:581–586. doi: 10.1016/0955-0674(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146:333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert DM, Malashkevich VN, Kim PS. Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues. J Mol Biol. 1998;284:859–865. doi: 10.1006/jmbi.1998.2214. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Eliason WK, Brunger AT, Jahn R. Identification of a minimal core of the synaptic SNARE complex sufficient for reversible assembly and disassembly. Biochemistry. 1998a;37:10354–10362. doi: 10.1021/bi980542h. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Otto H, Eliason WK, Jahn R, Brunger AT. Structural changes are associated with soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor complex formation. J Biol Chem. 1997;272:28036–28041. doi: 10.1074/jbc.272.44.28036. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998b;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig KM, Rice LM, Pollock E, Brunger AT. Folding intermediates of SNARE complex assembly. Nat Struct Biol. 1999;6:117–123. doi: 10.1038/5803. [DOI] [PubMed] [Google Scholar]
- Fujita Y, et al. Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron. 1998;20:905–915. doi: 10.1016/s0896-6273(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Heuser JE, Jahn R. Neurotransmitter release: four years of SNARE complexes. Curr Opin Neurobiol. 1997;7:310–315. doi: 10.1016/s0959-4388(97)80057-8. [DOI] [PubMed] [Google Scholar]
- Harbury PA. Springs and zippers: coiled coils in SNARE-mediated membrane fusion. Structure. 1998;6:1487–1491. doi: 10.1016/s0969-2126(98)00147-6. [DOI] [PubMed] [Google Scholar]
- Katz L, Hanson PI, Heuser JE, Brennwald P. Genetic and morphological analyses reveal a critical interaction between the C-termini of two SNARE proteins and a parallel four helical arrangement for the exocytic SNARE complex. EMBO J. 1998;17:6200–6209. doi: 10.1093/emboj/17.21.6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne P, Kondejewski LH, Houston ME, Jr, Sonnichsen FD, Lix B, Skyes BD, Hodges RS, Kay CM. Preferential heterodimeric parallel coiled-coil formation by synthetic Max and c-Myc leucine zippers: a description of putative electrostatic interactions responsible for the specificity of heterodimerization. J Mol Biol. 1995;254:505–520. doi: 10.1006/jmbi.1995.0634. [DOI] [PubMed] [Google Scholar]
- Lehman K, Rossi G, Adamo JE, Brennwald P. Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J Cell Biol. 1999;146:125–140. doi: 10.1083/jcb.146.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler BM, McGinnis W, Gehring WJ. Molecular cloning of lethal(2)giant larvae, a recessive oncogene of Drosophila melanogaster. EMBO J. 1985;4:1551–1557. doi: 10.1002/j.1460-2075.1985.tb03816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- Nair J, Muller H, Peterson M, Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson KL, Munson M, Miller RB, Filip TJ, Fairman R, Hughson FM. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p. Nat Struct Biol. 1998;5:793–802. doi: 10.1038/1834. [DOI] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Oakley MG, Kim PS. Protein dissection of the antiparallel coiled coil from Escherichia coli seryl tRNA synthetase. Biochemistry. 1997;36:2544–2549. doi: 10.1021/bi962391t. [DOI] [PubMed] [Google Scholar]
- O'Shea EK, Rutkowski R, Kim PS. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HR. SNAREs and the secretory pathway: lessons from yeast. Exp Cell Res. 1999;247:1–8. doi: 10.1006/excr.1998.4356. [DOI] [PubMed] [Google Scholar]
- Rossi G, Salminen A, Rice LM, Brunger AT, Brennwald P. Analysis of a yeast SNARE complex reveals remarkable similarity to the neuronal SNARE complex and a novel function for the C terminus of the SNAP-25 homolog, Sec9. J Biol Chem. 1997;272:16610–16617. doi: 10.1074/jbc.272.26.16610. [DOI] [PubMed] [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Scales SJ, Chen YA, Yoo BY, Patel SM, Doung Y C, Scheller RH. SNAREs contribute to the specificity of membrane fusion. Neuron. 2000;26:457–464. doi: 10.1016/s0896-6273(00)81177-0. [DOI] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993a;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993b;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: a new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimbs T, Mostov K, Low SH, Hofmann K. A model for structural similarity between different SNARE complexes based on sequence relationships. Trends Cell Biol. 1998;8:260–262. doi: 10.1016/s0962-8924(98)01285-9. [DOI] [PubMed] [Google Scholar]
- Weis WI, Scheller RH. Membrane fusion: SNARE the rod, coil the complex. Nature. 1998;395:328–329. doi: 10.1038/26354. [DOI] [PubMed] [Google Scholar]