Abstract
Proteolytic cleavage of TNF receptor 1 (TNFR1) generates soluble receptors that regulate TNF bioactivity. We hypothesized that the mechanism of TNFR1 shedding might involve interactions with regulatory ectoproteins. Using a yeast two-hybrid approach, we identified ARTS-1 (aminopeptidase regulator of TNFR1 shedding) as a type II integral membrane protein that binds to the TNFR1 extracellular domain. In vivo binding of membrane-associated ARTS-1 to TNFR1 was confirmed by coimmunoprecipitation experiments using human pulmonary epithelial and umbilical vein endothelial cells. A direct relationship exists between membrane-associated ARTS-1 protein levels and concordant changes in TNFR1 shedding. Cells overexpressing ARTS-1 demonstrated increased TNFR1 shedding and decreased membrane-associated TNFR1, while cells expressing antisense ARTS-1 mRNA demonstrated decreased membrane-associated ARTS-1, decreased TNFR1 shedding, and increased membrane-associated TNFR1. ARTS-1 neither bound to TNFR2 nor altered its shedding, suggesting specificity for TNFR1. Although a recombinant ARTS-1 protein demonstrated selective aminopeptidase activity toward nonpolar amino acids, multiple lines of negative evidence suggest that ARTS-1 does not possess TNFR1 sheddase activity. These data indicate that ARTS-1 is a multifunctional ectoprotein capable of binding to and promoting TNFR1 shedding. We propose that formation of a TNFR1–ARTS-1 molecular complex represents a novel mechanism by which TNFR1 shedding is regulated.
Introduction
TNF is a multifunctional cytokine that mediates pleiotropic biological processes, including inflammation, immunoregulation, cytotoxicity, antiviral actions, and the transcriptional regulation of numerous genes (1). Induction of TNF biological activity is mediated via binding to two distinct cell-surface receptors, the 55-kDa TNF receptor 1 (TNFR1, CD120a) and the 75-kDa TNFR2 (CD120b). Binding of TNF to TNFR1 induces receptor trimerization with activation of signal transduction cascades via aggregation of intracellular domains and recruitment of adapter or docking proteins (2). Recruitment and binding of the adapter protein TRADD (TNFR-associated death domain) via death domain association produces a platform for the recruitment of additional signaling molecules (3). TRADD recruitment of Fas-associated death domain mediates caspase activation and apoptosis, while recruitment of receptor-interacting protein and TNFR-associated factor 2 activates NF-κB and AP-1 signaling pathways that mediate the transcription of proinflammatory, immunoregulatory, and antiapoptotic genes.
TNFR1-mediated events can be modified by proteolytic cleavage and shedding of cell-surface TNF receptors. Soluble TNF receptors function as TNF-binding proteins that compete with cell-surface TNF receptors, thereby decreasing TNF bioactivity (4). TNF receptor shedding may also decrease the number of cell-surface receptors available for ligand binding. Soluble TNF receptors can also reversibly bind to and stabilize soluble trimeric TNF ligand (5). Subsequent dissociation of TNF from soluble TNF receptors may replenish the soluble TNF ligand pool, with the complex serving as a reservoir for release of TNF when levels are low. Therefore, soluble TNF receptors may serve a buffering function, attenuating TNF bioactivity when levels are elevated and reconstituting TNF when levels have declined (5).
In this study, we sought to identify proteins regulating TNFR1 ectodomain shedding. We reasoned that such proteins might be identified via a yeast two-hybrid approach using the extracellular domain of human TNFR1 as a bait fusion protein to screen a human lung cDNA library. Using this approach, we have identified, cloned, and characterized ARTS-1 (aminopeptidase regulator of TNFR1 shedding) as a member of the aminopeptidase family that directly binds to the TNFR1 extracellular domain and promotes TNFR1 shedding by both human epithelial and endothelial cells. We demonstrate that ARTS-1 does not function as a TNFR1 sheddase, but rather as a TNFR1-binding protein that promotes shedding via a direct interaction with the TNFR1 extracellular domain.
Methods
Yeast two-hybrid screening, cDNA cloning, and Northern blotting.
Yeast two-hybrid screening was performed on a human lung cDNA library (Clontech Laboratories Inc., Palo Alto, California, USA) using a Matchmaker System 2 (Clontech Laboratories Inc.) and a GAL4BD-TNFR1 extracellular domain fusion protein. Human ARTS-1 cDNA clones were isolated from a UniZAP XR phage cDNA library (Stratagene, La Jolla, California, USA) generated from NCI-H292 cells stimulated with 1 μM PMA for 24 hours and subjected to double-stranded automated fluorescent sequencing. Full-length ARTS-1 cDNA was generated by PCR of human lung poly(A+) mRNA (Clontech Laboratories Inc.) using Pfu Turbo (Stratagene) and the following primers: 5′-GCAAGAAGATGGTGTTTCTGCCCCTC-3′ (nucleotides 80–105) and 5′-TTACATACGTTCAAGCTTT-TCACT-3′ (nucleotides 2,890–2,913). Sequence analysis, including Kyte-Doolittle hydropathy prediction, was performed using MacVector 7.0 software (Accelrys, Burlington, Massachusetts, USA). The location of the putative hydrophobic transmembrane α-helical domain was predicted using several web-based analysis programs (MEMSAT2 [http://bioinf.cs.ucl.ac.uk/psipred/], SOSUI [http://sosui.proteome.bio.tuat.ac.jp.sosuiframe0.html], TMAP [http://www.mbb.ki.se/tmap/index.html], TMpred [http://www.ch.embnet.org/software/TMPRED_form.html], and TopPred2 [http://bioweb.pasteur.fr/seganal/interfaces/toppred.html]). A 32P-labeled probe was generated via random priming of gel-purified, full-length ARTS-1 cDNA and hybridized against a multiple tissue Northern blot (Clontech Laboratories Inc.) according to the manufacturer’s recommendations.
Anti–ARTS-1 serum generation.
A 17-amino-acid peptide (RGRNVHMKQEHYMKGSD), corresponding to amino acids 538–554 of the proposed ARTS-1 extracellular domain, was selected based upon its antigenicity and lack of homology with other protein sequences via BLAST homology search. New Zealand white rabbits were immunized with this peptide to generate anti–ARTS-1 serum (Research Genetics, Huntsville, Alabama, USA).
ARTS-1 immunoprecipitation and immunoblotting.
Human bronchial epithelial cells were obtained by fiberoptic bronchoscopy of normal research volunteers who had provided informed consent. The protocol for harvesting bronchial epithelial cells via bronchial brushings was approved by the National Heart, Lung, and Blood Institute Institutional Review Board for the protection of human subjects. Human bronchial epithelial cell lines were purchased from the American Type Culture Collection (Rockville, Maryland, USA). Primary cultures of normal human bronchial epithelial cells, umbilical vein endothelial cells (HUVECs), and fibroblasts were purchased from Clonetics Corp. (San Diego, California, USA). Crude membrane and cytosolic fractions were prepared from the NCI-H292 human pulmonary mucoepidermoid carcinoma cell line and HUVECs. Cells were isolated by scraping and disrupted by sonicating twice (for 10 seconds each time) in 50 mM Tris-HCl (pH 7.2), 150 mM NaCl (pH 7.2), 0.1% Triton X-100, and Complete protease inhibitor (Roche Diagnostics Corp., Indianapolis, Indiana, USA), followed by centrifugation at 1,000 g for 5 minutes to remove nuclei and debris. Cell lysates were centrifuged at 100,000 g for 1 hour and the crude membrane pellet was suspended by sonicating three times (for 2 seconds each time) in lysis buffer. For immunoprecipitation experiments, 200 μg of cell membrane fractions were incubated with 20 μg of a murine anti-human TNFR1 or TNFR2 monoclonal antibody (R&D Systems Inc., Minneapolis, Minnesota, USA), or with 1 μl of anti–ARTS-1 immune or preimmune serum. Incubation took place overnight at 4°C, followed by immunoprecipitation with protein A/G beads (Pierce Chemical Co., Rockford, Illinois, USA). Proteins were resolved via SDS-PAGE, electroblotted onto nitrocellulose membranes, and incubated with either ARTS-1 immune or preimmune serum diluted 1:20,000 or anti-TNFR1 or anti-TNFR2 monoclonal antibodies at a concentration of 2 μg/ml. Signals were detected by chemiluminescence using horseradish peroxidase–conjugated secondary antibodies. For immunoblotting experiments, 20 μg of cell membrane proteins per sample was used. For competitive immunoblotting experiments, 1 μl of anti–ARTS-1 serum was incubated with 1 mg of either the RGRNVHMKQEHYMKGSD peptide or BSA for 2 hours before immunoblotting. A goat anti-human TNF-α converting enzyme (TACE) polyclonal IgG antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) was used at a concentration of 1 μg/ml for TACE immunoblotting experiments.
ARTS-1 confocal immunofluorescence microscopy.
Nonfixed, nonpermeabilized bronchial brushings of normal human airway epithelial cells and frozen sections of normal human bronchi were used. Tissue samples of normal human bronchi were provided by the Cooperative Human Tissue Network (University of Virginia, Charlottesville, Virginia, USA), which is funded by the National Cancer Institute. Slides were blocked with 10% goat serum in PBS and incubated with antibodies diluted in 1.5% goat serum in PBS. The following antibodies were used: anti–ARTS-1 and preimmune rabbit serum (1:1,500 dilution), anti-TNFR1 extracellular domain (2 μg/ml murine monoclonal IgG2b; Santa Cruz Biotechnology Inc.), murine IgG2b isotype control (2 μg/ml; R&D Systems Inc.), Alexa Fluor 488 F(ab′)2 fragment of goat anti-rabbit IgG, and Alexa Fluor 633 F(ab′)2 fragment of goat anti-mouse IgG (4 μg/ml; Molecular Probes Inc., Eugene, Oregon, USA). Confocal immunofluorescence microscopy was performed using a Zeiss LSM 510 confocal microscope and 40× and 100× Plan-Neofluar objectives, respectively, for frozen sections of human bronchi and cytospins of bronchial brushings. Alexa Fluor 488 and Alexa Fluor 633 were imaged sequentially at a data depth of 8 bits using 488-nm and 633-nm excitation wavelength and LP 505 and LP 650 emission filters, with pinholes adjusted to produce an axial thickness of 4.0–5.0 μM.
Expression, purification, and characterization of recombinant ARTS-1.
The cDNA sequence for the putative ARTS-1 extracellular domain (amino acids 30–941) was cloned into the pGEX-6P-1 plasmid (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). The glutathione s-transferase–ARTS-1 (GST–ARTS-1) fusion protein was isolated from the insoluble fraction of transformed BL21 E. coli by denaturation with 6 M urea in PBS and refolded by serial dialysis against PBS containing decreasing urea concentrations. The GST–ARTS-1 fusion protein was purified using a glutathione Sepharose 4B affinity column; purity was assessed by Coomassie brilliant blue–stained SDS-PAGE gels and by fast-performance liquid chromatography (FPLC) analysis using a Superose 6 HR 10/30 gel filtration column (Amersham Pharmacia Biotech).
A 20-amino-acid TNFR1 peptide substrate (TKLCLPQIENVKGTEDSGTT) containing the major and minor cleavage sites for TNFR1 shedding was synthesized by Sigma-Genosys (The Woodlands,Texas, USA) (4). The TNFR1 peptide substrate was mixed with GST–ARTS-1 at a molar ratio of 4:1 in 50 μl of 50 mM Tris-HCl and 120 mM NaCl (pH 7.5) for 2 hours at either room temperature or 37°C. Samples were analyzed by HPLC (Hewlett-Packard, Palo Alto, California, USA) using a Jupiter C-18 column (Phenomenex Inc., Torrance, California, USA). Similarly, GST–ARTS-1 was incubated for 2 hours with a recombinant human TNFR1-Fc fusion protein (R&D Systems Inc.) containing the TNFR1 extracellular domain fused to the Fc region of human IgG1. Endopeptidase activity was assessed by TNFR1 immunoblotting.
To assess whether GST–ARTS-1 possessed nonspecific endopeptidase activity, 5 μg of GST–ARTS-1 was incubated separately with 10 μg of either human albumin, BSA, rabbit myosin heavy chain, or human transferrin (Sigma-Aldrich, Milwaukee, Wisconsin, USA) overnight at 37°C. Samples were resolved by SDS-PAGE and stained with Coomassie brilliant blue.
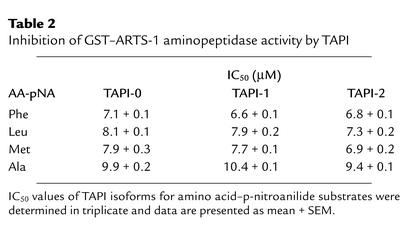
Aminopeptidase activity of recombinant GST–ARTS-1 fusion protein was determined as the rate of amide bond hydrolysis of amino acid–p-nitroanilide substrates (Bachem, Torrance, California, USA) under conditions of constant enzyme activity throughout the assay. Amino acid–p-nitroanilides (final concentrations 0.25–8 mM) were incubated at room temperature with 24 pmol of GST–ARTS-1 fusion protein in 200 μl of 50 mM Tris-HCl and 120 mM NaCl (pH 7.5) for 1 hour. Reactions were terminated by addition of 800 μl of 1.4 M sodium acetate (pH 4.4). Amide bond hydrolysis was quantified by p-nitroanilide absorbance at 380 nm and corrected for spontaneous hydrolysis of the substrate by subtracting the absorbance of control incubations that were terminated at time zero. Kinetic constants were determined by Lineweaver-Burk analysis using six concentrations of each amino acid–p-nitroanilide substrate with triplicate assays. Correlation coefficients for each line were greater than 0.997. For TNF-α protease inhibitor (TAPI) inhibition studies, selected amino acid–p-nitroanilides (4 mM) were incubated at room temperature for 1 hour with 15 pmol of GST–ARTS-1 and either TAPI-0, TAPI-1, or TAPI-2 (6.25–100 μM).
ARTS-1 cell lines.
cDNAs encoding the full-length ARTS-1 coding sequence in the sense orientation and ARTS-1 bases 61–213 in the antisense orientation were cloned into the pTarget mammalian expression vector (Promega Corp., Madison, Wisconsin, USA). The ARTS-1 catalytic site mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene). NCI-H292 cells were stably transfected using FuGENE 6 (Roche Diagnostics Corp.) and maintained under selective pressure with geneticin (500 μg/ml). Cell lines were established from clones that had been generated by limiting dilutions and selected based upon enhanced or suppressed ARTS-1 membrane-associated protein expression, as determined by immunoblotting. HUVECs were transiently transfected for 4 days prior to collection of cell culture supernatants. TNFR1 and TNFR2 shedding into cell culture supernatants over a 24-hour period was assayed using a sandwich ELISA with a sensitivity of 7.8 pg/ml (R&D Systems Inc.). TAPI-0, TAPI-1, and TAPI-2 were purchased from Peptides International Inc. (Louisville, Kentucky, USA). Statistical analysis was performed using a paired Student t test with a Bonferroni correction for multiple comparisons. A P value of less than 0.05 was considered significant.
Total RNA was extracted using the RNeasy Maxi Kit (Qiagen Inc., Valencia, California, USA), and ribonuclease protection assays were performed with the RPA III kit (Ambion Inc., Austin, Texas, USA) using 60 μg of total RNA per sample. Antisense TNFR1 and GAPDH riboprobes were labeled using the DIG RNA labeling kit and detected with the DIG luminescent detection kit (Roche Diagnostics Corp.) The TNFR1 template, which was used to generate the 124-bp protected fragment corresponding to nucleotides 356–478 of TNFR1 mRNA (GenBank accession number NM001065), was verified by sequencing, while the GAPDH template used to generate the 316-bp protected fragment was purchased from Ambion Inc.
Subcellular fractionation.
NCI-H292 cells were scraped in ice-cold lysis buffer (10 mM Tris at pH 7.5, 1 mM EDTA, 0.25 M sucrose, 0.8 mM AEBSF, 50 μg/ml antipain, 1.5 μM aprotinin, 83 μM chymostatin, 5.6 μM E-64, 2 μM leupeptin, 5 μM pepstatin, and 10 μM phosphoramidon) and lysed by 15 passes through a 25-gauge needle. Crude membranes were collected from postnuclear supernatants by centrifugation at 100,000 g for 1 hour, resuspended in lysis buffer, and underlaid with a discontinuous gradient containing 0.5 M, 0.86 M, 1.15 M, and 1.4 M sucrose in TE buffer. Following centrifugation at 175,000 g for 1.75 hours, the fractions and interfaces were collected from the top, and proteins were precipitated with 10% trichloroacetic acid and resuspended in gel loading buffer. Immunoblotting was performed as previously described, using monoclonal antibodies directed against TNFR1 (Santa Cruz Biotechnology Inc.), β-catenin, and GM130 (both from Becton, Dickinson and Co., Franklin Lakes, New Jersey, USA). Forty micrograms of protein per fraction was used on immunoblots to detect TNFR1 and β-catenin, while 200 μg of protein per fraction was used in studies to detect GM130.
Results
Identification and molecular cloning of ARTS-1.
A yeast two-hybrid screen of a human lung cDNA library was conducted to identify proteins interacting with the extracellular domain of the 55-kDa TNFR1 (amino acids 26–216, situated between the leader and transmembrane domains) (4). From 7.1 × 105 transformants screened, 33 positive clones were identified and sequenced. Clone L26C-53A contained a 2,355-bp insert with an open reading frame of 631 amino acids encoding a consensus zinc metalloprotease catalytic site. This clone corresponded to ARTS-1 base pairs 1,044–3,082, located within the ARTS-1 extracellular domain. Clone L26C-53A was used to screen a cDNA library prepared from a PMA-stimulated, NCI-H292 pulmonary mucoepidermoid carcinoma cell line. Four clones were identified that overlapped the L26C-53A sequence but were not full-length. One clone contained the putative 5′ UTR (bp 1–1,777) and three contained the putative 3′ UTR (bp 2,181–4,845). The remainder of the cDNA sequence was confirmed by PCR with human lung cDNA, using primers spanning bases 1,684–2,280. Sequences of both strands of all overlapping regions from the L26C-53A insert (bp 1,044–3,082), the four NCI-H292 cDNA library clones, and the PCR product consisting of bp 1,684–2,280 were identical.
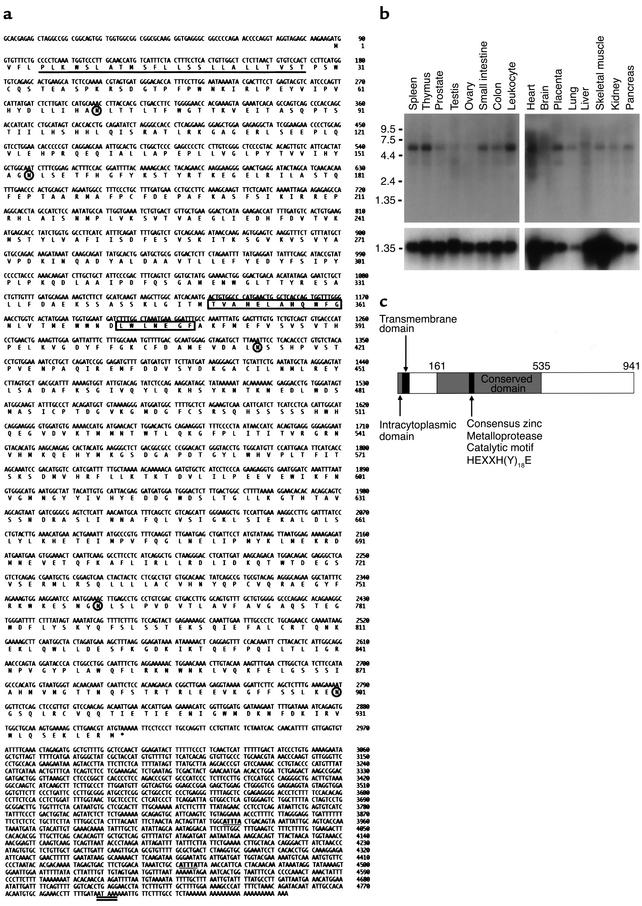
The full-length human ARTS-1 cDNA (Figure 1a) contains 4,845 nucleotides, including a 2,823-bp open reading frame that encodes a protein of 941 amino acids with a calculated molecular weight of 107,227 and an estimated pI of 6.0. The first in-frame ATG codon (nucleotide 88) follows an in-frame stop codon and matches the –3 and +4 nucleotides of the consensus Kozak sequence, consistent with a strong initiator codon (6). Sequence analysis revealed five potential N-glycosylation sites. The 3′ untranslated region contained two ATTTA mRNA destabilization motifs and a consensus polyadenylation signal. As shown in Figure 1b, on Northern blot analysis of poly(A+) mRNA from human tissues, a 5.7-kb ARTS-1 mRNA transcript is widely distributed, with highest levels in spleen, thymus, leukocyte, heart, and placenta.
Figure 1.
Characterization of ARTS-1 mRNA and protein. (a) ARTS-1 nucleotide and amino acid sequences. The full-length (4,845 bp) ARTS-1 cDNA includes a 2,823-bp open reading frame encoding a 941-amino-acid protein. The putative transmembrane domain (amino acids 5 and 28) is underlined, the consensus zinc metalloprotease catalytic motif HEXXH(Y)18E is boxed, five potential N-glycosylation sites are circled, two mRNA destabilization motifs in the 3′ UTR are in bold and underlined, and the putative polyadenylation signal is double underlined. The ARTS-1 sequence data were submitted to GenBank under accession number AF222340. (b) Tissue distribution of ARTS-1 mRNA expression. Northern blot analysis of mRNA from multiple human tissues hybridized with 32P-labeled ARTS-1 cDNA is shown in the top panel, and GAPDH mRNA is shown below. (c) ARTS-1 protein structure. The ARTS-1 protein is predicted to be a type II integral membrane protein with a very short, amino-terminal intracytoplasmic domain, followed by a hydrophobic transmembrane α-helical domain. Located within the large 913-amino-acid extracellular domain is a 375-amino-acid region containing the consensus zinc metalloprotease catalytic motif HEXXH(Y)18E, which is highly conserved among aminopeptidase family members.
ARTS-1 is predicted to be a type II integral membrane protein with a single hydrophobic transmembrane α-helical domain located between amino acids 5 and 28 (Figure 1, a and c) and a very short intracellular amino-terminal domain. Within the 913-amino-acid ARTS-1 extracellular domain is a highly conserved 375-amino-acid region (Figure 1c) that contains the consensus zinc metalloprotease catalytic motif HEXXH(Y)18E, as well as a GAMEN motif, both of which are highly conserved among zinc metalloproteases of the gluzincin aminopeptidase family (7, 8). ARTS-1 shares significant sequence identity and similarity with many aminopeptidase family members. Furthermore, our characterization of ARTS-1 as a type II integral membrane protein is consistent with the structure of other membrane-associated aminopeptidases, such as placental leucine aminopeptidase, aminopeptidase A, aminopeptidase N, insulin-regulated aminopeptidase, and thyrotropin-releasing hormone degrading enzyme. As has been demonstrated for aminopeptidase N, the putative ARTS-1 transmembrane domain may serve as a signal for membrane insertion as well as a membrane-spanning domain (9).
ARTS-1 is a type II integral membrane protein.
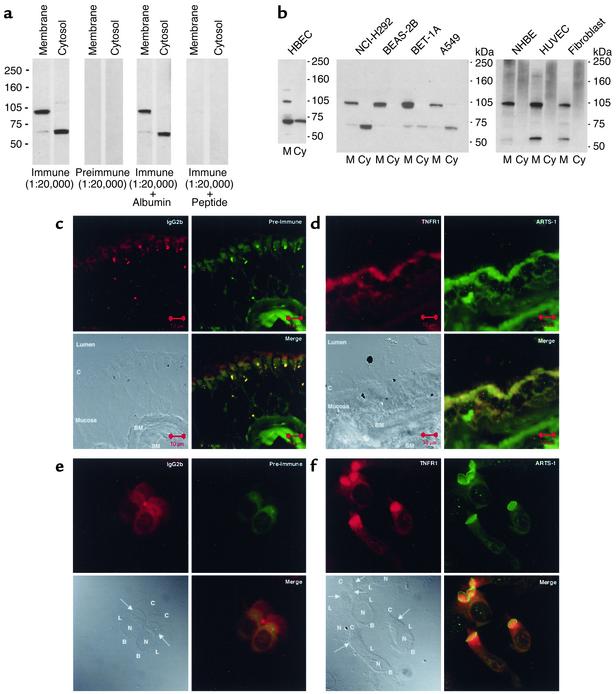
Anti–ARTS-1 serum was used to demonstrate that ARTS-1 is expressed as a membrane-associated protein. The anti–ARTS-1 antibody was demonstrated to be specific for ARTS-1 based on immunoblots of NCI-H292 membrane and cytosolic proteins that demonstrated no signal with preimmune serum and competitive inhibition after preincubation of the immune serum with the ARTS-1 peptide used for immunization (Figure 2a).
Figure 2.
Characterization of ARTS-1 as a type II integral membrane protein. (a) Specificity of anti–ARTS-1 serum. Immunoblots were performed on membrane and cytosolic fractions from NCI-H292 cells using anti–ARTS-1 immune or preimmune serum. Competitive inhibition experiments were conducted by preincubation of anti–ARTS-1 immune serum with either BSA or the peptide epitope against which the anti–ARTS-1 immune serum was raised. (b) ARTS-1 is a membrane-associated protein. Membrane (M) and cytosolic (CY) protein fractions of human bronchial epithelial cells (HBECs) obtained via bronchial brushings (left panel), human bronchial epithelial cell lines (NCI-H292, BEAS-2B, BET-1A, and A549) (center panel), and primary cultures of normal human bronchial epithelial cells (NHBEs), HUVECs, and fibroblasts (right panel) were separated by SDS-PAGE, transferred to nitrocellulose membranes, and reacted with anti–ARTS-1 immune serum. (c–f) Colocalization of membrane-associated ARTS-1 and TNFR1 in human bronchial epithelial cells. Confocal immunofluorescence laser microscopy was performed on nonfixed, nonpermeabilized frozen sections of normal human bronchi (c and d) and on nonfixed, nonpermeabilized cytospin preparations of normal human bronchial epithelial cells obtained via bronchial brushings (e and f) using a murine IgG2b isotype control and preimmune serum (c and e) and anti-TNFR1 and anti–ARTS-1 antibodies (d and f). An annotated differential interference contrast image is shown in the bottom left panels. Arrows denote the apical cell membrane. C, cilia; BM, basement membrane; SM, submucosa; N, nucleus; L, lateral cell membrane; B, basal cell membrane.
Immunoblots of human bronchial epithelial cells obtained via bronchial brushings and multiple human bronchial epithelial cell lines (NCI-H292, BEAS-2B, BET-1A, and A549) revealed that ARTS-1 was expressed as a 100-kDa membrane-associated species (Figure 2b, left and center panels). Minor 132-kDa and 68-kDa membrane-associated species were also present. The 68-kDa ARTS-1 species was also detected in cytosolic fractions. Similarly, ARTS-1 was expressed as a 100-kDa membrane-associated species and a minor 59-kDa species in membrane fractions of primary cultures of normal human bronchial epithelial cells, vascular endothelial cells (HUVECs), and fibroblasts (Figure 2b, right panel). These immunoblots demonstrate that ARTS-1 is expressed as a 100-kDa membrane-associated protein in multiple cell types.
Confocal immunofluorescence microscopy was performed to characterize ARTS-1 as a type II integral membrane protein and to colocalize the expression of the extracellular domains of membrane-associated ARTS-1 and TNFR1. Frozen sections of normal human bronchi and cytospins of human bronchial epithelial cells that were neither fixed nor permeabilized were used. As shown in Figure 2, d and f, ARTS-1 was localized to the apical cell membrane of ciliated human bronchial epithelial cells. Furthermore, ARTS-1 was most prominently visualized at the junction of the apical and lateral membranes (Figure 2d). These data are consistent with the conclusion that ARTS-1 is a type II integral membrane protein because the epitope against which the anti–ARTS-1 serum was raised is located in the putative ARTS-1 extracellular domain, and the anti–ARTS-1 antibody should bind only extracellular proteins in preparations that are not fixed or permeabilized. The TNFR1 extracellular domain was also expressed at the apical cell membrane of ciliated human bronchial epithelial cells and colocalized with ARTS-1. As shown in Figure 2, e and f, no signal was detected with preimmune serum or the IgG2b isotype control. These experiments demonstrate that the ARTS-1 and TNFR1 extracellular domains are colocalized at the apical cell membranes of human bronchial epithelial cells.
Characterization of ARTS-1 aminopeptidase activity.
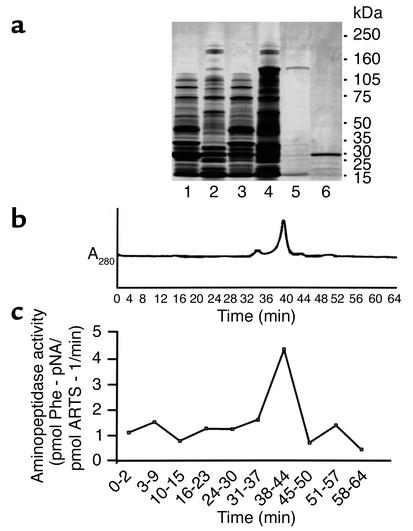
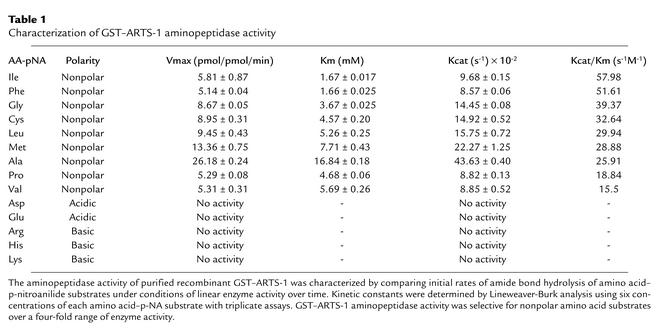
An enzymatically active recombinant GST–ARTS-1 fusion protein was expressed in BL21 E. coli and purified via binding to a glutathione affinity column (Figure 3, a–c). The aminopeptidase activity of purified recombinant GST–ARTS-1 was characterized by comparing initial rates of hydrolysis of different amino acid–p-nitroanilide substrates under conditions of linear enzyme activity over time. As shown in Table 1, recombinant GST–ARTS-1 aminopeptidase activity was selective for nonpolar amino acid substrates over a fourfold range of enzyme activity. Isoleucine–p-nitroanilide was the preferred substrate based on kcat/Km values, followed by Phe > Gly > Cys > Leu > Met > Ala > Pro > Val. Recombinant GST–ARTS-1 had no activity against either acidic (Asp or Glu) or basic (Arg, His, or Lys) amino acid–p-nitroanilides. Furthermore, GST–ARTS-1 aminopeptidase activity was inhibited by the hydroxamic acid–based zinc metalloprotease inhibitor TAPI. The IC50 values of TAPI isoforms for several amino acid substrates are shown in Table 2. Recombinant GST–ARTS-1 therefore possesses aminopeptidase activity that is selective for nonpolar amino acid substrates.
Figure 3.
Characterization of GST–ARTS-1 aminopeptidase activity. (a) Generation of GST–ARTS-1. Soluble and insoluble protein fractions were isolated from BL21 E. coli transformed with empty pGEX-6P-1 (Lane 1, soluble fraction; Lane 2, insoluble fraction) or ARTS-1 pGEX-6P-1 (Lane 3, soluble fraction; Lane 4, insoluble fraction). Proteins were subjected to SDS-PAGE and stained with Coomassie brilliant blue. Purified GST–ARTS-1 fusion protein from the insoluble fraction is shown as a predominant 130-kDa band in Lane 5, and the 26-kDa purified control GST tag is shown in Lane 6. (b) FPLC analysis of purified recombinant GST–ARTS-1 fusion protein revealed a major peak that eluted at approximately 40 minutes. (c) Assay of GST–ARTS-1 aminopeptidase activity. FPLC fractions were assessed for aminopeptidase activity using a phenylalanine–p-nitroanilide (Phe-pNA) substrate. Phenylalanine aminopeptidase activity was present in pooled fractions eluting from 38 to 44 minutes, which correlated with the major FPLC peak.
Table 1.
Characterization of GST–ARTS-1 aminopeptidase activity
Table 2.
Inhibition of GST–ARTS-1 aminopeptidase activity by TAPI
In vivo binding of membrane-associated ARTS-1 to TNFR1.
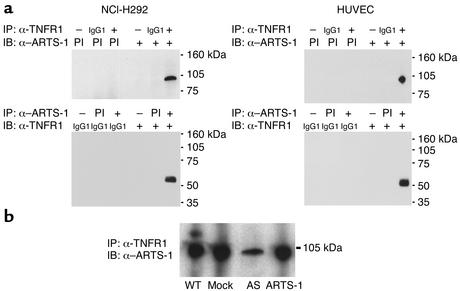
Immunoprecipitation experiments were performed to assess whether an in vivo interaction exists between ARTS-1 and TNFR1 in both a human bronchial epithelial cell line (NCI-H292) and HUVECs. As shown in Figure 4a, immunoprecipitation of cell membranes with an anti-TNFR1 monoclonal antibody pulled down the 100-kDa ARTS-1 species. In the reciprocal experiment, immunoprecipitation with anti–ARTS-1 serum pulled down 55-kDa TNFR1, which is consistent with binding of ARTS-1 to a full-length, uncleaved TNFR1. Furthermore, these data support our hypothesis that ARTS-1 binds to, but does not cleave, the TNFR1 ectodomain. Therefore, these experiments demonstrate that the 100-kDa membrane-associated ARTS-1 species binds di-rectly to membrane-associated TNFR1 in vivo.
Figure 4.
In vivo binding of ARTS-1 to TNFR1 in human epithelial and endothelial cells. (a) ARTS-1 binds to TNFR1 in vivo. Coimmunoprecipitation experiments were performed on membrane proteins from NCI-H292 cells (left) and HUVECs (right). As shown in the top panels, immunoprecipitations were performed with either an anti-TNFR1 monoclonal antibody (+) or a murine IgG1 isotype control (IgG1) and immunoblotted with either anti–ARTS-1 preimmune (PI) or immune (+) serum. As shown in the bottom panels, reciprocal coimmunoprecipitations were performed with either anti-ARTS-1 preimmune (PI) or immune (+) serum and immunoblotted with either an anti-TNFR1 monoclonal antibody (+) or a murine IgG1 isotype control (IgG1). IP indicates the antibody used for immunoprecipitation and IB indicates the antibody used for immunoblotting. (b) Effect of ARTS-1 protein expression on in vivo binding of ARTS-1 to TNFR1 in NCI-H292 cell lines. Membrane proteins of wild-type NCI-H292 cells, mock-transfected cells, and ARTS-1 cell lines were immunoprecipitated with an anti-TNFR1 monoclonal antibody and immunoblotted with anti–ARTS-1 serum.
ARTS-1 promotes TNFR1 ectodomain shedding.
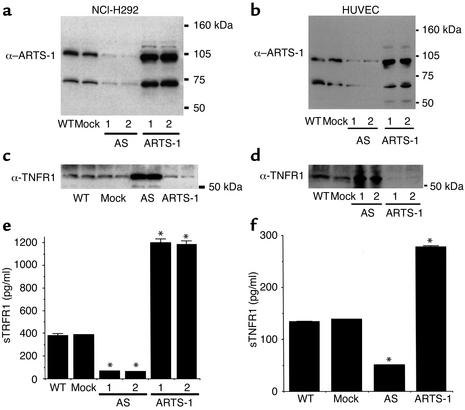
To determine whether ARTS-1 promotes TNFR1 shedding, we generated stably transfected cell lines expressing ARTS-1 cDNA in the sense or antisense orientation. As shown in Figure 5a, cell lines were cloned based upon levels of ARTS-1 protein as determined by immunoblotting of cell membrane fractions. Cell lines expressing antisense ARTS-1 mRNA contained less ARTS-1 protein than did mock-transfect-ed cell lines, while cell lines overexpressing ARTS-1 protein contained more membrane-associated ARTS-1 than did mock-transfected cell lines. Similar differen-ces in amounts of membrane-associated ARTS-1 protein levels were found in transiently transfected HUVECs (Figure 5b).
Figure 5.
ARTS-1 promotes TNFR1 shedding from human epithelial and endothelial cells. (a) ARTS-1 protein expression by NCI-H292 cell lines. Immunoblots were performed on membrane fractions from wild-type NCI-H292 cells (WT) or cells stably transfected with either empty pTarget (Mock), or pTarget encoding either sense (ARTS-1) or antisense (AS) ARTS-1 coding sequence. Samples 1 and 2 are from representative clonal lines. (b) ARTS-1 protein expression by transiently transfected HUVECs. Proteins were prepared the same way as for a. Samples 1 and 2 are from two representative transient transfections. (c) Effect of ARTS-1 protein expression on membrane-associated TNFR1 levels in ARTS-1 cell lines. Immunoblots of membrane fractions were performed in duplicate with an anti-TNFR1 antibody. (d) Effect of ARTS-1 protein expression on membrane-associated TNFR1 levels in HUVECs. Immunoblots of membrane fractions of transiently transfected HUVECs were performed with an anti-TNFR1 antibody. Samples 1 and 2 are from two representative transient transfections. (e) Effect of ARTS-1 on TNFR1 shedding from ARTS-1 cell lines. The amounts of sTNFR1 present in cell culture supernatants from two antisense (AS) and two sense ARTS-1 cell lines over a 24-hour period were determined by ELISA (n = 5). *P < 0.05 as compared to mock transfected cells. (f) Effect of ARTS-1 on TNFR1 shedding from HUVECs. The amount of sTNFR1 present in cell culture supernatants from transiently transfected HUVECs over a 24-hour period was determined by ELISA (n = 5). *P < 0.05 as compared with mock-transfected cells.
Immunoprecipitation experiments were performed to further characterize the NCI-H292 ARTS-1 cell lines. As shown in Figure 4b, immunoprecipitates of membrane proteins from cell lines expressing antisense ARTS-1 with an anti-TNFR1 monoclonal antibody contained less ARTS-1 protein than did those from mock-transfected cell lines or cell lines overexpressing ARTS-1, consistent with decreased synthesis of ARTS-1 protein. No increase in ARTS-1 protein was detected in the immunoprecipitates of cell lines expressing full-length ARTS-1 with an anti-TNFR1 monoclonal antibody, which reflects increased TNFR1 shedding related to overexpression of ARTS-1.
Experiments were performed to assess whether ARTS-1 protein expression correlates with changes in membrane-associated TNFR1 levels. As shown in Figure 5, c and d, membrane-associated TNFR1 was increased in NCI-H292 cells and HUVECs expressing antisense ARTS-1 mRNA, consistent with a reduction in TNFR1 ectodomain shedding. Similarly, in cells overexpressing ARTS-1, membrane-associated TNFR1 was decreased, consistent with an increase in TNFR1 ectodomain shedding. This demonstrates that a reciprocal relationship exists between changes in membrane-associated ARTS-1 and membrane-associated TNFR1 protein levels.
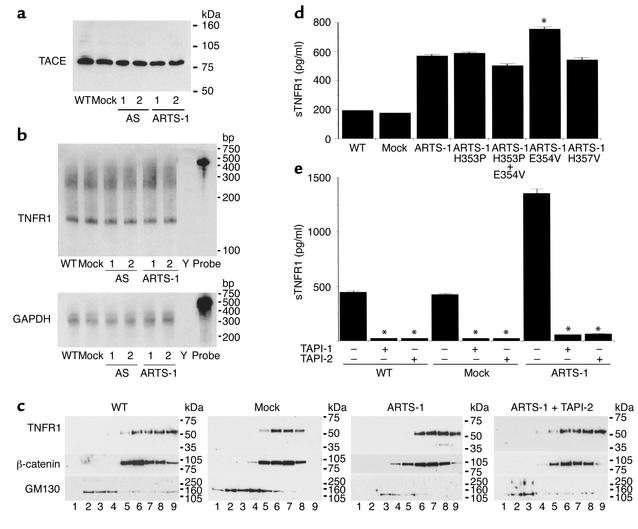
The effect of ARTS-1 protein expression on TNFR1 ectodomain shedding into cell culture supernatants from human epithelial and endothelial cells was also evaluated. As shown in Figure 5, e and f, supernatants from NCI-H292 cells and HUVECs overexpressing full-length ARTS-1 contained more soluble TNFR1 (sTNFR1), and supernatants from cells expressing antisense ARTS-1 contained less sTNFR1, than did supernatants from mock-transfected cells. Taken together, these experiments demonstrate that the amount of membrane-associated ARTS-1 is directly correlated with the amount of TNFR1 ectodomain shedding in both human epithelial and endothelial cells. Furthermore, the effect of ARTS-1 protein expression on TNFR1 ectodomain shedding was not a consequence of either altered TACE protein expression (Figure 6a) or altered TNFR1 mRNA levels (Figure 6b). In addition, subcellular fractionation experiments were performed to assess whether ARTS-1 alters TNFR1 protein sorting. As shown in Figure 6c, neither overexpression of ARTS-1 protein nor treatment with 25 μM of TAPI, a hydroxamic acid–based metalloprotease inhibitor of TACE, altered TNFR1 localization in NCI-H292 cells. In these experiments, TNFR1 primarily colocalized with β-catenin, an E-cadherin–binding protein that resides in the plasma membrane (10).
Figure 6.
Characterization of TNFR1 shedding from ARTS-1 cell lines. (a) ARTS-1 expression does not alter TACE protein levels. Immunoblots were performed on membrane fractions of ARTS-1 cell lines, as described in the legend to Figure 5a, and reacted with an anti-TACE antibody. Samples 1 and 2 are from representative clone cell lines. (b) ARTS-1 expression does not alter TNFR1 mRNA levels. Ribonuclease protection assays were performed on total RNA isolated from ARTS-1 cell lines. Probe, undigested riboprobe; Y, yeast tRNA negative control. (c) ARTS-1 expression does not alter TNFR1 subcellular localization. Crude membrane fractions from ARTS-1 cell lines, with or without TAPI-2 (25 μm) treatment, were centrifuged through a discontinuous sucrose gradient. TCA-precipitated proteins were immunoblotted with antibodies against TNFR1, β-catenin, and GM130. Discontinuous sucrose gradient fractions are as follows: Lane 1, 0.25 M; lane 2, 0.25/0.5 M interface; lane 3, 0.5 M; lane 4, 0.5/0.86 M interface; lane 5, 0.86 M; lane 6, 0.86/1.15 M interface; lane 7, 1.15 M; lane 8, 1.15/1.4 M interface; lane 9, pellet. (d) Increased TNFR1 shedding is preserved in ARTS-1 catalytic site mutants. Cell culture supernatants were collected after 24 hours and the amount of sTNFR1 present was determined by ELISA (n = 5). *P < 0.02 compared with ARTS-1. (e) TAPI inhibits ARTS-1–mediated increases in TNFR1 shedding. ARTS-1 cell lines were treated for 24 hours with TAPI-1 or TAPI-2 (25 μM). The amount of sTNFR1 present in cell culture supernatants was determined by ELISA and compared with untreated cells (n = 5). * P < 0.05.
ARTS-1 does not bind to or promote TNFR2 ectodomain shedding.
Immunoprecipitation was performed to assess whether ARTS-1 binds to TNFR2 in vivo. Using membrane fractions from human pulmonary epithelial cells and HUVECs, anti-TNFR2 antibodies failed to coimmunoprecipitate ARTS-1, whereas anti-TNFR1 antibodies successfully coimmunoprecipitated 100-kDa ARTS-1 protein (data not shown). Similarly, ARTS-1 antibodies failed to coimmunoprecipitate TNFR2 from membrane fractions of human pulmonary epithelial cells and HUVECs (data not shown). Membrane-associated TNFR2 was demonstrated in both human pulmonary epithelial cells and HUVECs by immunoprecipitation and immunoblotting (data not shown). There was no significant difference in TNFR2 shedding from HUVECs into culture supernatants between mock-transfected cells (65.3 ± 2.7 pg/ml, n = 5) and cells transiently transfected with antisense ARTS-1 mRNA (61.5 ± 0.6 pg/ml, n = 5, P value not significant) or full-length sense ARTS-1 mRNA (67.5 ± 3.3 pg/ml, n = 5, P value not significant). These experiments demonstrate that the ability of ARTS-1 to bind to and promote receptor shedding is specific to TNFR1 and does not extend to TNFR2.
ARTS-1 does not possess TNFR1 sheddase activity.
To assess whether ARTS-1 possesses TNFR1 sheddase activity, purified GST–ARTS-1 was incubated with a 20-amino-acid TNFR1 peptide substrate containing the major and minor ectodomain cleavage sites for TNFR1 (TKLCLPQIENVKGTEDSGTT). No endopeptidase activity was demonstrated using this model system (data not shown). Similarly, recombinant GST–ARTS-1 had no endopeptidase activity against a human TNFR1-Fc fusion protein containing the entire TNFR1 extracellular domain, and no detectable nonspecific endopeptidase activity against human albumin, BSA, rabbit myosin heavy chain, or human transferrin (data not shown). These experiments suggest that ARTS-1 does not possess endopeptidase activity and cannot cleave the TNFR1 ectodomain under these conditions.
To further assess whether ARTS-1 protein can catalyze TNFR1 shedding, four stably transfected NCI-H292 cell lines were generated with full-length ARTS-1 catalytic site mutants containing either single or double amino acid substitutions at key residues in the consensus zinc metalloprotease catalytic site HEXXH(Y)18E. The two histidines (H353 and H357) and the second glutamic acid (E376) in the consensus metalloprotease catalytic site participate in zinc binding, whereas the first glutamic acid (E354) mediates catalysis (11, 12). Mutation of either of the first two histidines within the zinc metalloprotease catalytic motif abolishes both catalytic activity and zinc binding, whereas mutation of the first glutamic acid abolishes enzymatic activity (11–13). The ARTS-1 catalytic site mutants contained the following mutations generated by site-directed mutagenesis: H353P; E354V; H353P and E354V; and H357V. There was no significant decrease in sTNFR1 levels in cell culture supernatants from NCI-H292 cell lines overexpressing ARTS-1 catalytic site mutants compared with cell lines overexpressing full-length ARTS-1 at 24 hours (Figure 6d). Furthermore, overexpression of the E354V mutant resulted in increased TNFR1 shedding. These experiments demonstrate that the increase in TNFR1 shedding associated with overexpression of ARTS-1 does not require an intact ARTS-1 zinc metalloprotease catalytic site. Instead, we hypothesize that the formation of a TNFR1–ARTS-1 molecular complex may facilitate ectodomain cleavage by the TNFR1 sheddase. Additional experiments were performed to assess whether the ability of ARTS-1 to promote TNFR1 shedding can be inhibited by TAPI, an inhibitor of ARTS-1 and TACE zinc metalloprotease activities, as well as TNFR1 shedding (14). TAPI-1 and TAPI-2 (25 μM) completely inhibited the increases in TNFR1 shedding into cell culture supernatants from cell lines overexpressing ARTS-1 (Figure 6e), thereby demonstrating the requirement for zinc metalloprotease activity. Taken together, these experiments suggest that ARTS-1 does not directly catalyze TNFR1 ectodomain shedding, but may instead promote the zinc metalloprotease activity of a TNFR1 sheddase such as TACE.
Discussion
In this study, we report the identification, cloning, and characterization of human ARTS-1. We propose that ARTS-1 directly binds to the TNFR1 extracellular domain and thereby facilitates TNFR1 shedding. This is supported by our demonstration of (a) in vivo binding of 100-kDa membrane-associated ARTS-1 to TNFR1 in human epithelial and endothelial cells; (b) an interaction between the extracellular domains of ARTS-1 and TNFR1 in the yeast two-hybrid system; (c) a direct correlation among increased ARTS-1 expression, increased TNFR1 ectodomain shedding, and decreased membrane-associated TNFR1; and (d) a direct correlation among decreased ARTS-1 expression, decreased TNFR1 shedding, and increased membrane-associated TNFR1. Furthermore, the ability of ARTS-1 to bind and promote receptor shedding is specific to TNFR1; ARTS-1 had no effect on TNFR2. Taken together, these data demonstrate that binding of ARTS-1 to TNFR1 promotes TNFR1 ectodomain cleavage and shedding from both human epithelial and endothelial cells. We propose that the formation of a TNFR1–ARTS-1 molecular complex represents an important and novel mechanism by which TNFR1 shedding is enhanced.
We have identified ARTS-1 as a type II integral membrane protein with a large extracellular domain containing a consensus zinc metalloprotease catalytic motif. Therefore, the ARTS-1 extracellular domain is correctly positioned to bind the TNFR1 extracellular domain and facilitate its proteolytic cleavage and subsequent shedding. These findings are consistent with our yeast two-hybrid data demonstrating an interaction between the extracellular domains of ARTS-1 and TNFR1 and the confocal immunofluorescence microscopy experiments demonstrating colocalization of the extracellular domains of ARTS-1 and TNFR1 in the apical cell membranes of human ciliated bronchial epithelial cells. In addition, the ARTS-1 zinc metalloprotease catalytic motif is correctly positioned to function as an ectoenzyme, thereby mediating the selective cleavage of nonpolar amino-terminal amino acids. Our characterization of ARTS-1 as a functional aminopeptidase, with activity that is selective for nonpolar amino acid substrates, is consistent with the recent cloning and identification of the soluble aminopeptidases (adipocyte-derived leucine aminopeptidase and puromycin-insensitive leucyl-specific aminopeptidase) that are essentially identical to ARTS-1 and demonstrate substrate specificity for leucine and methionine as well as for peptide hormones such as angiotensin II and kallidin (15–17). In contrast to these leucine-specific aminopeptidases, which were localized to cytoplasmic and vesicular compartments, we identified membrane-associated ARTS-1 by its novel ability to bind directly to and promote TNFR1 shedding.
Although ARTS-1 is an active zinc metalloprotease, our data demonstrate that it regulates TNFR1 ectodomain cleavage in a noncatalytic fashion. Our conclusion that ARTS-1 does not possess TNFR1 sheddase activity is based on several lines of negative evidence. First, GST–ARTS-1 did not cleave a TNFR1 ectodomain peptide substrate containing the known TNFR1 cleavage sites or a TNFR1 fusion protein containing the entire TNFR1 extracellular domain. Second, increased TNFR1 shedding was observed with overexpression of ARTS-1 catalytic site mutants, suggesting that an active ARTS-1 zinc catalytic site is not required for upregulation of TNFR1 shedding. Third, coimmunoprecipitation of TNFR1 with an anti–ARTS-1 antibody revealed full-length TNFR1, suggesting that ARTS-1 can bind to, but not cleave, TNFR1. Lastly, GST–ARTS-1 had no demonstrable nonspecific endopeptidase activity against several candidate protein substrates. These data lead us to conclude that ARTS-1 does not function as a TNFR1 sheddase. Instead, we hypothesize that the ARTS-1 extracellular domain binds to the TNFR1 extracellular domain and functions as an extracellular TNFR1 regulatory protein that promotes TNFR1 shedding.
Members of the metalloprotease-disintegrin (ADAM) family of zinc metalloproteases, including ADAM 9 (MDC9 or meltrin-γ), ADAM 10 (kuzbanian), ADAM 17 (TACE), and ADAM 19 (meltrin-β), have been identified as sheddases for cytokine and growth factor receptors and ligands, as well as for adhesion molecules and the amyloid precursor protein (18–20). ADAM 17 or TACE is of particular interest since it has been reported to possess TNFR1 sheddase activity. TACE was initially demonstrated to catalyze the ectodomain cleavage and shedding of TNF-α, as well as TGF-α, L-selectin, and TNFR2 (21–23). TACE was subsequently reported to catalyze the shedding of TNFR1 and IL-1RII ectodomains, based on the demonstration of increased TNFR1 and IL-1RII shedding following reconstitution of TACE-deficient cell lines (24). Although TACE can function as a TNFR1 sheddase, we demonstrate that the ability of ARTS-1 to enhance TNFR1 shedding is not mediated by increased TACE protein expression. Although it is possible that ARTS-1 modulates TACE activity by regulating removal of the prodomain and thereby processing the protein to a mature form, no differences in TACE maturation were noted by immunoblotting. In addition, further experiments will be needed to assess whether ARTS-1 promotes the shedding of other targets of TACE activity, such as TNF-α.
In summary, we have identified, cloned, and characterized ARTS-1 as a multifunctional protein capable of binding to and promoting shedding of the TNFR1 ectodomain. ARTS-1 also possesses selective aminopeptidase activity toward nonpolar amino-terminal residues. We hypothesize that formation of a TNFR1–ARTS-1 molecular complex may promote ectodomain cleavage by the TNFR1 sheddase. Therefore, the ability of membrane-associated ARTS-1 to augment TNFR1 shedding represents a novel mechanism by which TNF bioactivity can be regulated.
Acknowledgments
We thank Joel Moss, James Shelhamer, and Martha Vaughan for their helpful insights and critical review of the manuscript, and James Shelhamer for his help in the design and execution of the FPLC experiments. We also thank the 8 East Clinical Center nursing staff for their assistance with fiberoptic bronchoscopies and patient care.
Footnotes
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: TNF receptor (TNFR); TNFR-associated death domain (TRADD); aminopeptidase regulator of TNFR1 shedding (ARTS-1); human umbilical vein endothelial cell (HUVEC); soluble TNFR1 (sTNFR1); TNF-α converting enzyme (TACE); glutathione s-transferase (GST); fast-performance liquid chromatography (FPLC); TNF-α protease inhibitor (TAPI).
References
- 1.Vilcek J, Lee TH. Tumor necrosis factor. New insights into the molecular mechanisms of its multiple actions. J Biol Chem. 1991;266:7313–7316. [PubMed] [Google Scholar]
- 2.Smith CA, Farrah T, Goodwin RG. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell. 1994;76:959–962. doi: 10.1016/0092-8674(94)90372-7. [DOI] [PubMed] [Google Scholar]
- 3.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 4.Nophar Y, et al. Soluble forms of tumor necrosis factor receptors (TNF-Rs). The cDNA for the type I TNF-R, cloned using amino acid sequence data of its soluble form, encodes both the cell surface and a soluble form of the receptor. EMBO J. 1990;9:3269–3278. doi: 10.1002/j.1460-2075.1990.tb07526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D. Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J Exp Med. 1992;175:323–329. doi: 10.1084/jem.175.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 7.Laustsen PG, et al. The complete amino acid sequence of human placental oxytocinase. Biochim Biophys Acta. 1997; 1352:1–7. doi: 10.1016/s0167-4781(97)00036-5. [DOI] [PubMed] [Google Scholar]
- 8.Keller SR, Scott HM, Mastick CC, Aebersold R, Lienhard GE. Cloning and characterization of a novel insulin-regulated membrane aminopeptidase from Glut4 vesicles. J Biol Chem. 1995;270:23612–23618. doi: 10.1074/jbc.270.40.23612. [DOI] [PubMed] [Google Scholar]
- 9.Look AT, Ashmun RA, Shapiro LH, Peiper SC. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Invest. 1989;83:1299–1307. doi: 10.1172/JCI114015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Someya A, et al. ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc Natl Acad Sci USA. 2001;98:2413–2418. doi: 10.1073/pnas.051634798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vallee BL, Auld DS. Short and long spacer sequences and other structural features of zinc binding sites in zinc enzymes. FEBS Lett. 1989;257:138–140. doi: 10.1016/0014-5793(89)81805-8. [DOI] [PubMed] [Google Scholar]
- 12.Devault A, et al. Expression of neutral endopeptidase (enkephalinase) in heterologous COS-1 cells. Characterization of the recombinant enzyme and evidence for a glutamic acid residue at the active site. J Biol Chem. 1988;263:4033–4040. [PubMed] [Google Scholar]
- 13.Wang J, Cooper MD. Histidine residue in the zinc-binding motif of aminopeptidase A is critical for enzymatic activity. Proc Natl Acad Sci USA. 1993;90:1222–1226. doi: 10.1073/pnas.90.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mullberg J, et al. A metalloprotease inhibitor blocks shedding of the IL-6 receptor and the p60 TNF receptor. J Immunol. 1995;155:5198–5205. [PubMed] [Google Scholar]
- 15.Hattori A, Matsumoto H, Mizutani S, Tsujimoto M. Molecular cloning of adipocyte-derived leucine aminopeptidase highly related to placental aminopeptidase/oxytocinase. J Biochem. 1999;125:931–938. doi: 10.1093/oxfordjournals.jbchem.a022371. [DOI] [PubMed] [Google Scholar]
- 16.Hattori A, et al. Characterization of recombinant human adipocyte-derived leucine aminopeptidase expressed in chinese hamster ovary cells. J Biochem. 2000;128:755–762. doi: 10.1093/oxfordjournals.jbchem.a022812. [DOI] [PubMed] [Google Scholar]
- 17.Schomburg L, Kollmus H, Friedrichsen S, Bauer K. Molecular characterization of a puromycin-insensitive leucyl-specific aminopeptidase, PILS-AP. Eur J Biochem. 2000;267:3198–3207. doi: 10.1046/j.1432-1327.2000.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlondorff J, Blobel CP. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J Cell Sci. 1999;112:3603–3617. doi: 10.1242/jcs.112.21.3603. [DOI] [PubMed] [Google Scholar]
- 19.Schlondorff J, Lum L, Blobel CP. Biochemical and pharmacological criteria define two shedding activities for TRANCE/OPGL that are distinct from the tumor necrosis factor alpha convertase. J Biol Chem. 2001;276:14665–14674. doi: 10.1074/jbc.M010741200. [DOI] [PubMed] [Google Scholar]
- 20.Blobel CP. Remarkable roles of proteolysis on and beyond the cell surface. Curr Opin Cell Biol. 2000;12:606–612. doi: 10.1016/s0955-0674(00)00139-3. [DOI] [PubMed] [Google Scholar]
- 21.Black RA, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-alpha from cells. Nature. 1997;385:729–733. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 22.Moss ML, et al. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997;385:733–736. doi: 10.1038/385733a0. [DOI] [PubMed] [Google Scholar]
- 23.Peschon JJ. An essential role for ectodomain shedding in mammalian development. Science. 1998;282:1281–1284. doi: 10.1126/science.282.5392.1281. [DOI] [PubMed] [Google Scholar]
- 24.Reddy P, et al. Functional analysis of the domain structure of tumor necrosis factor-alpha converting enzyme. J Biol Chem. 2000;275:14608–14614. doi: 10.1074/jbc.275.19.14608. [DOI] [PubMed] [Google Scholar]