Abstract
SEC62 encodes an essential component of the Sec-complex that is responsible for posttranslational protein translocation across the membrane of the endoplasmic reticulum in Saccharomyces cerevisiae. The specific role of Sec62p in translocation was not known and difficult to identify because it is part of an oligomeric protein complex in the endoplasmic reticulum membrane. An in vivo competition assay allowed us to characterize and dissect physical and functional interactions between Sec62p and components of the Sec-complex. We could show that Sec62p binds via its cytosolic N- and C-terminal domains to the Sec-complex. The N-terminal domain, which harbors the major interaction site, binds directly to the last 14 residues of Sec63p. The C-terminal binding site of Sec62p is less important for complex stability, but adjoins the region in Sec62p that might be involved in signal sequence recognition.
INTRODUCTION
The analysis of protein translocation across the membrane of the endoplasmic reticulum (ER) in the yeast S. cerevisiae has revealed two distinct pathways to target proteins to the membrane and at least two different channels to guide them across (Matlack et al., 1998). The decision as to which targeting pathway to use, and through which channel to translocate is determined by the composition of the signal sequence located at the N terminus of the translocated protein. First believed to be interchangeable, it was later recognized that signal sequences differ in the kinetics of their translocation and in the selection of the different targeting and translocation components (Bird et al., 1987; Hann and Walter, 1991; Johnsson and Varshavsky, 1994b; Ng et al., 1996). This was an unexpected finding because a general hydrophobicity is the essential feature that is shared by all signal sequences. The signal sequence initiates the translocation of the polypeptide by binding to a signal sequence receptor (Walter and Lingappa, 1986). The different signal sequences can distinguish between two different receptor systems. The more hydrophobic signal sequences seem to be channeled cotranslationally, whereas the less hydrophobic sequences are translocated posttranslationally (Ng et al., 1996). During cotranslational translocation, the signal sequence is recognized early after its synthesis by the signal recognition particle (SRP), and transferred to the SRP receptor with the ribosome still attached. The SRP is released from the nascent chain after GTP hydrolysis and the signal sequence is transferred to the Sec61 heterotrimer, the actual channel across the membrane (Simon and Blobel, 1991; Crowley et al., 1994; Hanein et al., 1996; Beckmann et al., 1997; Rapiejko and Gilmore, 1997). A second signal sequence recognition event by the trimeric Sec61 complex follows shortly before the initiation of translocation (Jungnickel and Rapoport, 1995). During posttranslational translocation the interaction between signal sequence and receptor is thought to take place at a later stage in the synthesis of the nascent chain. Instead of being recognized by a cytosolic component, the signal sequence might bind directly to a receptor at the membrane of the ER. The identity of this signal sequence receptor is still a matter of debate. It is either the heptameric Sec-complex that is composed of the trimeric Sec61p complex and the tetrameric Sec62/Sec63p complex, solely the tetrameric Sec62/Sec63p complex, or even a third and still unknown component. It was convincingly shown by cross-linking analysis that the heptameric Sec-complex specifically binds signal sequences (Matlack et al., 1997; Plath et al., 1998). Whether this binding is similar to the second recognition step that occurs during cotranslational translocation, or constitutes the only recognition event during posttranslational translocation, is not clear.
Sec62p and Sec63p are the only components of the tetrameric Sec-complex that are essential. Sec72p can be deleted without major consequences for the yeast, whereas the deletion of Sec71p leads only to impaired growth (Feldheim et al., 1992, 1993). The understanding of the role of Sec63p in translocation is aided by its association with Kar2p, a member of the family of Hsp70 heat shock proteins located in the lumen of the ER. Sec63p and Kar2p are required for an ATP-dependent step after the initial binding of the signal sequence to the heptameric Sec-complex (Sanders et al., 1992; Brodsky and Schekman, 1993; Lyman and Schekman, 1995, 1997). The complex is proposed to bind the signal sequence or the nascent chain on the luminal side of the membrane and to provide directionality to the translocation process (Matlack et al., 1999). Besides its well-established role in translocation, Sec63p fulfills additional roles in karyogamy and nuclear import (Ng and Walter, 1996; Brizzio et al., 1999).
The functions of Sec62p in translocation are not defined. Sec62p is found close to certain signal sequences during translocation (Lyman and Schekman, 1997; Matlack et al., 1997; Dünnwald et al., 1999). It consists of two membrane-spanning regions that direct its N- and C-terminal domain into the cytosol of the cell (Deshaies and Schekman, 1989, 1990).
In this work we use the Split-Ubiquitin (Ub) technique to undertake a structural and functional dissection of Sec62p. The split-Ub method is based on the ability of Nub and Cub, the N- and C-terminal halves of ubiquitin, to assemble into a quasi-native Ub (Johnsson and Varshavsky, 1994a). Ub-specific proteases (UBPs), which are present in all eukaryotic cells, recognize the reconstituted Ub, though not its halves, and cleave the Ub moiety off a reporter protein, which has been linked to the C terminus of Cub. The release of the reporter serves as an indicator for the reconstitution of Ub. Two mutations were engineered into Nub to reduce its affinity to Cub and thereby suppress the spontaneous reassembly of the Ub-peptides. Nua and Nug carry an alanine or a glycine in position 13 of Nub. Nug has a lower affinity for Cub than Nua and both have a still lower affinity for Cub than Nui, the wild-type version carrying an isoleucine in this position. In these cases efficient reassociation is only seen if the two Ub-peptides are located in proximity to each other (Johnsson and Varshavsky, 1994a). The split-Ub assay has been shown to detect the stable in vivo interaction between soluble proteins, between membrane proteins, and the transient interaction between substrate and transporter during protein translocation (Stagljar et al., 1998; Dünnwald et al., 1999; Wellhausen and Lehming, 1999; Wittke et al., 1999) Here we show that Sec62p contains an N- and a C-terminal binding site for the Sec-complex and a functionally important region immediately following the second transmembrane element that might directly be involved in signal sequence recognition. The corresponding binding site for the N-terminal domain of Sec62p on the Sec-complex is assigned to the last 14 carboxy-terminal residues of Sec63p.
MATERIALS AND METHODS
Construction of Test Proteins
The SEC62 ORF was amplified via polymerase chain reaction (PCR) using yeast genomic DNA as a template and inserted between the PGAL1-, the PMET25-, or the PCUP1-promoter and the Dha module to create SEC62-Dha or mutants thereof in the pRS314, pRS315, or pRS316 vectors. The Nub-ORF-Dha constructs were assembled from the PCUP1-Nub-cassette and a PCR fragment containing the ORF or part of the ORF of the desired gene to finally reside in the vector pRS314 or pRS313. A BamHI site was used to bring the Nub in frame with the PCR product (Johnsson and Varshavsky, 1994a). The SalI site was used to bring the PCR product in frame with the Dha module (Wittke et al., 1999). To construct sec62-1-Dha the same PCR procedure was used but with genomic DNA of the strain RSY529 (sec62-1) as a template (Table 1). SEC62 constructs retaining the natural stop codon were obtained by PCR, using primer combinations as described (Dünnwald et al., 1999). Nub-GUK1-Dha and Nub-GUK1-ha were obtained by PCR of genomic DNA and primers to create a BamHI site at the 5′ and a SalI site at the 3′ end to allow the in-frame insertion of the PCR product between the Nub- and the -Dha or -ha module, respectively.
Table 1.
Yeast strains
| Strain | Relevant genotype | Source/comment |
|---|---|---|
| JD53 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 | Dohmen et al. (1995) |
| NJY79RU | MATa/α his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 SEC63/SEC63-CUB-RURA3∷pRS305 | Wittke et al. (1999) |
| NJY125 | MATa/α his3-Δ200/his3-Δ200 leu2-3,112/leu2-3,112 lys2-801/lys2-801 trp1-Δ63/trp1-Δ63 ura3-52/ura3-52 SEC62/SEC62∷KAN+ | Derivative of JD53; this work |
| NYJ126 | MATα his3-Δ200 leu2-3,112 lys2-801 trp1-Δ63 ura3-52 SEC62∷KAN+ PCUP1FLAG-SEC62∶pRS316 | Derivative of JD53; this work |
| RSY 529 | MATα his4 leu2-3,112 ura3-52 sec62-1 | Deshaies and Schekman (1989) |
To construct F-SEC63ΔN244, a fragment containing the last 1257 base paris (bp) of the SEC63 ORF and 172 bp of 3′ untranslated DNA was amplified from yeast genomic DNA and inserted via an XhoI and a KpnI site behind the sequence coding for the Flag-epitope. The construct was under the control of the PGAL1-promoter or the PCUP1-promoter and resided in the plasmid pRS416 or pRS314. F-SEC63ΔN244ΔC47 was created by PCR with F-SEC63ΔN244 as a template by inserting a stop codon 3′ to the PstI site in the SEC63 sequence. The sequence 3′ of the PstI site reads as follows: CTGCAGTGTAGCTCGAGGAGGTGTATT. The Pst1 site is underlined and the stop codon is in bold letters. F-SEC63ΔN244ΔC14 was constructed by cutting the SEC63 ORF of F-SEC63ΔN244 with ClaI, filling the overhanging 3′ ends with Klenow polymerase, and ligating the thus created blunt ends. The resulting frame shift after residue 649 reads DTIRIQKLKMMNHQNRYK. The last wild-type residue of Sec63p is marked in bold. F-FPR1-63C14 and F-FPR1-63 C47 were constructed by inserting a PCR fragment of the complete ORF of FPR1 into the EcoRI-ClaI or the EcoRI-PstI cut plasmid containing F-SEC63ΔN244to replace the ORF of SEC63 except the last 49 or 14 residues, respectively.
More detailed information on the constructs and their generation is available upon request. DNA sequences were determined by the MPIZ DNA facility on PE Biosystems Abi Prism 377 and 3700 sequencers by using BigDye-terminator chemistry. Oligonucleotides were purchased from Metabion (Martinsried, Germany).
Immunoblotting
Cell extraction for immunoblotting was performed essentially as described (Johnsson and Varshavsky, 1994b). Proteins were fractionated by 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted on nitrocellulose membranes (Schleicher & Schüll, Dassel; Germany), by using a semidry transfer system (Hoeffer, Pharmacia Biotech, San Francisco, CA). Blots were incubated with a monoclonal anti-ha antibody (Babco, Richmond, CA), an anti-Flag antibody (Eastman Kodak, New Haven, CT) or a polyclonal anti-Sec61p antibody (a gift from T. Rappoport, Harvard Medical School). Bound antibody was visualized with horseradish peroxidase-coupled rabbit anti-mouse or goat anti-rabbit antibody (Bio-Rad, Hercules, CA) by using the chemiluminescence detection system (Pierce, Rockford, IL). The amount of stained protein was quantified with the aid of the lumi-imager system (Boehringer, Mannheim, Germany).
Coimmunoprecipitation
JD53 cells expressing the plasmid-borne F-Sec63ΔN244 or mutants thereof and a plasmid containing Sec62ΔC125-Dha were cultured in 300 ml of 2% dextrose (SD) medium to an OD600 of 0.8–1.0. Cells were harvested and resuspended in 1 ml of lysis buffer (50 mM NaCl, 1 mM EDTA, 50 mM sodium-HEPES, pH 7.5) containing a protease inhibitor mix. Cells were lysed by vortexing with glass beads in 1 ml of buffer, extracts were cleared by centrifugation, and supernatants were incubated with anti-ha antibodies coupled to agarose beads (Babco, Berkeley, CA) overnight at 4°C or anti-Flag antibodies for 1 h (Kodak, Rochester, NY) followed by Protein A agarose (Boehringer) overnight at 4°C. The beads were washed four times with lysis buffer and boiled in 1 volume of 2× SDS sample buffer (20% glycerol, 100 mM Tris-Cl, pH 6.8, 4% SDS, 4% mercapto-ethanol) followed by 12.5% SDS-PAGE and immunoblotting with anti-Flag or anti-ha antibody, respectively.
JD53 cells containing SEC62-Dha or mutants thereof on a plasmid were grown in 300 ml of SD-medium to an OD600 of 0.8–1.0. Cells were extracted in 1 ml of buffer (250 mM HEPES-KOH, pH 7.5, 25 mM KOAc, 5 mM MgOAc, 5 mM EDTA, 5% glycerol, 10 mM dithiothreitol [DTT], plus a protease inhibitor mix) to prepare microsomes by glass bead vortexing. Microsomes were frozen in liquid N2 and stored at −80°C in buffer (50 mM HEPES-KOH, pH 7.5, 10% glycerol, 2 mM DTT) plus a protease inhibitor mix. Saponin (Sigma, Deisenhofen, Germany) and Digitonin (Fluka Chemie AG, Buchs, Switzerland) extractions were essentially as described (Görlich et al., 1992). Equivalents (1600) of the membranes as defined by Görlich et al. (1992) were used for each immunoprecipitation with anti-ha-coupled agarose beads. Sec61p was detected by immunoblotting with rabbit polyclonal antibody (Finke et al., 1996).
Pulse-Chase Analysis
Saccharomyces cerevisiae cells expressing the Nub- and Cub-fusions or the Dha-fusions were grown at 30°C in 10 ml of SD medium to an OD600 of ∼1, and labeled for 5 min with Redivue Promix-[35S] (Amersham, Buckinghamshire, United Kingdom), followed by immunoprecipitation with the anti-ha monoclonal antibody or a polyclonal anti-carboxypeptidase (CPY) antibody essentially as described but with the following modification (Johnsson and Varshavsky, 1994). The N-ethylmaleimide-treated cells were spun and boiled in 200 μl of buffer (30 mM DTT, 90 mM sodium-HEPES, pH 7.5, 2% SDS). Lysis buffer (800 μl) was added and supernatants were cleared by centrifugation and subjected to immunoprecipitation. Gels were fixed and the dried gels were exposed and scanned by using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Growth Assay, Competition Assay
Yeast rich (YPD) and synthetic minimal media with SD or 2% galactose (SG) followed standard recipes. S. cerevisiae cells were grown at 30°C in liquid selective media containing uracil. Cells were diluted in water and 4 μl was spotted on agar plates selecting for the presence of the fusion constructs but lacking uracil. The same dilutions were spotted on plates containing uracil to check for cell numbers. The plates were incubated at 30°C for 3–5 d unless mentioned otherwise. To obtain cell numbers for the semiquantitative competition, ∼10,000 cells of an overnight culture were plated on SG medium selecting for the fusion constructs and lacking uracil. Colonies were counted after 7 d of incubation at 30°C.
Deletion of SEC62, Plasmid Shuffle
The open reading frame of SEC62 was replaced by the dominant kanr marker in the diploid JD51 essentially as described (Güldener et al., 1996). The PCR primer used for the construction of the kanr disruption cassette annealing 5′ of the SEC62 ORF reads as follows: GGAGAAGAGTGGGCTTTTATAATTGCAGTTGAATGCAGTAC-CAGCTGAAGCTTCGTA. The PCR primer annealing 3′ of the SEC62 ORF reads as follows: GTATATTAAAGCCGGCCGGAAATTGAGTAATAATAACCGCTAGGCCACTAGTGGATC. Transformed yeast cells were selected for kanr integration by Geneticin (Life Technologies, Paisley, Scotland) and the deletion was verified by diagnostic PCR. The diploid yeast cells (NJY125) were transformed with a plasmid expressing the Flag-bearing F-Sec62p and containing the URA3 as the metabolic marker (Table 1). The diploids were sporulated and tetrads were dissected by using standard yeast methods. Spores were selected by growth on Geneticin and analyzed for the absence of the chromosomal SEC62 by PCR and absence of the protein by immunoblotting with antibodies against Sec62p and the presence of the F-Sec62p by replicaplating on Ura- and immunoblotting with anti-Flag antibody. SEC62-Dha or mutants thereof were transformed on a TRP1 plasmid into NJY126 and the transformants were checked for the expression of the proteins by immunoblotting with anti-ha antibody (Table 1). The cells were cultured for 3 d on SD-trp containing uracil and 106 cells were streaked on plates containing 1 mg/ml 5-fluoroorotic acid (5-FOA) (WAK-Chemie, Bad Soden, Germany) and 50 μg/ml uracil. After 2 d of growth at 25°C, single colonies were picked and restreaked onto the same medium. Single colonies were analyzed by immunoblotting and PCR for the absence of the Flag-Sec62p and the presence of the desired Sec62p derivative.
RESULTS
Sec62p Contains Multiple Binding Sites for the Sec-Complex
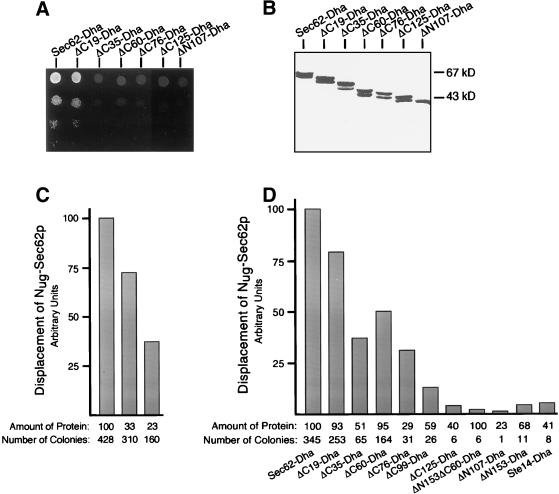
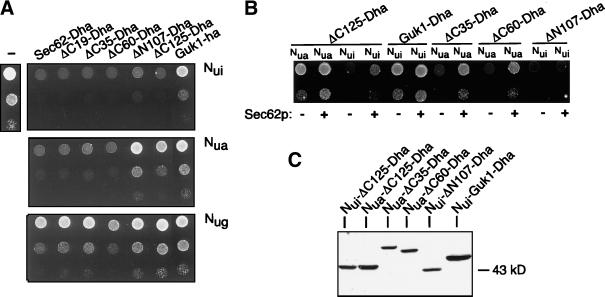
We defined the domains of Sec62p that are important for its association within the Sec-complex by testing several deletion mutants of Sec62p in an in vivo competition assay. The assay is based on the split-Ub technique and measures the displacement of Nug-Sec62p from the Sec-complex that contains a Cub-RUra3p (CRUp) extended Sec63p. Due to the presence of both Nug-Sec62p and Sec63CRUp in one complex, RUra3p is efficiently cleaved and degraded by the N-end-rule. The cells are phenotypically ura- and do not grow on plates lacking uracil (Wittke et al., 1999). Expressing Sec62p or any of its mutants still able to enter the Sec-complex in addition displaces Nug-Sec62p from its complex with Sec63CRUp. As a consequence, less reassembly of Nub-Cub will occur and the Ura3p activity of the uncleaved Sec63CRUp will enable the cells to grow on SD-ura (Figure 1). We constructed a series of Sec62p mutants that carried deletions at the N or C terminus (Figure 2). The constructs are extended at their C terminus by the dihydrofolate reductase gene carrying an ha-tag (Dha) (Johnsson and Varshavsky, 1994b). The Dha module allowed to estimate the relative amount of the different Sec62p constructs in the cell by immunoblotting by using an anti-ha antibody. Because all tested Sec62p constructs were expressed from the inducible PGAL1-promoter, no competition was expected on glucose-containing medium and indeed none of the cotransformed cells grew on SD medium lacking uracil (Wittke et al., 1999). When the experiment was performed on medium containing galactose, all Sec62-Dha constructs were found to be expressed, but good growth on SG-ura was only observed for the cells expressing the full-length Sec62-Dha or a mutant of Sec62p lacking the last 19 carboxy-terminal residues (ΔC19-Dha; Figures 2 and 3A). To increase the resolution of the assay, we plated 10,000 cells on medium containing galactose but lacking uracil and counted all colonies after 7 d. The number of colony-forming cells was compared with the number obtained by plating the strain that expressed the ER membrane protein Ste14-Dha from the PGAL1-promoter, or a strain containing the empty plasmid. An average of <10 colonies per 10,000 cells was counted for both strains. The number of colonies in this assay should depend on the affinity of the Sec62-Dha construct for the Sec-complex and on its cellular concentration. A positive correlation between number of colonies and the cellular amount of Sec62-Dha was confirmed by expressing SEC62-Dha from the PMET25-promoter under three different methionine concentrations. The higher the methionine concentration in the medium the less Sec62-Dha is made. As a consequence the numbers of colonies that are formed on medium lacking uracil decrease (Figure 3C). Gel electrophoresis of cell extracts and quantification by immunodetection revealed that not all truncations of Sec62p were equally abundant (Figure 3B). We therefore adjusted the numbers of colonies that were induced by the expression of the different constructs by the estimated concentrations of the fusion proteins in the cell. The amount of Sec62-Dha was arbitrarily set to 100 (Figure 3D). The binding of Sec62p to the Sec-complex is reduced when the C terminus is shortened by 19 (ΔC19-Dha) or 35 residues (ΔC35-Dha). Binding is slightly improved or remains roughly constant when a further 25 (ΔC60-Dha) or 41 (ΔC76-Dha) residues are deleted from the C terminus of ΔC35-Dha. The affinity of Sec62p for the Sec-complex continues to decrease when the second membrane-spanning element is removed to create ΔC99-Dha (Figure 3D).
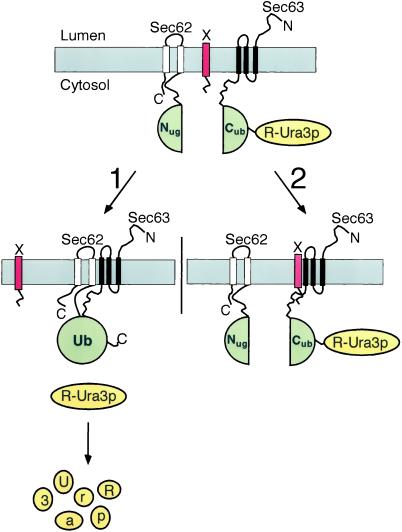
Figure 1.
Using the split-Ub technique to identify the binding sites of Sec62p for the Sec-complex by an in vivo competition assay. Nug-Sec62p, Sec63-Cub-RUra3p, and a fragment of Sec62p (X, red bar) are expressed in one cell. Pathway 1: when X does not or only weakly interacts with Sec63p, the Nug-labeled Sec62p is allowed to bind to the Cub-labeled Sec63p (both Ub-halves are in green). The induced proximity between Nug and Cub leads to their efficient reassociation. The assembled Ub is recognized by the UBPs, and the RUra3p reporter (yellow) is cleaved and subsequently degraded. The cells are phenotypically ura−. Pathway 2: X binds to Sec63p and displaces Nug-Sec62p from its complex. The increased distance between the Nug and Cub inhibits their reassociation. The RUra3p reporter remains linked to Cub and is not degraded. The cells are phenotypically ura+.
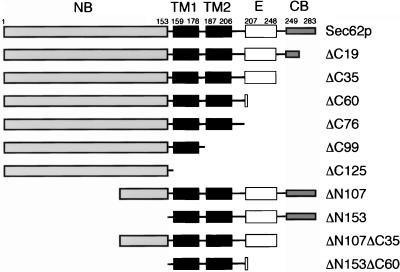
Figure 2.
Deletion constructs of SEC62 used in this study. The cytosolic N-terminal domain (NB, ░⃞), the two transmembrane spanning segments (TM1 and TM2, ▪), and the cytosolic C-terminal domain consisting of a functionally important region (E, □) and a C-terminal binding domain (CB, ░⃞) are emphasized. The names of the constructs indicate how many residues were deleted from either the N (ΔN) or the C terminus (ΔC) of Sec62p. The corresponding Nub-fusions of these constructs carry the Nub-module attached to the N terminus of the protein. Unless mentioned otherwise, all constructs are extended at their C terminus by DHFR-ha (-Dha).
Figure 3.
The N-terminal domain of Sec62p harbors the major interaction site. (A) Cells expressing Nug-Sec62p and Sec63CRUp were transformed with one of the PGAL1-controlled constructs shown in Figure 2, and 105, 104, 103, and 102 cells were spotted onto medium containing galactose but lacking uracil, histidine, leucine, and tryptophan to select for the presence of the plasmids. Growth was recorded after 4 d at 30°C. (B) Cells that were assayed for binding were extracted with buffer containing 1% Triton X-100, and the proteins were separated by 12.5% SDS-PAGE and incubated with anti-ha antibody after transfer onto nitrocellulose. The closely spaced doublets of bands seen for Sec62p and its C-terminal truncations arise very probably by an alternative initiation from a second methionine in position 10 of Sec62p. (C) Cells expressing Nug-Sec62p and Sec63CRUp were transformed with SEC62-Dha under the control of the PMET25-promoter, and 104 cells were plated on medium containing 0, 35, or 70 μM of methionine and lacking uracil, tryptophan, histidine, and leucine to select for the presence of the plasmids. Colonies were counted after 7 d of growth at 30°C. The number of colonies (the average of three experiments) was arbitrarily set to 100 for the cells grown without methionine, and the numbers of the cells grown in the presence of methionine were adjusted accordingly. The amount of protein was estimated after cell extraction and immunblotting by quantitative chemiluminescence (the average of three experiments is shown in arbitrary units). (D) Same as in C but cells were expressing Sec62-Dha or one of the deletion constructs under the control of the PGAL1-promoter, and the cells were plated on medium containing galactose. The number of colonies (the average of five experiments) was divided by the cellular amount of Dha-fusion protein as estimated by two independent immunoblotting experiments and quantitative chemiluminescence with the help of the lumi imaging system. The obtained ratio was arbitrarily set to 100 for Sec62-Dha, and the ratios of all the other tranformants were adjusted accordingly.
Truncating the cytosolically exposed N-terminal domain of Sec62p fully prevents the competition of the corresponding Sec62p fragments (ΔN107-Dha and ΔN153-Dha; Figure 3, A and D). The binding of the N-terminally truncated mutants is indistinguishable from the binding of the Sec62p fragment that simultaneously lacks the N- and the C-terminal interaction sites (ΔN153ΔC60-Dha; Figure 3D). We conclude that Sec62p bears at least two binding sites that are both important for the association of Sec62p within the Sec-complex. The first segment is contributed by the N-terminal 107 residues and the second site is located between residues 249 and 283 of Sec62p (Figure 2). A third binding site is probably contributed by the second membrane-spanning element (see DISCUSSION). We noted that Sec62-Dha, as judged by the number of colony-forming cells (345), already shows reduced binding to the Sec-complex compared with the unmodified Sec62p (840).
The N-terminal binding domain once expressed without the two membrane-spanning elements (ΔC125-Dha) does not displace Nug-Sec62p from the Sec-complex (Figure 3D). The majority of this protein is found in the soluble fraction after extracting the cells without detergents (Figure 9). The lack of competition of ΔC125-Dha is therefore most likely due to the high local concentration of Nug-Sec62p at the membrane and its additional C-terminal binding site(s).
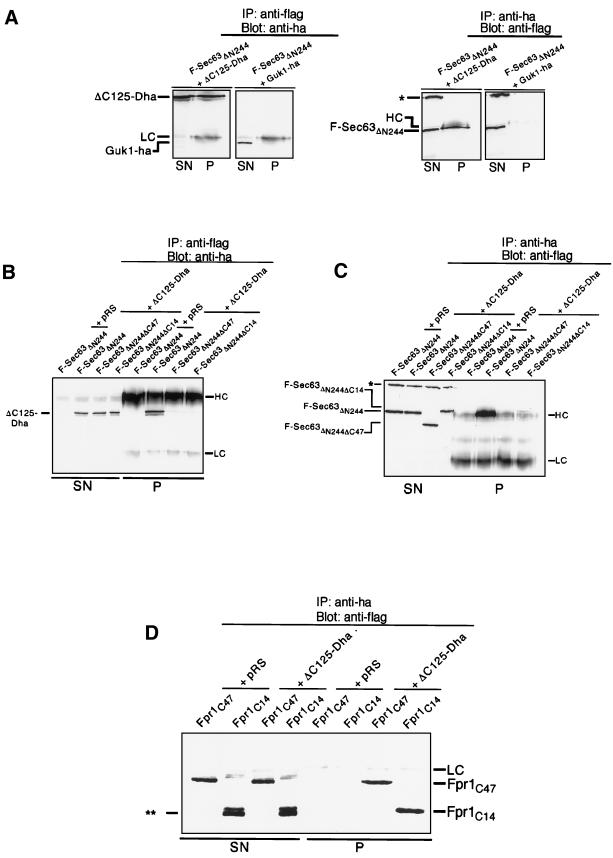
Figure 9.
The N-terminal domain of Sec62p binds to the acidic C-terminal segment of Sec63p. (A) Cells containing ΔC125-Dha and the flag-tagged Sec63ΔN244 (F-Sec63ΔN244) or Guk1-ha and F-Sec63ΔN244 were subjected to immunoprecipitation with anti-flag antibody to precipitate F-Sec63ΔN244 or with anti-ha to precipitate ΔC125-Dha or Guk1-ha. Supernatants and pellets of the immunoprecipitation were analyzed by 12.5% SDS-PAGE and immunoblotting with anti-ha antibody or anti-flag antibody. (B) The acidic C terminus of Sec63p is needed for the strong binding to the N-terminal domain of Sec62p. Cells containing an empty plasmid and F-Sec63ΔN244, ΔC125-Dha and F-Sec63ΔN244, ΔC125-Dha and F-Sec63ΔN244ΔC47, or ΔC125-Dha and F-Sec63ΔN244ΔC14 were subjected to immunoprecipitation with anti-flag antibody. Supernatants (SN) and pellets (P) were analyzed after 12.5% SDS-PAGE with anti-ha antibody. (C) Cells were exactly treated and analyzed as described under B except that the immunoprecipitation was performed with the anti-ha antibody and the analysis of the reaction was carried out with anti-flag antibody. (D) The acidic peptide derived from the C terminus of Sec63p binds to the N-terminal domain of Sec62p. Fpr1p containing the last 49 (Fpr1C49) or 14 (Fpr1C14) residues of Sec63p at its C terminus and the Flag-epitope at its N terminus was coexpressed together with ΔC125-Dha or an empty plasmid. Cells were extracted and subjected to anti-ha immunoprecipitation. The analysis of the immunoprecipitation was carried out after 12.5% SDS-PAGE with the anti-flag antibody. Ponceau staining of the blot revealed a 1:1 stochiometry of the coprecipitated ΔC125-Dha and Fpr1C49/Fpr1C14 and no further detectable protein (our unpublished observations). ∗∗ marks a proteolytic degradation product of Fpr1C14. Light chains (LC), heavy chains (HC), and a cross-reacting band of the flag-antibody (∗) are indicated.
To verify the results of the competition assay and to confirm the role of the N-terminal domain as an independent interaction site we tested the binding of a subset of the different Sec62p-fragments by introducing Nub-fusions of these constructs into cells carrying Sec63CRU. Nub-Sec62p, or mutants thereof that still bind, increase the local concentration between Nub and Cub and induce the cleavage of the RUra3p from Cub. As a consequence the growth of the cells on medium lacking uracil is impaired (Figure 1). Binding between Sec62p and Sec63p in the Sec-complex is tight enough for Nug-Sec62p to inhibit the growth of the cells on SD-ura (Wittke et al., 1999) (Figure 3A). Binding was recorded by the growth of the transformants on plates lacking uracil. Cells containing Nug-Sec62-Dha, Nug-ΔC19-Dha, Nug-ΔC35-Dha, or Nug-ΔC60-Dha display slightly reduced growth on SD-ura, whereas the cells containing Nug-ΔC125-Dha, Nug-ΔN107-Dha, or Nug-Guk1-ha grow unimpaired (Figure 4A). Derivatives of Guk1p, the cytosolic guanylate kinase of the yeast, were included in this assay to display the behavior of proteins that are not attached to membrane of the ER.
Figure 4.
Direct split-Ub interaction assay. (A) Cells containing Sec63CRU and the Nui-, Nua-, or Nug-derivatives of the different Sec62-Dha proteins, Guk1-ha, or an empty plasmid were spotted on SD-plates lacking uracil, leucine, and tryptophan to select for the presence of the Nub- and Cub-constructs. Growth was scored after 4 d at 30°C. (B) Cells coexpressing Sec62p (+) under the PGAL1-promoter or an empty plasmid (−), the different Nub-constructs under the PCUP1-promoter, and Sec63CRU under control of its own promoter were spotted onto galactose medium lacking uracil and tryptophan, leucine, and histidine to select for the presence of the plasmids. Growth was recorded after 4 d at 30°C. (C) Detection of Nui- and Nua-fusion proteins that were used for the competition assay shown in B. Cells were grown in medium containing galactose but lacking leucine, histidine, and tryptophan. The proteins were detected by the ha-antibody after cell extraction and 12.5% PAGE.
The expression of the Nua-constructs of Sec62-, ΔC19-, ΔC35-, and ΔC60-Dha inhibit the growth of the cells. Nua-ΔC125-Dha also impairs the growth, whereas the expression of Nua-ΔN107-Dha or Nua-Guk1-ha have no significant effect (Figure 4A). All Nui-constructs interfere with the growth of the Sec63CRUp-containing cells on SD-ura (Figure 4A). Here the strong inhibition by Nui-ΔC125-Dha indicates interaction because Nui-Guk1-ha and all other tested Nui-fusions of cytosolic proteins did only slightly impair the growth of Sec63CRUp-containing cells (Figure 4A; Wittke et al., 1999). The displacement of the Nua-fusions and of Nui-ΔC125-Dha from the Sec-complex through the simultaneous overexpression of Sec62p was shown by the improved growth of the cells containing an extra copy of a PGAL1-driven Sec62p on SG-ura (Figure 4B). The protein analysis of cell extracts by immunoblotting confirmed that all Nui- and Nua-constructs used in this assay were equally well expressed (Figure 4C). The successful competition therefore confirms the specificity of these interactions. In contrast to the other Nub-Sec62-Dha constructs, the proximity between Nui-ΔN107-Dha and Sec63CRU is not impaired by the overexpression of Sec62p (Figure 4B). The shared residence of both proteins in the membrane of the ER is therefore the likely cause of the measured proximity (Wittke et al., 1999). We conclude that the cytosolic N-terminal domain of Sec62p directly binds to the membrane bound Sec-complex.
Deletion of the N- or the C-Terminal Binding Site of Sec62p Reduces but Does Not Abolish the Binding to Sec61p
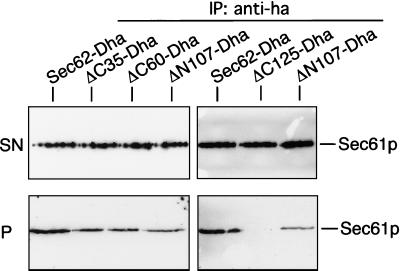
We performed coimmunoprecipitations of the different Sec62-Dha with Sec61p in wild-type cells to test whether the previously described binding sites are also responsible for attaching Sec62p to the trimeric Sec61p-complex (Figure 5). Binding is seen for the full-length Sec62-Dha, and further reduced binding can be observed for the C-terminally truncated ΔC35-Dha, ΔC60-Dha and the N-terminally truncated ΔN107-Dha. ΔC125-Dha, the N-terminal cytosolic domain, does not bind to Sec61p under these conditions (Figure 5). This experiment differs in its outcome from the assays that are based on Sec63CRUp as a sensor of interactions. Although the deletion the N-terminal domain has a more severe effect on the measured proximity to Sec63p than the deletion of the C-terminal binding site, the different Sec62p derivatives display roughly the same interaction with Sec61p.
Figure 5.
The N-terminal binding domain of Sec62p is not essential for the interaction with Sec61p. Digitonin-extracted membranes of wild-type cells expressing Sec62-Dha, ΔC35-Dha, ΔC60-Dha, ΔN107-Dha, or ΔC125-Dha were subjected to anti-ha immunoprecipitation. Two independent experiments of the Sec62-Dha- and the ΔN107-Dha-Sec61p–coprecipitation are shown. Immunoprecipitates were separated on 12.5% SDS-PAGE and treated with anti-Sec61p antibody. A portion of the supernatants (SN) and the pellets (P) of the immunoprecipitations are displayed on the upper and lower row, respectively.
The Major Interaction Domains Are Not Essential for the Functions of Sec62p
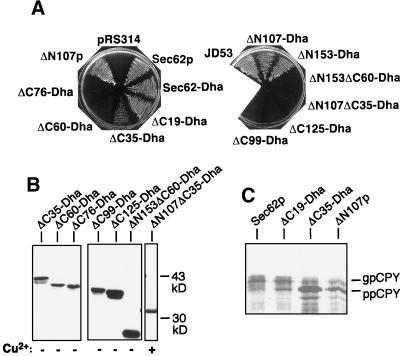
To test the functionality of the different SEC62 constructs, we performed plasmid shuffle experiments. The transformation with a functional derivative of SEC62 allows the NJY126 cells to lose a plasmid that simultaneously contains SEC62 and URA3 (Table 1). As a consequence the cells can grow on 5-FOA–containing medium. Both constructs lacking the N-terminal binding domain (ΔN107-Dha, ΔN153-Dha, ΔN107p) code for functional molecules (Figure 6A). The C-terminally truncated molecules ΔC19-Dha and ΔC35-Dha can replace Sec62p, although the ΔC35-Dha–induced plasmid loss is already less efficient. Because single deletions of either the N- or the C-terminal Sec-binding sites leave Sec62p still functional, we tested a derivative lacking both binding domains (ΔN107ΔC35-Dha). Although the protein can be detected by immunoblotting with anti-ha antibodies before selection on 5-FOA (Figure 6B), it does not confer 5-FOA resistance (Figure 6A). We conclude that the N- and C-terminal binding sites anchor a functionally important domain into the Sec-complex. This putative effector domain is defined by the construct ΔC60-Dha. Although carrying the intact N-terminal binding site and retaining a weak association with Sec61p, this protein is the first in the series of C-terminally truncated molecules that is not functional (ΔC76-, ΔC99-, ΔC125-, ΔN153ΔC60-Dha; Figure 6, A and B) (Deshaies and Schekman, 1990).
Figure 6.
Test for functionality. (A) Different constructs of SEC62 were transformed into a ΔSEC62 strain that contained SEC62 on a URA3 bearing plasmid. The transformants were applied onto plates containing 5-FOA to select for SEC62-URA3 plasmid loss and 100 μM copper sulfate to ensure full expression of the constructs. Growth was recorded after 3 d at 25°C. Growth of cells attests the functionality of the transformed construct. (B) Protein levels of the constructs that did not or only poorly allow the plasmid loss on 5-FOA were tested before 5-FOA selection by protein extraction, 12.5% SDS-PAGE, and immunodetection with anti-ha antibody. (C) Test for CPY translocation. Cells lacking the chromosomal SEC62 but containing the plasmid-borne Sec62p, ΔC19-Dha, ΔC35-Dha, or ΔN107p instead were pulsed for 5 min with [35S]methionine and subjected to immunoprecipitation with anti-CPY antibodies. Equal counts were loaded onto a 12.5% SDS-PAGE gel, and protein was visualized by phosphorimaging. Running position of the translocated form of CPY (gpCPY) and its untranslocated form (ppCPY) are indicated.
Measuring the efficiency of ER translocation is a more sensitive assay for evaluating the influence of mutations on the role of Sec62p (Ng et al., 1996). The translocation of the Sec62p-dependent substrate CPY was compared between strains that carried the plasmid-borne SEC62, ΔC19-Dha, ΔC35-Dha, or ΔN107 instead of the chromosomal SEC62 (Deshaies and Schekman, 1989; Ng et al., 1996). Accumulation of the cytosolic form of CPY and the concomitant decrease of its translocated fraction after a short [35S]methionine pulse revealed a major translocation defect in cells carrying ΔC35-Dha, which was less pronounced in cells carrying ΔN107p and barely detectable in cells containing ΔC19-Dha (Figure 6C). The deletion of each binding domain clearly impairs the role of Sec62p in translocation.
Proximity of Sec62p to Signal Sequences Is Not Abolished by Deleting the Major N-Terminal Sec-binding Domain
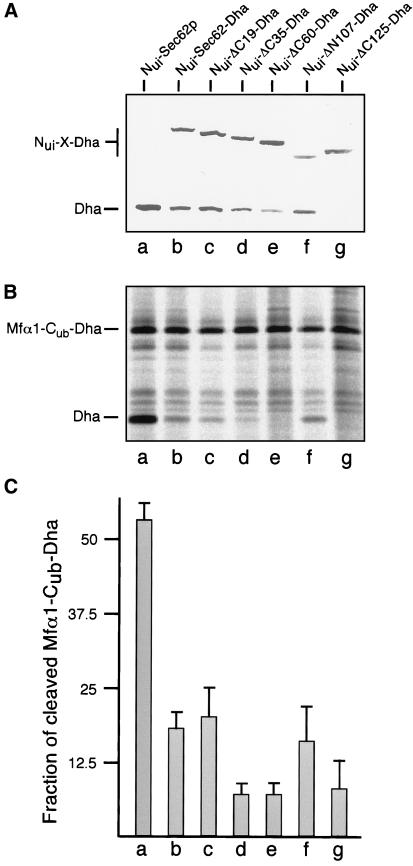
To understand the role of the effector domain of Sec62p, we measured the interaction between different Nub-labeled Sec62-Dha constructs and a signal sequence bearing Cub-Dha fusion protein. In this configuration, the split-Ub assay is capable of monitoring the short-lived proximity between substrate and transporter during protein translocation across the membrane of the ER (Dünnwald et al., 1999). Here proximity is measured by the amount of cleaved Dha that accumulates in the cytosol of the cell. As previously shown, Nui-ΔC60-Dha displays no significant interaction with the Mfα-Cub-Dha translocation substrate (Dünnwald et al., 1999). To find out whether this lack of proximity is caused by ΔC60-Dha's impaired binding to the Sec-complex or by the absence of the functionally important domain, we had to compare the proximity of a subset of the different Sec62p mutants to the Mfα-Cub-fusion. The Nub-fusion proteins were coexpressed with Mfα-Cub-Dha and the cleaved Dha quantified by immunoblotting (Figure 7A). Nui-Sec62p, Nui-Sec62-Dha, and Nui-ΔC19-Dha induce cleavage of the Mfα-Cub-Dha. In accordance with the binding assays, cleavage of Mfα-Cub-Dha is already significantly reduced in cells that carry Nui-Sec62-Dha or Nui-ΔC19-Dha (Figure 7A). Further deletions at the C terminus abolish the specific proximity of Sec62p to the signal sequence. Nui-ΔC35-Dha and Nui-ΔC60-Dha induce only background cleavage of the Mfα-Cub-Dha, which probably arises by both proteins being concentrated in the membrane of the ER (Figure 7A). Compared with Nui-ΔC60-Dha and Nui-ΔC35-Dha, the amount of cleaved Dha that is induced by Nui-ΔN107-Dha increases roughly twofold (Figure 7A). Nui-ΔC125-Dha induces no significant cleavage of the translocation substrate (Figure 7A). Similar results were obtained by pulse labeling the cells with [35S]methionine and immunoprecipitating cleaved and uncleaved Mfα-Cub-Dha (Figure 7, B and C). The ratios of cleaved to uncleaved protein that are induced by the different Nui-fusion proteins roughly correlate with the functionality of the corresponding Sec62-Dha constructs (Figure 7C). ΔN107-Dha is functional but less is incorporated into the Sec-complex than ΔC35-Dha or ΔC60-Dha. Yet Nui-ΔN107-Dha shows a twofold higher ratio of cleaved to uncleaved Mfα-Cub-Dha (Figure 7C).
Figure 7.
The in vivo proximity between a signal sequence and Sec62p and its derivatives. (A) Cells coexpressing the translocation substrate Mfα1-Cub-Dha and Nui-Sec62p (a), Nui-Sec62-Dha (b), Nui-ΔC19-Dha (c), Nui-ΔC35-Dha (d), Nui-ΔC60-Dha (e), Nui-ΔN107-Dha (f), or Nui-ΔC125-Dha (g) were subjected to immunoblot analysis with anti-ha antibody after protein extraction and 12.5% SDS-PAGE. X denotes Sec62-Dha and its truncated derivatives. Cleaved Dha indicates proximty. The uncleaved Mfα1-Cub-Dha is not detected by this steady-state analysis (Dünnwald et al., 1999). (B) Same as in A but cells were pulse labeled for 5 min with [35S]methionine before protein extraction and anti-ha immunoprecipitation. (C) Quantification of five independent experiments as shown in B. The counts of the cleaved Dha and the uncleaved Mfα1-Cub-Dha were quantified by phosphorimaging of the dried gels to calculate the ratio of cleaved to uncleaved translocation substrate.
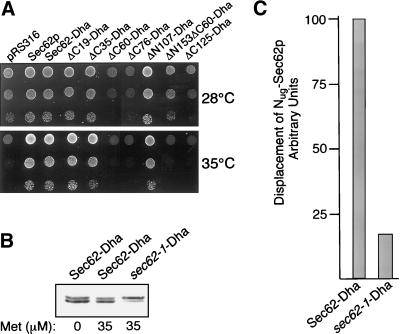
A Single Mutation in the N-Terminal Domain Impairs Binding of sec62–1p to Sec-Complex
While studying the topology of Sec62p, Deshaies and Schekman (1990) discovered that certain mutants of Sec62p showed a dominant-negative effect on the growth of cells carrying the sec62-1 allele at the permissive temperature and speculated that the N-terminal part of Sec62p is involved in the interaction with other components of the Sec-complex. By expressing the constructs that were generated during this work in the sec62-1-carrying strain, we could show that all Sec62p derivatives that display this dominant toxic effect have a common denominator. The constructs that possess the intact N-terminal Sec-binding domain and lack the effector domain (ΔC60, ΔC76, ΔC125) do not allow growth at the restrictive temperature (Figure 8A, 35°C) and impair growth at the permissive temperature (Figure 8A, 28°C). A SEC62 construct that lacks both the N-terminal binding domain and the effector domain although not functional does not interfere with the growth of the sec62-1 containing cells at 28°C (ΔN153ΔC60).
Figure 8.
Sec62-1 contains a mutation that interferes with the binding to the Sec-complex. (A) Different SEC62 constructs were transformed into the yeast RSY529 (sec62-1), and 105, 104, and 103 cells were spotted at the restrictive (35°C) and the permissive (28°C) temperature onto medium containing 100 μM copper sulfate to ensure full expression of the constructs. Growth was recorded after 3 d. (B) Amount of Sec62-Dha was adjusted to the amount of sec62-1-Dha by expressing the mutated copy from the PGal1-promoter and the native copy from the PMet25-promoter. Cells containing either of the two constructs and Nug-SEC62 and SEC63CRU were grown in galactose medium containing different concentrations of methionine. After cell extraction and 12.5% PAGE, the nitrocellulose-transferred proteins were treated with anti-ha antibody. The amount of protein was estimated by lumi-imaging and is approximately equal when the cells are grown at 35 μM methionine and 2% galactose. (C) Cells (104) described in B were plated on medium containing galactose and 35 μM of methione but lacking uracil, tryptophan, histidine, and leucine. The average number of colonies of three counts was set to 100 for the cells containing Sec62-Dha and adjusted correspondingly for sec62-1-Dha.
Because the sec62-1 allele might reveal more about the exact position of the Sec-binding site, we amplified the sequence of sec62-1 and sequenced its reading frame. We detected a single missense mutation that leads to the exchange of a glycine against an aspartate in position 46 of Sec62p. This mutation falls into a cluster of otherwise positively charged residues. As expected the corresponding sec62-1-Dha construct allowed the NJY126 cells to lose the SEC62 plasmid relatively efficiently at 25°C but only very inefficiently at 35°C (our unpublished observations). To test whether this mutation reduces the binding of Sec62p to the Sec-complex, we introduced the sec62-1-Dha construct into cells containing Nug-Sec62p and Sec63CRUp. We found no effective competition at either 25 or 30°C (our unpublished results). Again 10,000 cells were plated onto SD-ura and the colonies were counted after 7 d at 25°C. The numbers were compared with the number of colonies obtained by cells expressing the wild-type Sec62-Dha (Figure 8C). In this experiment the cellular amount of Sec62-Dha was adjusted to the lower levels of sec62-1-Dha by expressing the native protein from the PMET25- and the mutated protein from the PGAL1-promoter (Figure 8B). As judged by our assay, sec62-1-Dha shows at 25°C already a sixfold weaker binding to the Sec-complex than the protein carrying the intact N-terminal domain (Figure 8C).
The N Terminus of Sec62p Directly Interacts with Acidic C-Terminal Tail of Sec63p
Although the competition assay revealed the multiple binding sites of Sec62p to the Sec-complex, this type of assay cannot identify the corresponding binding partner among the many subunits of the Sec-complex. We considered the possibility, that the stretch of positive residues in the N-terminal domain of Sec62p might directly interact with the negatively charged cytosolic tail-domain of Sec63p (residues 245–663) (Ng and Walter, 1996). The Flag-tagged C-terminal domain of Sec63p (F-Sec63ΔN244) was therefore coexpressed with ΔC125-Dha in yeast cells. Cells were extracted without the use of detergent and the extracts cleared by centrifugation. The major fraction of both fusion proteins remained in the supernatant. The following immunoprecipitation with anti-Flag antibodies specifically yielded ΔC125-Dha, whereas immunoprecipitation with anti-ha antibodies specifically yielded F-Sec63ΔN244 (Figure 9A). No interaction was observed between F-Sec63ΔN244 and Guk1-ha, which served as a control protein for testing the specificity of the immunoprecipitation protocol (Figure 9A). The experiment thus demonstrates that Sec62p and Sec63p interact via their N- and C-terminal cytosolic domains. The last 47 residues of Sec63p harbor 25 negatively and no positively charged residues. Deleting this tail in the corresponding Flag-labeled F-SEC63ΔN244ΔC47 construct destroys its binding to ΔC125-Dha in the coimmunoprecipitation experiments (Figure 9, B and C). Replacing the last 14 residues, including eight negatively charged residues of Sec63ΔN244 by an unrelated and slightly longer sequence that contains six positive charges (F-SEC63ΔN244ΔC14), greatly reduces the binding to the N-terminal domain of Sec62p (Figure 9, B and C). Note that a trace of ΔC125-Dha–bound F-SEC63ΔN244ΔC14 could still be detected on the blots (Figure 9C). Because deleting parts of a protein can influence the structure of the remainder of the molecule, we transferred the last 47 residues and the last 14 residues from the C terminus of Sec63p to the C terminus of Fpr1 that carried the Flag epitope at its N terminus to create FPR1C47 and FPR1C14. Fpr1p, the cytosolic FK506 binding protein of the yeast, has no role in protein translocation and does not bind to the Sec-complex. Coimmunprecipitation experiments confirmed a direct interaction between the two Sec63p-derived peptides and the N-terminal domain of Sec62p (Figure 9D). In the case of FPR1C14 we detect a closely spaced doublet of proteins on the blots by the anti-Flag antibody. Only the upper band is precipitated by the N-terminal domain (Figure 9D). As judged by the shift in the running behavior during denaturing gel electrophoresis, the faster migrating band must have lost most if not all of the attached Sec63p peptide by proteolysis. The inability of this proteolytic product to bind is therefore an additional control for the specificity of the detected interaction between ΔC125-Dha and the intact FPR1C14. We conclude that the last 14 residues of Sec63p constitute the major interaction site for the N-terminal domain of Sec62p.
DISCUSSION
The tetrameric Sec62/Sec63p complex endows the Sec61p translocation channel with a specificity toward certain signal sequences and provides the directionality of posttranslational protein translocation (Matlack et al., 1999). The specific role of Sec62p in translocation has long been enigmatic. This article describes structural features of Sec62p that point to a more active role of Sec62p in signal sequence recognition and targeting.
The Modular Structure of Sec62p: Multiple Binding Sites for the Sec-Complex
We used a split-Ub–based competition assay to follow the effect of deletions in Sec62p on its association within the Sec-complex. The advantage of this assay compared with techniques that rely on cell extraction and solubilization is that the harsh conditions of solubilization and their influence on the actual measurements are avoided. The readout of this assay, cell survival, is very indirect. However, the positive correlation between the amount of the protein that is used as the competitor and the number of cells that survive on the selective media confirmed this approach (Figure 3C). Because the correlation is not strictly linear one has to regard the linear adjustment that we performed to better compare the different deletion constructs as a first approximation (Figure 3D). Furthermore, we attached the Dha-moiety to the C terminus of all our constructs to quantitatively compare the amounts of fusion proteins made in the split-Ub based assays. We have shown that this moiety impairs the binding characteristics of Sec62p to some extent. This restricted our analysis to a qualitative comparison of the Dha-modified Sec62p derivatives.
Using this assay we found that Sec62p uses at least two binding sites for its association with the Sec-complex. The two sites are located at opposite ends of the molecule. The first interaction site is at the N terminus. The second binding site maps to the cytosolic C terminus of Sec62p (Figure 3). Removal of either domain causes a defect in translocation, the effect being stronger for the removal of the C-terminal site (Figure 6). However, only the simultaneous deletion of both binding sites renders the protein completely inactive (Figure 6). We conclude that Sec62p's major role in translocation is not to serve as a docking factor for other proteins of the Sec-complex but that the two binding sites anchor a functionally important domain into the Sec-complex.
A stretch of 41 residues that follows the second membrane-spanning sequences and extends into the cytosol is an essential part of this domain. A comparison between the C-terminally truncated ΔC60-Dha, which lacks 25 of these residues, and the N-terminally truncated ΔN107-Dha with regard to their proximity to a signal sequence and the Sec-complex points to the role of this domain. ΔC60-Dha still associates with the Sec-complex, yet is nonfunctional and has lost its specific proximity to the Mfα-signal sequence (Deshaies and Schekman, 1990; Dünnwald et al., 1999; this work). In contrast to ΔC60-Dha, ΔN107-Dha lacks the N-terminal Sec63p binding site. Yet ΔN107-Dha is functional, and shows an albeit reduced proximity to the signal sequence (Figures 3, 6, and 7). We performed an Sec61p coprecipitation with both Sec62p-mutants to test whether they differ in their binding to Sec61p. However, both mutants display roughly the same weak affinity to Sec61p (Figure 5). Based on this comparison we propose that the region of Sec62p that immediately follows the second transmembrane segment contributes actively to the recognition of signal sequences during posttranslational protein translocation. The experiments, however, cannot distinguish whether this domain is part of a signal sequence-binding pocket or more indirectly primes Sec61p to bind the signal sequence. The N-terminal borders of this putative effector domain are not yet exactly defined. Whether the two membrane-spanning sequences are still part of this functionally important region remains to be tested.
The C-terminal Sec-complex binding site overlaps with the so defined effector domain. Binding decreases once 19 (ΔC19-Dha) or 35 (ΔC35-Dha) residues are deleted from the C terminus (Figure 2). In contrast to ΔC19-Dha, ΔC35-Dha is only partially functional and shows in our assay no detectable or only a very reduced interaction with a signal sequence (Figures 6 and 7). The most prominent feature of the 16 residues that distinguishes ΔC19 from ΔC35 is a stretch of positive residues that is also seen in Sec62p proteins from other species (Meyer et al., 2000; Tyedmers et al., 2000).
No further decrease in binding affinity seems to occur upon removal of 25 (ΔC60) or 41 (ΔC76) residues. A significant reduction in binding is first seen when the second membrane-spanning element is deleted (ΔC99-Dha; Figure 3). Whether this effect is indicative for an additional third binding site to the Sec-complex or reflects an inefficient incorporation into the membrane has not been investigated. However, a similar Sec62p truncation that was fused at its C terminus to invertase showed the correct topology and a stable association with the membrane (Deshaies and Schekman, 1990). The partners for the C-terminal binding sites in the Sec-complex are still unknown.
The Sec62p-Sec63p Interface
The binding site on Sec63p for the N-terminal domain of Sec62p is located at the very C terminus. The last 35 residues of Sec63p constitute an imperfect duplication of a 17-residue peptide. Interestingly, only the carboxy-terminal repeat shows strong binding to the N-terminal domain of Sec62p. The last 14 carboxy-terminal residues of this repeat are sufficient for binding to the N-terminal domain of Sec62p (Figure 9). Of these 14 residues, eight are either Glu or Asp. A similar acidic segment is seen at the C terminus of Sec63p from worms and humans, indicating that the interaction between Sec63p and Sec62p is evolutionarily conserved (Meyer et al., 2000; Tyedmers et al., 2000).
A first clue about the exact localization of the corresponding binding site in the N-terminal domain of Sec62p was derived from the observation of Deshaies and Schekman (1990) that certain Sec62p fragments are toxic in the presence of the sec62-1 allele (Deshaies and Schekman, 1990). Indeed we could localize a single mutation in the N-terminal domain of the sec62-1 allele that has a major effect on the binding to Sec63p (Figure 8). The nature and position of the exchange are very suggestive because glycine 46 is replaced by aspartate. This position falls into a cluster of positive charges that could drive the interaction between Sec62p and the negatively charged C-terminal peptide of Sec63p. Converting the central Gly into the negatively charged Asp might then lead to a repulsion of the two molecules. This interpretation requires that the exchange does not disturb the structure of the N-terminal domain. In an application of a newly established technique, we could demonstrate that the first 153 residues of Sec62p have a distinct structure, and furthermore that the conformation of this structure is altered, and probably more unfolded by the amino acid exchange in this position (Raquet, Eckert, and Johnsson, unpublished data). This makes the interpretation of this particular mutation not invalid though less straightforward. The detection of this altered conformation helps to explain how a mutation in a nonessential domain can cause a ts-phenotype. The mutation in this domain not only inhibits binding to Sec63p, but initiates the destruction of the complete protein. Once the amount of the protein falls below a critical level, translocation across the ER cannot be sustained and the cells die.
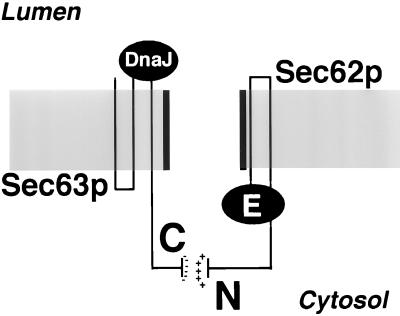
We propose that the functional significance of the interaction between the N-terminal domain of Sec62p and the C terminus of Sec63p is to tightly align the effector domains of both proteins across the membrane. The flow of translocated polypeptides from the cytosolic domain of Sec62p to the luminal DnaJ domain of Sec63p occurs via the translocation pore (Figure 10). Interestingly, the tight interaction between Sec62p and Sec63p is important for efficient translocation, but not essential (Figure 6; Ng and Walter, 1996).
Figure 10.
Summary of the molecular dissection of Sec62p. The N-terminal domain of Sec62p (N) and the acidic C-terminal segment of Sec63p (C) align the putative effector domain of Sec62p (E) and the DnaJ domain of Sec63p on opposite sites of the membrane across the channel formed by Sec61p. In combination with the trimeric Sec61p, signal sequence binding is achieved by the E domain of Sec62p. The nascent chain is transferred across the channel to the DnaJ domain of Sec63p, which in combination with the luminal heat shock protein 70 Kar2p binds to the nascent chain to keep it from sliding back into the cytosol.
In addition to serving as the main interacting partner for Sec62p, the C-terminal tail of Sec63p is required for membrane fusion during karyogamy. Sec62p seems to play no role in this process (Ng and Walter, 1996; Brizzio et al., 1999). We speculate that the tetrameric Sec62/Sec63p complex is in a dynamic equilibrium. Its tetrameric form serves translocation and the C-terminal peptide of Sec63p is complexed and neutralized by the N-terminal binding domain of Sec62p. During mating a fraction of the tetrameric Sec62/Sec63p complex disassembles and the trimeric Sec63/Sec71/Sec72p complex exposes the tail peptide of Sec63p to catalyze a currently undefined step in the fusion of nuclear membranes. Sec63/Sec71/Sec72p could be separated from Sec62p by ion exchange chromatography under high salt (Brodsky and Schekman, 1993). The salt sensitivity of the complex can now be explained by the identification of the highly charged region in Sec63p as being responsible for the tight interaction with Sec62p. However, it remains to be proven whether such a core complex exists in living cells.
ACKNOWLEDGMENTS
We thank Gabi Fischer von Mollard, Walther Mothes, Tom Rapoport, Randy Schekman, and Thomas Sommer for the gift of yeast strains and antisera, and Silke Müller for excellent technical assistance. We thank Jörg H. Eckert, Nicole Lewke, and Richard Thompson for critically reading the manuscript. This work was supported by a grant to N.J. from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie (0311107).
REFERENCES
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Bird P, Gething MJ, Sambrook J. Translocation in yeast and mammalian cells: not all signal sequences are functionally equivalent. J Cell Biol. 1987;105:2905–2914. doi: 10.1083/jcb.105.6.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizzio V, Khalfan W, Huddler D, Beh CT, Andersen SSL, Latterich M, Rose MD. Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:609–626. doi: 10.1091/mbc.10.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. SEC62 encodes a putative membrane protein required for protein translocation into the yeast endoplasmic reticulum. J Cell Biol. 1989;109:2653–2664. doi: 10.1083/jcb.109.6.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. Structural and functional dissection of Sec62p, a membrane-bound component of the yeast endoplasmic reticulum protein import machinery. Mol Cell Biol. 1990;10:6024–6035. doi: 10.1128/mcb.10.11.6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen RJ, Stappen R, McGrath JP, Forrova H, Kolarov J, Goffeau A, Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Dünnwald M, Varshavsky A, Johnsson N. Detection of transient in vivo interactions between substrate and transporter during protein translocation into the endoplasmic reticulum. Mol Biol Cell. 1999;10:329–344. doi: 10.1091/mbc.10.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an endoplasmic reticulum membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Yoshimura K, Admon A, Schekman R. Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol Biol Cell. 1993;4:931–939. doi: 10.1091/mbc.4.9.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke K, Plath K, Panzner S, Prehn S, Rapoport TA, Hartmann E, Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Matlack KE, Jungnickel B, Plath K, Kalies KU, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994a;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Ubiquitin-assisted dissection of protein transport across membranes. EMBO J. 1994b;13:2686–2698. doi: 10.1002/j.1460-2075.1994.tb06559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell. 1999;97:553–564. doi: 10.1016/s0092-8674(00)80767-9. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Mothes W, Rapoport TA. Protein translocation: tunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- Matlack KE, Plath K, Misselwitz B, Rapoport TA. Protein transport by purified yeast Sec complex and Kar2p without membranes. Science. 1997;277:938–941. doi: 10.1126/science.277.5328.938. [DOI] [PubMed] [Google Scholar]
- Meyer H-A, Grau H, Kraft R, Kostka S, Prehn S, Kalies K-U, Hartmann E. Mammalian Sec61 is associated with Sec62 and Sec63. J Biol Chem. 2000;275:14550–14557. doi: 10.1074/jbc.275.19.14550. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Walter P. ER membrane protein complex required for nuclear fusion. J Cell Biol. 1996;132:499–509. doi: 10.1083/jcb.132.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Rapiejko PJ, Gilmore R. Empty site forms of the SRP54 and SR alpha GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell. 1997;89:703–713. doi: 10.1016/s0092-8674(00)80253-6. [DOI] [PubMed] [Google Scholar]
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380. doi: 10.1016/0092-8674(91)90455-8. [DOI] [PubMed] [Google Scholar]
- Stagljar I, Korostensky C, Johnsson N, te Heesen S. A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA. 1998;95:5187–5192. doi: 10.1073/pnas.95.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Lerner M, Bies C, Dudek J, Skowronek MH, Haas IG, Heim N, Nastainczyk W, Volkmer J, Zimmermann R. Homologs of the yeast Sec complex subunits Sec62p and Sec63p are abundant proteins in dog pancreas microsomes. Proc Natl Acad Sci USA. 2000;97:7214–7219. doi: 10.1073/pnas.97.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Lingappa VR. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Wellhausen A, Lehming N. Analysis of the in vivo interaction between a basic repressor and an acidic activator. FEBS Lett. 1999;453:299–304. doi: 10.1016/s0014-5793(99)00718-8. [DOI] [PubMed] [Google Scholar]
- Wittke S, Lewke N, Müller S, Johnsson N. Probing the molecular environment of membrane proteins in vivo. Mol Biol Cell. 1999;10:2519–2530. doi: 10.1091/mbc.10.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]