Abstract
The bidirectional nucleocytoplasmic transport of macromolecules is mediated by the nuclear pore complex (NPC) which, in yeast, is composed of ∼30 different proteins (nucleoporins). Pre-embedding immunogold-electron microscopy revealed that Nic96p, an essential yeast nucleoporin, is located about the cytoplasmic and the nuclear periphery of the central channel, and near or at the distal ring of the yeast NPC. Genetic approaches further implicated Nic96p in nuclear protein import. To more specifically explore the potential role of Nic96p in nuclear protein import, we performed a two-hybrid screen with NIC96 as the bait against a yeast genomic library to identify transport factors and/or nucleoporins involved in nuclear protein import interacting with Nic96p. By doing so, we identified the yeast nucleoporin Nup53p, which also exhibits multiple locations within the yeast NPC and colocalizes with Nic96p in all its locations. Whereas Nup53p is directly involved in NLS-mediated protein import by its interaction with the yeast nuclear import receptor Kap95p, it appears not to participate in NES-dependent nuclear export.
INTRODUCTION
The nuclear pore complex (NPC) is a large supramolecular assembly that spans the double membrane of the nuclear envelope (NE) and mediates bidirectional nucleocytoplasmic transport (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998). Using amphibian oocytes extensive electron microscopic analyses has unveiled the principal structural organization of the ∼125 MDa vertebrate NPC (Panté and Aebi, 1996; Stoffler et al., 1999). To a large extent the yeast NPC appears to be designed according to the same architectural principles except that its linear dimensions appear to be ∼15% smaller (Fahrenkrog et al., 1998) and its mass only amounts to ∼60 MDa (Rout and Blobel, 1993; Yang et al., 1998). Compared with vertebrate NPC three-dimensional (3-D) reconstructions (cf. Akey and Radermacher, 1993), 3-D reconstruction of the yeast NPC exhibits more tenuous cytoplasmic and nuclear ring moieties (Yang et al., 1998).
The vertebrate NPC is composed of in excess of 50 different proteins, termed nucleoporins (Nups), whereas the yeast NPC is thought to consist of ∼30–50 different nucleoporins (Doye and Hurt, 1997; Stoffler et al., 1999; Rout et al., 2000). To date, ∼20 vertebrate and ∼30 yeast nucleoporins, i.e., presumably all yeast nucleoporins (Rout et al., 2000), have been identified and characterized. Localization by immunogold-electron microscopy (immunogold-EM) in both vertebrate and yeast has revealed that these nucleoporins mainly reside at the cytoplasmic and the nuclear periphery of the NPC, many of them having dual locations in a near-symmetrical manner relative to the central plane of the NPC (Panté and Aebi, 1996; Stoffler et al., 1999; Rout et al., 2000).
Ions and small molecules can traverse the NPC by passive diffusion, whereas proteins, RNAs, and ribonucleo protein (RNP) particles are transported through the NPC by a signal-mediated mechanism. More specifically, the nuclear import of cargoes harboring a classical nuclear localization signal (NLS) is mediated by a soluble dimeric receptor consisting of an adaptor subunit, called importin α in vertebrates and Srp1p in yeast, and the actual receptor subunit, called importin β in vertebrates and Kap95p in yeast. The adaptor subunit recognizes the cargo's NLS, whereas the receptor subunit recognizes distinct nucleoporins and interacts with various transport factors (e.g., NTF2; Nehrbass and Blobel, 1996) as it escorts the cargo-adaptor complex from the cytoplasm through the NPC into the nucleus (Corbett and Silver, 1997; Fabre and Hurt, 1997; Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998). Other adaptors, e.g., the importin α-like snurportin involved in the import of U snRNPs, and receptors, e.g., the importin β-like transportin involved in import of hnRNP A1, have been identified and characterized, and the related transport pathways have been elucidated (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998). Similarly, nuclear export is mediated by the formation of a heterotrimeric complex consisting of the cargo harboring a nuclear export signal (NES), the export receptor, and Ran-GTP (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998). The first NES has been identified in the HIV-1 Rev protein together with its export receptor CRM1 (exportin): Both Rev and CRM1 are involved in the export of unspliced viral RNA (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998). Other export receptors have been identified, for example, exportin-t or TAP, whereas the export signals for many RNA export pathways, especially those for mRNA export, have remained elusive (Izaurralde and Adam, 1998; Mattaj and Englmeier, 1998; Ohno et al., 1998).
Nic96p is an essential nucleoporin in yeast: it has been identified by its interaction with Nsp1p which, in turn, is the first yeast nucleoporin that has been identified and molecularly characterized (Hurt, 1988; Nehrbass et al., 1990; Grandi et al., 1993). Affinity purification of ProtA-Nsp1p by IgG-Sepharose chromatography identified Nic96p as a copurifying constituent (Grandi et al., 1993). Additionally, mutations in NSP1 and NIC96 were found to be synthetically lethal (Grandi et al., 1995). Biochemically, both Nic96p and Nsp1p belong to one subcomplex of the yeast NPC, i.e. the Nsp1p complex, which also contains the nucleoporins Nup49p and Nup57p (Grandi et al., 1993, 1995; Schlaich et al., 1997). Nic96p resides about the cytoplasmic and the nuclear periphery of the central channel in a near-symmetrical manner, as well as near or at the distal ring of the nuclear basket (Fahrenkrog et al., 1998; Fahrenkrog et al., 2000). Hence, Nic96p and Nsp1p closely colocalize in all three sites (Fahrenkrog et al., 1998, 2000). Evidently, both nucleoporins, Nic96p and Nsp1p, are involved in protein import into the nucleus (Nehrbass et al., 1993; Grandi et al., 1995). However, although Nsp1p interacts with distinct soluble factors involved in nuclear import, i.e. importin β and Ran (Stochaj et al., 1998; Seedorf et al., 1999), the direct interaction of Nic96p with nuclear import factors has remained elusive.
To gain more insight into the specific role of Nic96p in nuclear protein import, e.g., to identify transport factors interacting with Nic96p, we performed a two-hybrid screen with NIC96 as the bait against a yeast genomic library. According to this assay, we found Nic96p not to interact with transport factors, but with the nucleoporin Nup53p, a nucleoporin that has recently been identified independently in a synthetic lethal screen with POM152p (Marelli et al., 1998). Nup53p is located at the cytoplasmic and the nuclear periphery of the central NPC framework, and at the nuclear basket. Although mutations of NUP53 cause no obvious structural alterations of the yeast NPC, they lead to defects in nuclear protein import. Molecularly, the role of Nup53p in NLS-mediated protein import involves its direct interaction with the NLS-receptor Kap95p. In contrast, NES-mediated nuclear export appears not to be impaired by the disruption of NUP53.
MATERIALS AND METHODS
Yeast Strains and Media
The yeast strains used in this study are listed in Table 1. All strains were grown at 30°C, unless otherwise stated. Media and genetic methods, including mating, sporulation, and tetrad dissection were as described elsewhere (Guthrie and Fink, 1991). Yeast cells were transformed by using the lithium acetate method (Gietz et al., 1992).
Table 1.
Yeast strains
| Strain | Genotype | Source/references |
|---|---|---|
| CG-1945 | Mata, ura3-52, his3-200, lys2-801, ade2-101, trp1-901, leu2-3, 112, gal4-542, gal80-538, LYS2∷GAL1-HIS3, URA3∷(GAL4 17mers)3-CYC1-lacZ, cyhr2 | Stan et al. (1994) |
| Y187 | Matα, ura3-52, his3, ade2-101, trp1-901, leu2-3, 112, met−, gal4Δ, gal80Δ, URA3∷GAL1-lacZ | Song et al. (1994) |
| BMA41 | Mata/α, ade2-1/ade2-1, leu2-3, 112/leu2-3, 112, ura3-1/ura3-1, trp1Δ/trp1Δ, his3-11, 15/his3-11, 15, can1-100/can1-100 | Baudin-Baullieu et al. (1997) |
| BMA41/1a | Mata, ade2-1, leu2-3, 112, ura3-1, trp1Δ, his3-11, 15, can1-100 | Baudin-Baullieu et al. (1997) |
| Δnup53 | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 | This study |
| ProtA-Nup53p | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 (pUN100-NOP1∷ProtA-TEV-NUP53) | This study |
| BMA41 (NLS-GFP) | Mata/α, ade2-1/ade2-1, leu2-3, 112/leu2-3, 112, ura3-1/ura3-1, trp1Δ/trp1Δ, his3-11, 15/his3-11, 15, can1-100/can1-100 (pADH-NLS-GFP-lacz) | This study |
| Δnup53 (NLS-GFP) | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 (pADH-NLS-GFP-lacZ) | This study |
| Δnup53 (NES-NLS-GFP) | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 (pADH-NLS-NES-GFP-GFP) | This study |
| BMA41/1a (NES-NLS-GFP) | Mata, ade2-1, leu2-3, 112, ura3-1, trp1Δ, his3-11, 15, can1-100 (pADH-NLS-NES-GFP-GFP) | This study |
| Δnup53 (Rev-GFP) | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 (pGal-Rev-GFP) | This study |
| BMA41/1a (Rev-GFP) | Mata, ade2-1, leu2-3, 112, ura3-1, trp1Δ, his3-11, 15, can1-100 (pGal-Rev-GFP) | This study |
| BMA41/1a (Yap1p-GFP) | Mata, ade2-1, leu2-3, 112, ura3-1, trp1Δ, his3-11, 15, can1-100 (pLDB419) | This study |
| Δnup53 (Yap1p-GFP) | Mata, ade2-1, leu2-3, ura 3-1, trp1Δ, his3-11, 15, can1-100, nup53∷TRP1 (pLDB419) | This study |
| xpo1-1 | ade2-1, ura3-1, his3-11,15, trp1-1, leu2-3, 112, can1-100, xpo1∷LEU2 (pKW456) | Stade et al. (1997) |
| xpo1-1 (Yap1p-GFP) | ade2-1, ura3-1, his3-11,15, trp1-1, leu2-3, 112, can1-100, xpo1∷LEU2 (pKW456, pLDB419) | This study |
| xpo1-1 (NES-NLS-GFP) | ade2-1, ura3-1, his3-11,15, trp1-1, leu2-3, 112, can1-100, xpo1∷LEU2 (pADH-NES-NLS-GFP-GFP) | This study |
| xpo1-1 (Rev-GFP) | ade2-1, ura3-1, his3-11,15, trp1-1, leu2-3, 112, can1-100, xpo1∷LEU2 (pGal-Rev-GFP) | This study |
| nup49-313 | Matα, ade2, ade3, his3, leu2, ura3, nup49∷TRP1 (pUN90-nup49-313) | Grandi et al. (1995) |
| nup49-313 (NLS-GFP) | Matα, ade2, ade3, his3, leu2, ura3, nup49∷TRP1 (pUN90-nup49-313) (pADH-NLS-lacZ-GFP) | This study |
Plasmids
The following yeast plasmids were used: pUN100-NOP1::ProtA-TEV (pNOPPATA1L; Hellmuth et al., 1998); YEp13-NIC96 (Grandi et al., 1993; kindly provided by Ed Hurt, Biochemie-Zentrum, Heidelberg, Germany); pAS2(ΔΔ) and pACT2 (Harper et al., 1993; Fromont-Racine et al., 1997); pPS815(pADH-NLS-GFP-lacZ), pPS1372(pADH-NLS-NES-GFP-GFP), pPS1494(pGAL-REV-GFP), kindly provided by Jennifer Hood and Pamela Silver (Dana Farber Institute, Boston, MA); pLDB419(pYAP1-GFP LEU2 2 μ), kindly provided by Anita Corbett (Emory University School of Medicine, Atlanta, GA) and Laura Davis (Brandeis University, Waltham, MA). pAS2-NIC96, a polymerase chain reaction (PCR) amplification of NIC96 open reading frame (ORF) extending from nucleotide +1 to +2590 inserted into BamHI-NcoI cut pAS2. The PCR product was exchanged against a BsmI fragment of genomic NIC96 from YEp13-NIC96. pACT2, 2 μ/LEU2 based yeast genomic library. pNOPPATA1L-NUP53, PCR product of NUP53 ORF extending from nucleotide +1 to +1428 inserted into NcoI-BamHI cut pNOPPATA1L.
Yeast Two-Hybrid Screen
The yeast two-hybrid screen, with NIC96 as the bait against a yeast genomic library (Table 1), was performed exactly as described (Fromont-Racine et al., 1997)
Gene Disruption and Recombinant Protein A Tagging of NUP53
NUP53 deletion constructs were prepared by replacing nucleotides −10 to +500 with the TRP1 selectable marker gene generated by PCR. nup53::TRP1 was transformed into the diploid BMA41 strain (Baudin-Baillieu et al., 1997) and selected on SD-W plates (Rothstein, 1991). Trp1+ transformants were characterized for correct integration of nup53::TRP1 at the NUP53 locus by PCR analysis. BMA41 diploid heterozygous for NUP53 were sporulated and subsequently dissected by tetrad analysis. For recombinant protein A tagging, the NUP53 gene was amplified by PCR, thereby generating an NcoI and a BamHI site at the 5′ and the 3′ end, respectively. The resulting PCR product was sequenced and inserted into NcoI-BamHI cut pNOPPATA1L. The resulting plasmid was transformed into the Δnup53 strain and selected on SD-LW plates (Gietz et al., 1992).
Immunogold-EM
Preparation and in situ immunolocalization of ProtA-Nup53p was performed by pre-embedding labeling yeast cells with an anti-protein A antibody directly conjugated to 8-nm colloidal gold as described previously (Fahrenkrog et al., 1998).
EM
To evaluate the morphology of the Δnup53 strain, yeast cells were transformed into spheroplasts, washed twice in 0.1 M potassium phosphate buffer, pH 6.5, and fixed in 2% glutaraldehyde for 1 h, all steps as described (Fahrenkrog et al., 1998). After 1-h postfixation in 1% osmium tetroxide, the yeast cells were processed for electron microscopy (Fahrenkrog et al., 1998). Thin-sections were cut on a Reichert Ultracut ultramicrotome (Reichert-Jung Optische Werke, Vienna, Austria) by using a diamond knife (Diatome, Biel, Switzerland). The sections were collected on collodion-coated copper grids and stained with 6% uranyl acetate for 1 h followed by 2% lead citrate for 2 min. Specimens were inspected and electron micrographs recorded with a Hitachi H-7000 transmission electron microscope (Hitachi Ltd., Tokyo, Japan) operated at an acceleration voltage of 100 kV.
Affinity Purification of ProtA-TEV-Nup53p
ProtA-TEV-Nup53p was affinity purified from a Δnup53 strain transformed with recombinant Nup53p that was amino-terminally tagged with two IgG binding domains of Staphylococcus aureus protein A followed by a cleavage site for the TEV protease, by IgG-Sepharose chromatography (Hellmuth et al., 1998; Senger et al., 1998). The lysis buffer used contained either 100 mM potassium acetate for the elution of Nic96p or 300 mM potassium acetate for the elution of Kap95p. The NPC-containing fraction (2 μl) and 100 μl of the eluate, respectively, were precipitated in acetone, resuspended in 20 μl of gel-loading buffer, and analyzed by SDS-PAGE, followed by Coomassie blue staining and Western blotting by using an anti-Nic96p antibody (Grandi et al., 1995) and an anti-Kap95p antibody (Koepp et al., 1996), respectively. Finally, the blot was stained by a secondary antibody conjugated with alkaline-phosphatase.
In Vivo Protein Import Assay
An in vivo protein import assay was performed as described (Shulga et al., 1996). In this assay, logarithmically growing yeast cells constitutively expressing NLS-green fluorescent protein (GFP) (i.e., Δnup53 [NLS-GFP]; Table 1) as a reporter cargo were treated with sodium azide and deoxyglucose to block NLS-mediated protein import. At steady state, this block yields an approximately even distribution of the NLS-GFP cargo in the cytoplasm and the nucleus by passive diffusion of NLS-GFP across the NPC. After removal of the metabolic inhibitors the cells recover immediately in fresh medium, and the NLS-GFP cargo is reimported into the nucleus in an active manner. The relative import rates in the mutant and the wild-type strain were compared by counting the cells that exhibited NLS-GFP nuclear accumulation as a function of time. For immunogold-EM, the assay was performed in the same way: after 0, 5, and 15 min of active reimport, the cells were fixed by addition of 2% paraformaldehyde, pH 6.5, and prepared for EM as described elsewhere (Fahrenkrog et al., 1998). Cells were labeled with a polyclonal anti-GFP antibody (a kind gift from Jennifer Hood and Pamela Silver) directly conjugated to 8-nm colloidal gold.
In Vivo Protein Export Assays
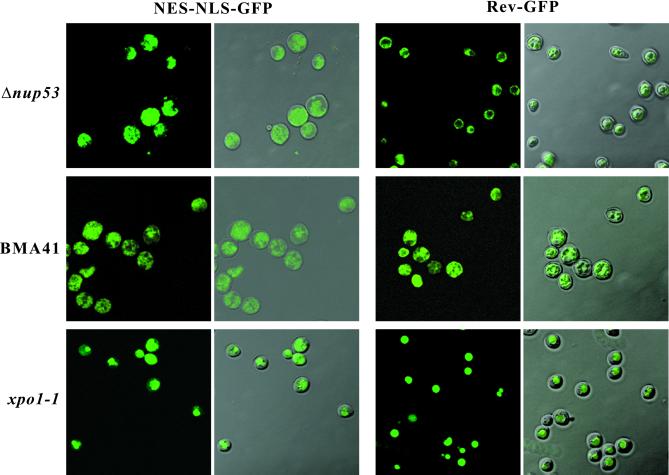
In a first protein export assay a GFP reporter was fused simultaneously to the NLS of the simian virus 40 large T antigen and the NES of the protein kinase inhibitor PKI (i.e. NES-NLS-GFP; Table 1; Stade et al., 1997). For this purpose, the Δnup53 and BMA41 and xpo1-1 control strain were transformed with a plasmid expressing the NES-NLS-GFP reporter. The cells were grown in selective media and the subcellular location of the reporter was determined by confocal laser scanning microscopy.
In a second protein export assay the Δnup53 and BMA41 and xpol1-1 control strain were transformed with a construct where a GFP reporter was fused to the HIV-1 Rev protein (i.e., Rev-GFP; Table 1; Taura et al., 1998), grown in selective medium containing glucose. After shifting the cells to galactose-containing medium for 4 h, the subcellular location of the Rev-GFP reporter was analyzed by confocal laser scanning microscopy.
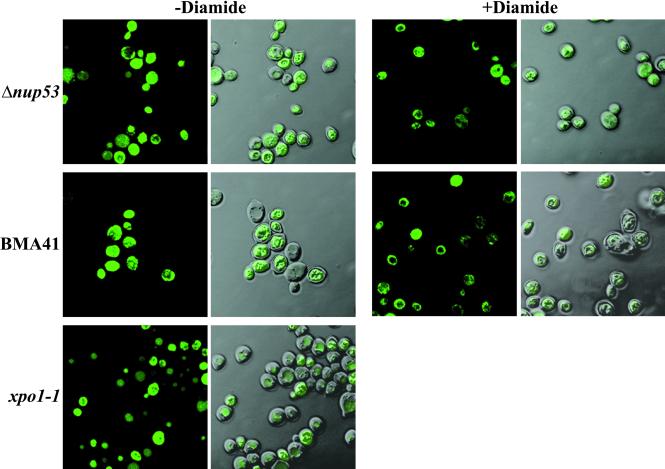
In a third protein export assay the Δnup53, BMA41, and xpo1–1 cells were transformed with the plasmid pLDB419 expressing the Yap1p-GFP reporter (Table 1; Yan et al., 1998). The cells were grown in selective medium and the subcellular location of Yap1p-GFP was determined under steady-state conditions or after treatment of cells with 1.5 mM diamide (i.e. to induce oxidative stress) for 15 h by confocal laser scanning microscopy.
RESULTS
Nic96p Interacts with the Yeast Nucleoporin Nup53p by a Two-Hybrid Screen
Based on the known involvement of Nic96p in nuclear protein import (Grandi et al., 1995) and its multiple locations about the cytoplasmic and the nuclear periphery of the central channel and near or at the distal ring of the nuclear basket (Fahrenkrog et al., 1998; Fahrenkrog et al., 2000), we set out to gain more insight into the molecular interactions Nic96p may experience while a cargo is traversing the NPC on its way from the cytoplasm into the nucleus. Hence, we carried out a yeast two-hybrid screen with full-length NIC96 (i.e., fused in frame to the GAL4 DNA binding domain) as the bait. To achieve this, the strain CG1945 containing the bait plasmid was mated to the strain Y187 containing FRY1 libraries (Table 1). One hundred and forty clones exhibited activation of the two reporter genes HIS3 and lacZ; 96 of these positive clones were sequenced and 8 of these were identified as the ORF YMR153w by a database search of the yeast genome. Recently, YMR153w has been identified to encode the yeast nucleoporin Nup53p by a synthetic lethality screen with POM152 (Marelli et al., 1998). The interactions between NIC96 and the prey NUP53 resided in two overlapping fragments in the N-terminal domain of NUP53, classifying this fusion as an A1 fusion (Fromont-Racine et al., 1997). Because NIC96 alone did not activate the transcription of the reporter genes, we conclude that the interaction between Nic96p and Nup53p is specific. A database search with NUP53 revealed putative homologous ORFs and ESTs in various species, e.g., human, mouse, Xenopus laevis, Caenorhabditis elegans, Drosophila melanogaster, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana, and Gossypium hirsutum, with the highest homologies among these ORFs residing in their central domains, thus indicating high conservation of these ORFs from yeast to higher eukaryotes and even plants (our unpublished results; Marelli et al., 1998).
Ultrastructural Localization of Nup53p by Immunogold-EM
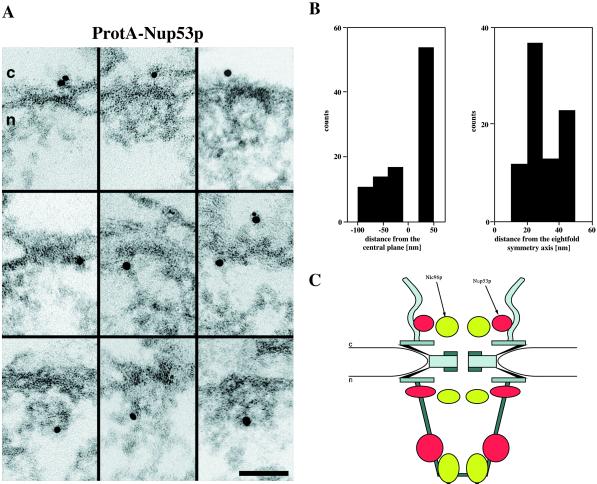
Nup53p has been previously localized to the nuclear rim by immunofluorescence microscopy and to both the cytoplasmic and the nuclear face of the NPC by immunogold-EM (Marelli et al., 1998; Rout et al., 2000). To more precisely localize Nup53p at the ultrastructural level within the yeast NPC to distinct NPC substructures, we performed immunogold-EM with the ProtA-Nup53p strain (Table 1). For this purpose, we constructed a yeast strain that is disrupted for NUP53 (i.e., Δnup53; see MATERIALS AND METHODS; Table 1). In this strain we then replaced its disrupted NUP53 gene by a plasmid-borne version of NUP53 fused to two IgG binding domains of the S. aureus protein A to the 5′ end of its ORF. Pre-embedding labeling of spheroplasted yeast cells with an anti-protein A antibody directly conjugated to 8-nm colloidal gold revealed that Nup53p resides at the cytoplasmic and the nuclear periphery of the central NPC framework (Figure 1A, top and middle), as well as at the fibrils forming the nuclear basket (Figure 1A, bottom). Quantification of the gold particle distribution (Figure 1B) with respect to the central plane of the NPC revealed that 55% of the gold particles were detected at distances of 25–50 nm from the central plane (30.2 ± 6.4 nm). Together with the corresponding radial distances of 30–50 nm (31.5 ± 7.9 nm) this locates Nup53p to the cytoplasmic face of the central framework rather than to the cytoplasmic fibrils. In the latter case the gold particles would have been detected at radial distances ranging from 0 to ∼60 nm due to the high flexibility of the cytoplasmic fibrils. In addition, 45% of all gold particles were depicted on the nuclear face of the NPC, with ∼40% at vertical distances of −20 to −40 nm (−24.3 ± 4.5 nm) and ∼60% at vertical distances of −50 to −100 nm (−68.3 ± 15.4 nm), corresponding to the nuclear periphery of the central framework and the nuclear basket fibrils, respectively. The gold particles found on the nuclear side of the NPCs were detected 1) at radial distances ranging from 30 to 50 nm (34.7 ± 12.1 nm), thus labeling the nuclear face of the central framework; and 2) at distances of 20–30 nm (26.5 ± 10.4 nm), thus labeling an epitope residing at the nuclear basket.
Figure 1.
Immunogold-EM localization of Nup53p in ProtA-Nup53p cells (i.e., Δnup53[ProtA-Nup53p] strain; Table 1). (A) Triton X-100-extracted spheroplasts were preimmunolabeled with a polyclonal anti-protein A antibody directly conjugated to 8-nm colloidal gold. Shown are selected examples of gold-labeled NPCs in cross-sections along the NE. The antibody labeled the cytoplasmic (top) and the nuclear (middle) periphery of the central framework, and the nuclear basket (bottom). c, cytoplasm; n, nucleus. (B) Quantitative analysis of the gold particle distribution associated with the NPCs in the Δnup53(ProtA-Nup53p) strain. For quantitative analysis 94 gold particles were scored. (C) Schematic representation of the close colocalization of Nic96p and Nup53p within the yeast NPC by their respective “location clouds” (Fahrenkrog et al., 2000). Accordingly, Nic96p is located about the cytoplasmic and the nuclear periphery of the central channel, and near or at the distal ring of the nuclear basket. Similarly, Nup53p resides on the cytoplasmic and the nuclear face of the central framework (i.e., close to or at the base of the cytoplasmic and the nuclear fibrils), and near the distal end of the nuclear basket fibrils. Scale bar, 100 nm (A).
Nup53p and Nic96p Closely Colocalize
As displayed schematically in Figure 1C, based on the previously published immunogold-EM localization of Nic96p (Fahrenkrog et al., 1998, 2000), both Nup53p and Nic96p exhibit three distinct locations within the yeast NPC. The corresponding “location clouds” are centered about the average distances (x, y) of each epitope of Nup53p and Nic96p normal to the central plane (y-axis) and perpendicular to the central eightfold symmetry axis (x-axis) of the NPC (Fahrenkrog et al., 2000). The radii of the elliptical location clouds are defined by the respective SDs from the mean of the distances from the central plane and the central eightfold symmetry axis. Although the location clouds of Nic96p and Nup53p do not exactly coincide, they closely colocalize. Overall, the Nup53p epitopes reside at higher radii compared with Nic96p. As illustrated schematically in Figure 1C, the Nup53p and Nic96p epitopes come close enough so that the two nucleoporins may indeed physically interact (see below) at all three distinct sites.
Nup53p and Nic96p Physically Interact
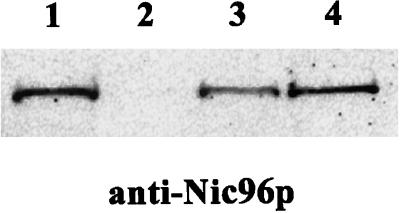
To confirm that Nup53p and Nic96p do indeed physically interact as suggested by the yeast two-hybrid screen, Nup53p was affinity-purified from yeast cells, and analyzed for copurifying components. To do so, recombinant Nup53p was amino-terminally tagged with two IgG binding domains of S. aureus protein A followed by a cleavage site for the TEV protease, and transformed into the Δnup53 strain (Table 1). ProtA-TEV-Nup53p was purified from this strain by IgG-Sepharose chromatography under nondenaturating conditions. Nup53p together with bound proteins was released from the IgG-Sepharose column upon incubation with TEV protease. The yeast cells after enzymatic removal of the cell wall followed by homogenization in lysis buffer containing 0.5% Tween 20 were further fractionated by centrifugation. The resulting supernatant containing the NPC fraction, which was applied to the IgG-Sepharose column, and the eluate of the column after digestion with TEV protease were analyzed by SDS-PAGE followed by Coomassie blue staining and by Western blotting with an anti-Nic96p antibody (Figure 2). Although it was not possible to identify any specific copurifying components from the Commassie blue stained gel (our unpublished results), Western-Blot analysis with a polyclonal anti-Nic96p antibody clearly demonstrated the specific interaction between Nup53p and Nic96p (Figure 2). Nic96p was present in the NPC-containing fraction of the yeast strain expressing ProtA-TEV-NUP53p (Figure 2, lane 3) as well as in the eluate from the IgG Sepharose beats after TEV protease digestion (Figure 2, lane 4). This interaction of Nic96p with Nup53p is specific because in the wild-type BMA41 control strain (Table 1) Nic96p could be detected in the NPC-containing fraction of this strain (Figure 2, lane 1), but not in the eluate after IgG-Sepharose chromatography (Figure 2, lane 2). Hence, Nic96p does not unspecifically bind to the IgG beads but rather via ProtA-TEV-NUP53p.
Figure 2.
Affinity purification of ProtA-TEV-Nup53p from Δnup53(ProtA-TEV-Nup53p) cells (Table 1) reveals interaction with Nic96p. ProtA-TEV-Nup53p was affinity-purified by IgG-Sepharose chromatography and Nup53p was released by TEV-mediated proteolytic cleavage. Western blot analysis with a polyclonal anti-Nic96p antibody reveals specific interaction of Nup53p with Nic96p. Lane 1, NPC-containing fraction of a wild-type control strain (BMA41); lane 2, eluate derived from the wild-type control strain; lane 3, NPC-containing fraction of the strain expressing ProtA-TEV-Nup53p; and lane 4, eluate derived from the ProtA-TEV-Nup53p.
nup53 Deletion Strains Are Viable and Exhibit No Obvious Structural Abnormalities of Their NPCs
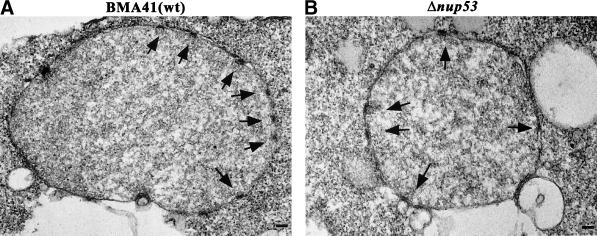
To examine the phenotype of the nup53 null strain (i.e. Δnup53; see MATERIALS AND METHODS; Table 1), a Trp1+ transformant containing the disrupted NUP53 gene was selected, sporulated, and tetrads were dissected. All four tetrads were viable, indicating that NUP53 is not essential. Trp+ haploids lacking NUP53 grow at similar rates than those observed in the presence of wild-type NUP53 at temperatures ranging from 15°C to 37°C (our unpublished results). To further characterize the phenotype of the Δnup53 null strain, we examined its morphology by thin-section electron microscopy. For this purpose, Δnup53 cells were grown in YPAD medium at 30°C and processed for EM. As documented in Figure 3, the morphology of the NE and the NPCs of the Δnup53 cells (Figure 3B) appear indistinguishable from those of the wild-type BMA41 control cells (Figure 3A). Additionally, in Δnup53 cells grown at 37°C the morphology of the NE and the NPCs also appear indistinguishable from those of the wild-type BMA41 cells grown under the same conditions (our unpublished results). These observations indicate that Nup53p is not required for specifying NPC assembly and structure, at least not to the extent that could be detected at the resolution level provided by embedding/thin sectioning EM.
Figure 3.
Thin-section electron micrographs of Δnup53 cells (B) and of a wild-type BMA41 control strain (A) (for the cell strains see Table 1). Cells were grown at 30°C, fixed, embedded, and processed for thin-section electron microscopy. The morphology of the nucleus, the NE, and the NPCs in the Δnup53 cells appears indistinguishable from that of the wild-type cells. Arrows mark the nuclear basket of the NPCs. Scale bars, 100 nm (A and B).
Disruption of NUP53 Attenuates Nuclear Protein Import
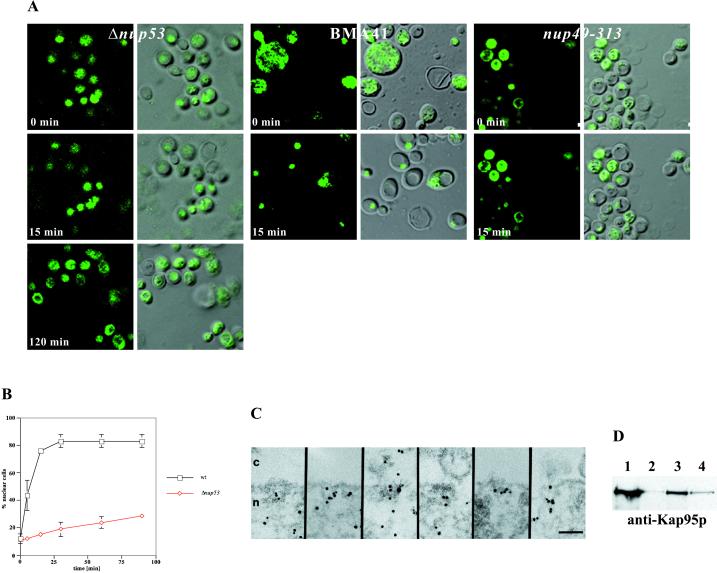
Next we set out to gain more insight into the possible functional role of Nup53p in nucleocytoplasmic transport. To test whether the Δnup53 strain is impaired in nuclear protein import, we performed an in vivo import assay (Shulga et al., 1996). In this assay, logarithmically growing yeast cells constitutively expressing an NLS-GFP reporter cargo (i.e. Δnup53 [NLS-GFP]; Table 1) were treated with sodium azide and deoxyglucose to block NLS-mediated nuclear protein import. At steady state, this block yields an even distribution of the GFP signal in the cytoplasm and the nucleus by passive diffusion of NLS-GFP across the NPC. After removal of the metabolic inhibitors by placing the cells in fresh glucose-containing medium, they recover from the block and the NLS-GFP is actively imported into the nucleus. The relative import rates in the Δnup53 and the wild-type BMA41 strain were compared by counting the cells that exhibited NLS-GFP nuclear accumulation as a function of time (Figure 4, A and B). As illustrated in Figure 4A, the Δnup53 cells displayed a strongly attenuated import rate of the NLS-GFP reporter (left) compared with that of the wild-type BMA41 strain (middle). A nup49–313 control strain that is known to be defective in NLS-dependent nuclear protein import accumulated the NLS-GFP reporter predominantly in the cytoplasm (Figure 4A, right). Quantification of the relative import rates for the BMA41 control strain and the Δnup53 strain revealed that for the BMA41 control strain a relative accumulation rate of GFP inside the nucleus of 13.5%/min, whereas the Δnup53 strain yielded an accumulation rate of only 3%/min (Figure 4B). However, protein import is not completely blocked in the Δnup53 strain because under steady-state conditions as well as after ∼5 h of removing the sodium azide/deoxyglucose block the NLS-GFP reporter does accumulate in the nucleus (our unpublished results). Immunogold-EM with a polyclonal anti-GFP antibody directly conjugated to 8-nm colloidal gold further revealed that the NLS-GFP reporter cargo preferentially accumulates in the nuclear basket (Figure 4C), indicating that although the NLS-GFP cargo can traverse the central pore it gets caught in the nuclear basket so that its release from the NPC into the nucleoplasm is strongly attenuated albeit not completely inhibited. As displayed in Table 2, quantitation of the gold particle distribution reveals a slower accumulation of the NLS-GFP reporter in the nucleus of the Δnup53 cells compared with that of the BMA41 control cells after drug removal and initiation of signal-mediated import of the NLS-GFP reporter. Taken together, the results obtained by immunogold-EM (Figure 4C and Table 2) are in good agreement with those obtained by immunofluorescence microscopy (Figure 4, A and B).
Figure 4.
Nup53p mediates nuclear import of a NLS-GFP reporter. (A) Intracellular localization of the NLS-GFP reporter in Δnup53, wild-type BMA41 control cells, and nup49-313 control cells (Table 1) after azide and deoxyglucose treatment (i.e., 0 min) and after recovery from the drug treatment in a glucose-containing medium (i.e., 15 and 120 min). Shown are confocal fluorescence micrographs (left) and coincident fluorescence/differential interference contrast images (right). (B) Quantification of the relative import rates in the Δnup53 and the wild-type BMA41 control cells by counting cells harboring nuclear fluorescence as a function of time. (C) Immunogold-EM reveals that the NLS-GFP reporter accumulates within the nuclear basket of the yeast NPCs in the Δnup53 cells (for quantification see Table 2). (D) Nup53p specifically interacts with the yeast nuclear import receptor Kap95p. For this purpose, ProtA-TEV-Nup53p was affinity purified by IgG-Sepharose chromatography and analyzed for copurifying components. Shown is a Western blot analysis with a polyclonal anti-Ka95p antibody revealing the specific interaction of Nup53p with Kap95p. Lane 1, NPC-containing fraction of a wild-type control strain (BMA41); lane 2, eluate derived from the wild-type control strain; lane 3, NPC-containing fraction of the strain expressing ProtA-TEV-Nup53p; and lane 4, eluate derived from the ProtA-TEV-Nup53p. Scale bar, 100 nm (C).
Table 2.
Protein import assay
| Yeast strain | Cytoplasmic∶nuclear ratio before drug treatment | Cytoplasmic∶nuclear ratio immediately after drug removal | Cytoplasmic∶nuclear ratio 5 min after drug removal |
|---|---|---|---|
| BMA41 (NLS-GFP) | 1∶4 | 1∶1 | 1∶5 |
| Δnup53 (NLS-GFP) | 1∶4 | 1∶2 | 1∶3 |
Quantitation of the distribution of 8-nm gold particles coupled to anti-GFP antibody within wild-type (i.e., BMA41) and mutant (i.e., Δnup53) cells (Table 1) after pre-embedding immuno-gold EM (see MATERIALS AND METHODS; for selected examples in a Δnup53 background, see Figure 4C). Twenty-two cells each were analyzed for the BMA41 (NLS-GFP) and the Δnup53 (NLS-GFP) strain. Quantitation was performed by a stereological counting method (Lucocq, 1992).
Nup53p Interacts with Kap95p
To address the question of whether Nup53p's involvement in NLS-dependent nuclear protein import is via a direct interaction with a nuclear import factor, we affinity-purified Nup53p from yeast cells by IgG-Sepharose chromatography under nondenaturing conditions, exactly as described above, and analyzed the copurifying constituents for transport factors. Western blot analysis was performed with antibodies directed against the yeast nuclear protein import receptor Kap95p (importin β) and the small GTPase Gsp1p (i.e. the yeast homologue of vertebrate Ran), respectively. As documented in Figure 4D, we found Nup53p to specifically interact with Kap95p. Kap95p could be detected in the NPC-containing fraction of the Δnup53 strain (Figure 4D, lane 3) and in the eluate after IgG-Sepharose chromatography (Figure 4D, lane 4). The NPC-containing fraction of the BMA41 control strain also included Kap95p (Figure 4D, lane1), but not so its eluate after IgG-Sepharose chromatography (Figure 4D, lane 2). Under the same isolation conditions no interaction with Gsp1p was depicted (our unpublished results).
Nup53p Does Not Participate in NES-mediated Nuclear Protein Export
The involvement of Nup53p in the import of NLS proteins (this study) and ribosomal proteins (Marelli et al., 1998) prompted us to also evaluate a potential role of Nup53p in nuclear protein export. For this purpose, three protein export assays were evaluated, all involving GFP as a fluorescent reporter. All three assays required conditions enabling the protein cargo to be imported into the nucleus, so that in a second step its export could be evaluated.
First, we performed a competition assay (Stade et al., 1997) in the Δnup53 and the BMA41 control strain by introducing a GFP reporter consisting of two GFP moieties, which was simultaneously fused to the NES of the protein kinase inhibitor and the NLS of the SV40 large T antigen (NES-NLS-GFP; Table 1). Because the observed NLS-mediated protein import defect in Δnup53 cells is relatively weak, we assume that such an NES-NLS-GFP reporter can be imported into the nucleus in a Δnup53 background, and that this assay will therefore provide insight into the potential role of Nup53p in NES-dependent protein export. As illustrated by confocal laser scanning microscopy in Figure 5 (left), in both the Δnup53 strain and the wild-type BMA41 the NES-NLS-GFP reporter accumulated in the cytoplasm in cells grown at 30°C because evidently under steady-state conditions the action of the NES dominates over that of the NLS, whereas in a xpo1-1 control strain, which is defect in nuclear protein export, the NES-NLS-GFP reporter accumulated inside the nucleus.
Figure 5.
NES-mediated protein export monitored in Δnup53 cells and in a wild-type BMA41 control strain (Table 1). An NLS-NES-GFP or a Rev-GFP fusion construct was expressed in Δnup53 or BMA41 cells. The localization of the GFP reporter was analyzed after growing the cells at 30°C revealing accumulation of the GFP signal in the cytoplasm, thus indicating that the presence of Nup53p is not necessary for NES-mediated protein export. Shown are confocal fluorescence and coincident fluorescence/differential interference contrast images to localize the GFP reporters relative to the cell periphery.
In a second export assay we transformed the Δnup53 and the BMA41 control strain with a construct where a GFP reporter was fused to the HIV-1 Rev protein (Table 1), which harbors both an NES and an NLS (Fischer et al., 1995; Wen et al., 1995). Expression of the Rev-GFP reporter in these cells was induced by shifting to galactose-containing medium and analyzed by confocal laser scanning microscopy. As documented in Figure 5 (right), in both the Δnup53 strain and the wild-type BMA41 the Rev-GFP reporter accumulated in the cytoplasm at 30°C, whereas the same reporter accumulated in the nucleus of the xpo1-1 control strain. Additionally, when the growth temperature of either the Δnup53 or the wild-type BMA41 cells was shifted to 37°C (i.e., for 2 h), the GFP signal of both the NES-NLS-GFP and the Rev-GFP reporter also accumulated in the cytoplasm (our unpublished results).
A third export assay involved localization of a Yap1p-GFP reporter. The Yap1p protein is a yeast activator protein-1-like transcription factor that activates genes that are required for the response to oxidative stress (Kuge and Jones, 1994; Kuge et al., 1997; Yan et al., 1998). Yap1p is normally cytoplasmic and translocates to the nucleus after addition of oxidants to the growth medium (Kuge et al., 1997). Yap1p harbors an NES-like sequence and its cytoplasmic location is dependent on Xpo1p/Crm1p (Yan et al., 1998). To test whether the location of Yap1p is altered in a Δnup53 background, we transformed the Δnup53 strain and a wild-type control (BMA41) with full-length Yap1p fused to GFP and determined the location of this reporter in the presence and absence of the oxidant diamide. As shown in Figure 6, the Yap1p-GFP is located in the cytoplasm of Δnup53 and BMA41 cells in the absence of diamide (left), but translocates to the nucleus in both Δnup53 and BMA41after incubation with diamide (right). In a xpo1-1 control, the Yap1p-GFP accumulates in the nucleus without treating the cells with diamide. These experiments demonstrate that the Yap1p-GFP reporter can enter the nucleus in a Δnup53 background under oxidative stress and hence can be actively exported by Xpo1p under steady-state conditions.
Figure 6.
Yap1p-GFP export is not impaired in a Δnup53 background. Yeast strains BMA41, Δnup53, or xpo1-1 (Table 1) were transformed with pLDB419(Yap1-GFP), grown in selective medium in the presence or absence of the oxidant diamide. GFP fusion proteins are visualized by confocal fluorescence and coincident fluorescence/differential interference contrast optics.
Hence, based on three protein export assays, i.e., involving NES-NLS-GFP, Rev-GFP, and Yap1p-GFP as synthetic export cargoes, we conclude that NES-mediated nuclear protein export is not impaired by the disruption of NUP53. The NES-NLS-GFP reporter is imported into the nucleus in a NLS-dependent manner via the importin α/β pathway (Stade et al., 1997; Taura et a., 1998), whereas the Rev-GFP reporter is imported into the nucleus by direct interaction of the Rev NLS with importin β (Truant and Cullen, 1999). Therefore, the export assays underscore the results from the nuclear protein import assay (see above; Figure 4), suggesting that the NLS-mediated import in the Δnup53 strain is not completely blocked but rather attenuated compared with the wild-type control strain BMA41.
DISCUSSION
To eventually arrive at a more structure-based understanding of the functional involvement of the NPC in nucleocytoplasmic transport it is necessary to identify and locate in 3-D the nucleoporins that do participate in distinct transport steps, and to determine how these nucleoporins interact with their neighbors and with transport factors. Toward this goal, we have identified and characterized the yeast nucleoporin Nup53p as a physical neighbor of Nic96p within the yeast NPC. Primary sequence analysis and secondary structure prediction have revealed that both Nup53p and Nic96p harbor heptad repeat segments and thus are potential coiled-coil–forming proteins (cf. Lupas et al., 1991). This finding, in turn, suggests that the interaction between these two nucleoporins is mediated by their coiled-coil domains. In fact, by the two-hybrid screen we found that it was the N-terminal domain of Nup53p consisting of two long coiled-coil stretches starting at amino acid 47 and 124, respectively, that is interacting with Nic96p. Moreover, we have documented that Nup53p is directly involved in NLS-dependent nuclear protein import by its specific interaction with Kap95p, yet its absence from the NPC does not appear to significantly interfere with NES-mediated nuclear protein export nor with NPC assembly and/or structural integrity.
Based on the multiple locations of Nic96p and Nup53p within the 3-D architecture of the NPC (Figure 1C) the spatial separation of nucleoporins that are evidently constituents of the central framework or of the cytoplasmic or nuclear fibrillar periphery of the NPC is not as stringent as it might be expected. Nic96p, for example, is located about the cytoplasmic and the nuclear periphery of the central channel in a near-symmetrical arrangement with respect to the central plane of the NPC, and near or at the distal ring of the nuclear basket (Figure 1C; Fahrenkrog et al., 1998, 2000; Stoffler et al., 1999; Rout et al., 2000). Because these multiple locations of Nic96p line the transport route of cargoes on their way into or out of the nucleus, they suggest that Nic96p directly participates in nucleocytoplasmic transport. In fact, temperature-sensitive nic96 mutants, although causing cytoplasmic accumulation of an NLS-containing reporter protein, do not exhibit an obvious export defect (Grandi et al., 1995). Nevertheless, as yet there has been no biochemical evidence for a direct interaction of Nic96p with factors involved in protein import, despite the interaction of Nic96p with Pse1p, the import receptor for the transcription factor Pho4p as recently demonstrated by fluorescence resonance energy transfer analysis (Damelin and Silver, 2000). In agreement with these earlier findings, we also failed to establish a direct interaction of Nic96p with protein import factors by the yeast two-hybrid system. Therefore, it is conceivable that the import defect observed in NIC96 mutant strains is a secondary effect, in the sense that Nic96p does not actually physically interact with the cargo complex. Instead, absence of Nic96p might cause loss or destabilization of the Nsp1p complex from the central framework of the NPC in temperature-sensitive nic96 mutants because Nic96p anchors the Nsp1p complex within the yeast NPC (Grandi et al., 1995; Schlaich et al., 1997; Bucci and Wente, 1998). Hence, these results demonstrate that location of a particular nucleoporin along the route followed by cargo in or out of the nucleus does not necessarily imply a direct role of this nucleoporin in nucleocytoplasmic transport.
Our immunogold-EM analysis has revealed three distinct locations of Nup53p within the 3-D architecture of the NPC (Figure 1). It is a constituent of the central framework of the NPC (i.e., in a near-symmetrical way at its cytoplasmic and nuclear periphery), and it resides at the nuclear basket, an NPC substructure that is directly involved in nucleocytoplasmic transport (cf. Bastos et al., 1996; Shah et al., 1998; Nakielny et al., 1999; Ullman et al., 1999). Therefore, our immunolocalization of Nup53p is consistent with previous publications, which localized Nup53p to both faces of the NPC, but not precisely to any NPC substructures (Marelli et al., 1998; Rout et al., 2000). Surprisingly, although Nup53p is part of the central framework of the yeast NPC, a nup53 null strain exhibited no obvious morphological alterations of its NPCs, at least not at the level of embedding/thin-sectioning EM. Structural alterations of the NPC or its spatial distribution within the NE, such as NPC clustering, are known from mutations in various nucleoporins, e.g., Nup85p and Nup145p (Siniossoglou et al., 1996). Both of these nucleoporins are constituents of the Nup84p complex (Siniossoglou et al., 1996) which, in turn, forms part of the cytoplasmic fibrils of the yeast NPC (Fahrenkrog et al., 1997; Stoffler et al., 1999; Siniossoglou et al., 2000). Therefore, it remains elusive why, on the one hand, mutations in nucleoporins that are part of the central framework do not necessarily cause structural defects of the NPC in the resulting mutant strain, whereas, on the other hand, mutations in nucleoporins that are constituents of the cytoplasmic fibrils or the nuclear basket cause such drastic structural alterations as NPC clustering.
The absence of any obvious structural alterations of the NPC in the Δnup53 strain and the physical interaction of Nup53p with the yeast nuclear protein import receptor Kap95p suggest that, different from Nic96p (see above), Nup53p does play a direct role in nuclear protein import. In this context, the interaction of a number of nucleoporins with importin β-like proteins appears to be mediated by phenylalanine–glycine (FG) repeats within the amino acid sequence of these nucleoporins (Seedorf et al., 1999; Stoffler et al., 1999). Although Nup53p harbors four separated FG sequence motifs within its amino acid sequence, it does clearly not represent an FG-repeat containing nucleoporin. Hence, whereas the interaction between Nup53p and Kap95p may indeed involve one or several of these FGs, there must be additional amino acids specifying this interaction.
Location of Nup53p at the nuclear basket (Figure 1A) and accumulation of an NLS-GFP reporter, most likely in the nuclear basket in a nup53 null strain (Figure 4C), support a model in which Nup53p is involved in a late step of nuclear protein import. However, because NUP53 is not essential and protein import is not completely inhibited in the Δnup53 strain (Figure 4B), other nucleoporins residing at the nuclear basket, e.g., Nup1p, the presumed yeast homologue of vertebrate Nup153 (Moroianu et al., 1997), must be able to at least partially substitute for Nup53p in its absence. This may be even more so in the case of cargo export, because NES-mediated protein export is not noticeably impaired by the disruption of NUP53 (Figures 5 and 6).
Taken together, we have structurally and functionally identified and characterized the yeast nucleoporin Nup53p that physically (i.e., both by a yeast two-hybrid screen and biochemically) interacts with the essential yeast nucleoporin Nic96p. Nup53p resides near-symmetrically (i.e. relative to the central plane of the NPC) at the cytoplasmic and the nuclear periphery of the central framework, and at the nuclear basket fibrils of the yeast NPC. Although deletion of NUP53 causes no obvious morphological defects, nuclear protein import is attenuated significantly in a nup53 null strain. Due to its specific interaction with the yeast import receptor Kap95p, Nup53p must play a direct functional role in nuclear protein import. In contrast, Nup53p does not appear to play a significant role in cargo export because its absence does not significantly interfere with NES-mediated protein export.
ACKNOWLEDGMENTS
We thank Drs. Jennifer Hood and Pamela Silver for providing several plasmids and the antibodies against GFP and Kap95p, Dr. Ed Hurt for providing the antibody against Nic96p, Drs. Anita Corbett and Laura Davis for the Yap1p-GFP plasmid, and Drs. Françoise Stutz and Karsten Weis for the xpo1-1 strain. We also thank Dr. Markus Dürrenberger for help with confocal laser scanning microscopy, Ursula Sauder for help with preparing samples for EM, Robert Wyss for help with Figure 1C, and Hedi Frefel and Marlies Zoller for expert photographic work. This work was supported by a research grant from the Human Frontier Science Program (HFSP), and by the Kanton Basel-Stadt and the M. E. Müller Foundation of Switzerland.
REFERENCES
- Akey CW, Radermacher M. Architecture of the Xenopus nuclear pore complex revealed by 3-dimensional cryo-electron microscopy. J Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R, Lin A, Enarson M, Burke B. Targeting and function in mRNA export of the nuclear pore complex protein Nup153. J Cell Biol. 1996;134:1141–1156. doi: 10.1083/jcb.134.5.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin-Baillieu A, Guillemet E, Cullin C, Lacroute F. Construction of a yeast strain deleted for the TRP1 promotor and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. doi: 10.1002/(SICI)1097-0061(19970330)13:4<353::AID-YEA86>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Bucci M, Wente SR. A novel fluorescence-based genetic strategy identifies mutants of Saccharomyces cerevisiae defective for nuclear pore complex assembly. Mol Biol Cell. 1998;9:2439–2461. doi: 10.1091/mbc.9.9.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett AH, Silver PA. Nucleocytoplasmic transport of macromolecules. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damelin M, Silver PA. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- Doye V, Hurt E. From nucleoporins to nuclear pore complexes. Curr Opin Cell Biol. 1997;9:401–411. doi: 10.1016/s0955-0674(97)80014-2. [DOI] [PubMed] [Google Scholar]
- Fabre E, Hurt EC. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu Rev Genet. 1997;31:277–313. doi: 10.1146/annurev.genet.31.1.277. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Aebi U, Panté N. Yeast nuclear pore complexes (NPCs): dissecting their molecular architecture. Mol Biol Cell. 1997;8:236a. [Google Scholar]
- Fahrenkrog B, Aris JP, Hurt EC, Panté N, Aebi U. Comparative localization of protein A tagged and endogenous yeast nuclear pore complex proteins by immunoelectron microscopy. J Struct Biol, 2000;129:295–305. doi: 10.1006/jsbi.2000.4223. [DOI] [PubMed] [Google Scholar]
- Fahrenkrog B, Hurt EC, Aebi U, Panté N. Molecular architecture of the yeast nuclear pore complex: localization of Nsp1p subcomplexes. J Cell Biol. 1998;143:577–588. doi: 10.1083/jcb.143.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Doye V, Hurt EC. Purification of NSP1 reveals complex formation with “GLFG”nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 1993;12:3061–3071. doi: 10.1002/j.1460-2075.1993.tb05975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 CdK-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt EC. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 1988;7:4324–4334. doi: 10.1002/j.1460-2075.1988.tb03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Adam SA. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interaction with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hyperperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucocq J. Quantitation of gold labeling and estimation of labeling efficiency with a stereological counting method. J Histochem Cytochem. 1992;40:1929–1936. doi: 10.1177/40.12.1453009. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marelli M, Aitchinson JD, Wozniak RW. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. J Cell Biol. 1998;143:1813–1830. doi: 10.1083/jcb.143.7.1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci USA, 1997;94:4451–4456. doi: 10.1073/pnas.94.18.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Blobel G. Role of the nuclear transport factor p10 in nuclear import. Science. 1996;272:120–122. doi: 10.1126/science.272.5258.120. [DOI] [PubMed] [Google Scholar]
- Nehrbass U, Fabre E, Dihlmann S, Herth W, Hurt EC. Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of nuclear pore protein NSP1. Eur J Cell Biol. 1993;62:1–12. [PubMed] [Google Scholar]
- Nehrbass U, Kern H, Mutvei A, Horstmann H, Marshallsay B, Hurt EC. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell. 1990;61:979–989. doi: 10.1016/0092-8674(90)90063-k. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Panté N, Aebi U. Molecular dissection of the nuclear pore complex. Crit Rev Biochem Mol Biol. 1996;31:153–199. doi: 10.3109/10409239609106583. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchinson JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. J Cell Biol. 1993;109:2641–2652. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaich NL, Häner M, Lustig A, Aebi U, Hurt EC. In vitro reconstitution of a heterotrimeric nucleoporin complex consisting of recombinant Nsp1p, Nup49p, and Nup57p. Mol Biol Cell. 1997;8:33–46. doi: 10.1091/mbc.8.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase. Mol Cell Biol. 1999;19:1547–1557. doi: 10.1128/mcb.19.2.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Roberts P, Gu ZY, Spitz L, Tabb MM, Nomura M, Goldfarb DS. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa, Leonard K, Müller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Song HY, Dunbar JD, Donner DB. Aggregation of a intracellular domain of the type 1 tumor necrosis factor receptor defined in the two-hybrid system. J Biol Chem. 1994;269:22492–22495. [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Stan R, McLaughlin MM, Cafferkey R, Johnson RK, Rosenberg M, Livi GP. Interaction between FKBP12-rapamycin and TOR involves a conserved serine residue. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- Stochaj U, Héjazi M, Belhumeur P. The small GTPase Gsp1p binds to the repeat domain of the nucleoporin Nsp1p. Biochem J. 1998;330:412–427. doi: 10.1042/bj3300421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Taura T, Krebber H, Silver PA. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Shah S, Powers MA, Forbes DJ. The nucleoporin Nup153 plays a critical role in multiple types of nuclear export. Mol Biol Cell. 1999;10:649–64. doi: 10.1091/mbc.10.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. J Cell Biol. 1995;133:4–14. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Yan C, Lee LH, Davis LI. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Rout MP, Akey C. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]