Short abstract
Twenty-nine novel human genes were predicted with putative coding regions sharing an open reading frame with the cynomolgus monkey, and the expression of 21 pairs of genes was confirmed by reverse transcription-coupled polymerase chain reaction.
Abstract
Background
The complete assignment of the protein-coding regions of the human genome is a major challenge for genome biology today. We have already isolated many hitherto unknown full-length cDNAs as orthologs of unidentified human genes from cDNA libraries of the cynomolgus monkey (Macaca fascicularis) brain (parietal lobe and cerebellum). In this study, we used cDNA libraries of three other parts of the brain (frontal lobe, temporal lobe and medulla oblongata) to isolate novel full-length cDNAs.
Results
The entire sequences of novel cDNAs of the cynomolgus monkey were determined, and the orthologous human cDNA sequences were predicted from the human genome sequence. We predicted 29 novel human genes with putative coding regions sharing an open reading frame with the cynomolgus monkey, and we confirmed the expression of 21 pairs of genes by the reverse transcription-coupled polymerase chain reaction method. The hypothetical proteins were also functionally annotated by computer analysis.
Conclusions
The 29 new genes had not been discovered in recent explorations for novel genes in humans, and the ab initio method failed to predict all exons. Thus, monkey cDNA is a valuable resource for the preparation of a complete human gene catalog, which will facilitate post-genomic studies.
Background
In 2001, it was announced that most of the human genome had been sequenced and that the complete sequence would be determined by 2003 [1,2]. As the first step in decoding the entire sequence of the human genome, we must identify the protein-coding regions in the human genome sequence. Predicting the genes in the human genome is one of the most substantial applications of the sequence data, but is also the most difficult. Prediction of protein-coding genes by computer algorithm (ab initio prediction) is accurate to some extent in organisms whose genome sequence has already been determined, for example the fruit fly Drosophila melanogaster and the nematode Caenorhabditis elegans. Similar attempts to predict human genes, however, have met with limited success because small exons are generally separated by long introns. In the fly, ab initio methods can correctly predict around 90% of individual exons, and all the coding exons in a gene in about 40% of genes [3], in contrast to about 70% and 20%, respectively, in humans [4,5]. Thus, prediction of genes in humans requires further experimental evidence, for example, from cDNAs and expressed sequence tags (ESTs) [6]. However, the uneven expression of various transcripts makes it difficult to isolate the cDNA of genes expressed as a very small proportion of the total transcripts, and it has not been possible to completely eliminate artifacts arising from cDNAs and ESTs derived from genomic DNA or partially spliced mRNAs [7,8]. Moreover, EST sequences are less informative about the boundary of an individual gene. It is also difficult to predict single-exon genes encoding small proteins, because it is impossible to determine easily whether the short open reading frames (ORFs) are actually translated into protein. In our previous study, we isolated approximately 20,000 clones from oligo-capped cDNA libraries of parts of the cynomolgus monkey brain (parietal lobe and cerebellum) and sequenced their 5' ends. We subsequently determined the entire sequence of 118 novel cDNAs, whose human orthologous cDNAs had not been entered in the public databases [9]. The cynomolgus monkey (Macaca fascicularis) is one of the species of Macaca, an Old World monkey. On the basis of DNA sequence comparison complemented by fossil evidence, the divergence of humans and Old World monkeys is estimated at about 25 million years ago [10]. On the basis of nucleotide-sequence similarity of a 10.9 kb globin genomic region in the previous study, the difference in nucleotide sequence between human and Macaca is 7% [11].
By using cynomolgus monkey brain, we were able to reduce the degradation of the mRNA, which is very fragile, during the construction of cDNA libraries, because, unlike human brain tissue, brain tissue can be removed intentionally from the anesthetized monkey and frozen immediately after extirpation. Since that study, we have isolated an additional 12,000 clones and determined the entire sequence of 673 novel cDNAs from frontal lobe, temporal lobe and medulla oblongata of the cynomolgus monkey brain. Moreover, comparison between the cynomolgus monkey cDNA sequence and the human genome draft sequence makes it possible to distinguish ORFs that actually encode proteins from spurious ORFs, because the high genomic conservation between human and Macaca means that protein-coding ORFs are likely to be maintained between the human and monkey sequences. In this study, we have predicted unidentified human genes from the human genome draft sequence by referring to cDNA sequences of cynomolgus monkey and have experimentally confirmed the expression of most of these genes. This method allowed us to identify novel human genes that had eluded other recent exploratory studies.
Results and discussion
In our previous study [9], we constructed two oligo-capped cDNA libraries of the cynomolgus monkey brain (cerebellum cortex, QccE; parietal lobe, QnpA). Subsequently, we constructed three more brain libraries from the frontal lobe (QflA), temporal lobe (QtrA) and medulla oblongata (QmoA), and sequenced the 5' ends of approximately 12,000 clones isolated from the three libraries. We then determined the entire sequence of 673 clones whose 5'-end sequences showed no significant similarities to sequences in the GenBank nr or EST databases and deposited them in the DDBJ/EMBL/GenBank nucleotide database (accession numbers: AB055250-055381, AB056322-056432, AB056799-056847, AB060202-060263, AB062934-063100, AB066511-066549). From these clones, we selected 90 that carried a putative coding region longer than 300 bp and showed no homology to mRNA sequences in the GenBank database by BLAST search (cut-off value: 1e-90), except for minimal overlap (less than 30% coverage of ORF). The 90 novel cDNAs are listed on our website [12]. Next, we tried to identify the sequences of the human genome sequence (1 April 2001 data) that corresponded to the novel cynomolgus monkey cDNAs by using the program BLAT at the University of California at Santa Cruz (UCSC) website [13]. The search yielded 78 cynomolgus monkey clones with orthologous sequences in the human genome (more than 90% similarity), but 14 of 78 clones showed partial matches with the genome sequence, making it impossible to obtain the entire sequence of the human orthologs. The remaining 12 clones that did not match any sequences in the human genome are probably located in the genomic region missing in the current draft sequence.
The Sim4 program was used to align each cynomolgus monkey cDNA sequence with the orthologous human genome sequence [14]. Although Sim4 was not designed to align sequences derived from two divergent species, the monkey cDNA sequence was similar enough to human to allow Sim4 to be used. Whenever Sim4 failed to align monkey cDNA sequence with human genome DNA sequence, sequences were compared by BLAST and the alignment corrected manually. In the intron sequences, the sequences GT at the 5' splice site and AG at the 3' site (the GT-AG pattern) and the GC-AG pattern were regarded as indicating splice sites, and corresponding regions in the human genome were concatenated to construct a hypothetical human cDNA sequence (Figure 1). Spurious cynomolgus monkey ORFs that might appear in the untranslated region (UTR) of longer transcripts were disrupted by several gap insertions in the hypothetical human cDNA; however, the hypothetical human cDNAs without an ORF corresponding to that of monkey could be used as probes for assignment of novel human genes in the human genome. Ultimately, 29 of the 64 pairs of ORFs in human and monkey cDNAs were found to be highly conserved, suggesting the existence of the human cDNAs predicted from the novel cynomolgus cDNAs (Table 1). When a few gap insertions interrupted the ORF of a hypothetical human cDNA, we assumed that the gap insertions were caused by errors in the human draft genome sequence. We therefore revised the part of human draft genome sequence concerned by alignment of corresponding EST sequences, or if necessary, sequenced the RT-PCR product for the corresponding part amplified from human brain RNA (described below). As these 29 human hypothetical cDNAs have not actually been sequenced, they could not be deposited in public databases but are available at our website [12].
Figure 1.
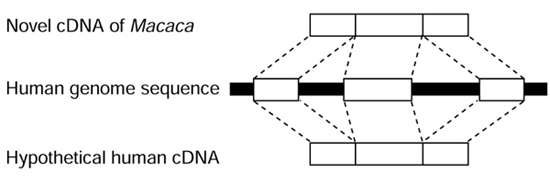
Construction of hypothetical human cDNA. The regions in human genome corresponding to the cynomolgus monkey cDNA were digested and concatenated in silico as putative exons, and the hypothetical novel human gene was predicted.
Table 1.
Summary of the 29 new genes
| Monkey accession number | Cynomolgus monkey (Macaca) clone | Location* | RT-PCR† | Length (amino acids) in Macaca‡ | Length (amino acids) in human§ | Number of exons¶ | Exon predicted# | Functional annotation (putative)** |
| AB055251 | QflA-10072 | 10p12.1 | + | 743 | 743 | 2 (1) | 1 (0) | Adenylate kinase |
| AB055256 | QflA-10350 | 9q34.3 | - | 308 | 276 | 5 (4) | 1 (1) | Unknown |
| AB055264 | QflA-10778 | 2q24.2 | - | 603 | 577 | 13 (13) | 6 (2) | Nuclear protein |
| AB056798 | QflA-11110 | 6q23.2 | + | 621 | 617 | 5 (5) | 3 (0) | Sugar transporter |
| AB055271 | QflA-11149 | 1p31.1 | - | 510 | 514 | 5 (4) | 2 (0) | Leucine-rich repeat protein |
| AB055273 | QflA-11186 | 10q26.2 | + | 624 | 625 | 8 (8) | 4 (1) | Unknown |
| AB055276 | QflA-11332 | 17q23.3 | + | 590 | 590 | 17 (14) | 10 (1) | Unknown |
| AB055278 | QflA-11381 | 11q22.3 | + | 197 | 197 | 1 (1) | 0 (1) | Unknown |
| AB055280 | QflA-11470 | 7q11.22 | - | 114 | 114 | 1 (1) | 0 (0) | Unknown |
| AB056389 | QflA-12365 | 10q26.3 | ++ | 368 | 369 | 12 (10) | 6 (2) | Serine/threonine protein kinase |
| AB055295 | QflA-12453 | 5q35.2 | - | 103 | 103 | 2 (1) | 0 (0) | Unknown |
| AB056800 | QflA-12512 | 15q15.2 | + | 653 | 653 | 20 (16) | 10 (2) | Glycoside hydrolase family 31 |
| AB056802 | QflA-12743 | 7q11.21 | ++ | 214 | 214 | 6 (4) | 2 (2) | Unknown |
| AB060227 | QflA-15038 | 9q34.13 | + | 327 | 327 | 2 (2) | 1 (1) | Unknown |
| AB056812 | QflA-15102 | 4p15.1 | + | 102 | 102 | 1 (1) | 0 (0) | Unknown |
| AB060245 | QflA-15249†† | 11p15.1 | + | 413 | 369 | 11 (9) | 8 (1) | Thyrosine phosphatase |
| AB056426 | QflA-15366 | 10q22.1 | + | 581 | 581 | 3 (2) | 2 (0) | Cysteine-rich protein with leucine-rich repeat |
| AB066513 | QmoA-10247 | 1q25.3 | + | 196 | 196 | 11 (6) | 1 (1) | G-protein signaling protein |
| AB063014 | QmoA-11221 | 6q14.1 | - | 430 | 430 | 7 (6) | 4 (2) | Nuclear protein |
| AB063019 | QmoA-11380 | Xq13.3 | + | 309 | 309 | 7 (1) | 0 (0) | Unknown |
| AB063029 | QmoA-11613 | 10p12.33 | ++ | 654 | 654 | 12 (11) | 5 (2) | Unknown |
| AB066529 | QmoA-11640 | 16p12.3 | - | 122 | 122 | 2 (1) | 0 (0) | Unknown |
| AB066540 | QmoA-12446 | 19q13.2 | - | 654 | 655 | 3 (3) | 0 (1) | Zinc-finger protein with KRAB domain |
| AB066542 | QmoA-12482 | 10p11.23 | + | 391 | 392 | 5 (2) | 1 (1) | Zinc-finger protein |
| AB063089 | QtrA-10552 | 11q22.3 | + | 664 | 664 | 16 (12) | 9 (2) | RNA-binding methyl transferase |
| AB060878 | QtrA-12612 | 9q32 | + | 238 | 238 | 5 (3) | 2 (1) | Nuclear protein |
| AB063095 | QtrA-13256 | 6q27 | + | 300 | 301 | 18 (12) | 9 (0) | Unknown |
| AB060262 | QtrA-14732 | 3p21.31 | ++ | 396 | 396 | 3 (3) | 3 (3) | Unknown |
| AB060922 | QtrA-14779†† | 1p31.1 | + | 432 | 536 | 12 (11) | 6 (1) | Adenylate kinase |
*Human chromosomal locationwas determined by computer homology search of the human genome working draft sequence at UCSC. †Results of RT-PCR analysis: +, single product; ++, multiple products; -, no product. QtrA-10552 and QtrA-13256 showed a distinct splicing pattern between human and Macaca. ‡Length of putative coding region deduced from cDNA sequence of Macaca. §Length of putative coding region deduced from human genome sequence corresponding to cDNA of Macaca. ¶Number of exons in the human hypothetical cDNA. The number of coding exons is in parentheses. #Number of exons correctly predicted by GenScan in the human genome. Partially predicted exons are in parentheses. **Function of putative protein was deduced by InterPro category search and BLAST homology search. ††ORF sequence was revised by sequencing the RT-PCR product.
To confirm the expression of these hypothetical human cDNAs and cynomolgus monkey cDNAs, we designed RT-PCR primers to amplify the coding regions of hypothetical human cDNAs and investigated expression by using total RNA from human and monkey brain. In total, 21 novel genes were shown to be expressed in both human and cynomolgus monkey brain (Figure 2). When the size of product was different from the expected length, we sequenced the RT-PCR product to confirm the splicing pattern. For four of the cDNAs, several DNA bands were detected as expression products besides the band of expected size, but we assumed that they were derived from an alternatively spliced mRNA.
Figure 2.
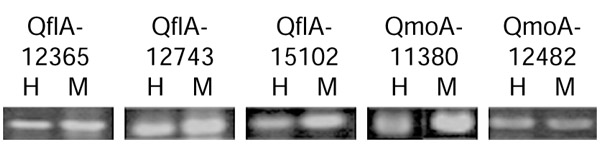
Expression of each gene by RT-PCR. The primer sets covering a putative protein-coding region were designed for amplification of transcripts from total brain RNA of human (H) and cynomolgus monkey (M). Of 29 primer pairs, 21 could amplify the transcript in both human and Macaca.
Interestingly, two genes showed a clearly different pattern of expression in humans and the cynomolgus monkey. Using the first primer set designed, the cDNAs of QtrA-10522 and QtrA-13256 appeared to be expressed only in monkey brain. However, with a primer set covering another pair of exons, expression of these genes in human and cynomolgus monkey was observed in both clones. These observations indicated some differences between humans and the cynomolgus monkey in the splicing pattern of the genes. The cynomolgus monkey cDNA for both genes carried additional exons that were spliced as introns in humans.
We could not amplify expected RT-PCR products of ORFs of eight genes. However, the conservation of ORFs between humans and the cynomolgus monkey strongly supported their being functional regions. Therefore, it is possible that the eight genes could not be amplified by RT-PCR because of technical problems, such as high GC content or inhibitory structure in the targeted region, or because the amount of transcript was too small to detect in the total RNA that we used for RT-PCR [15].
We then investigated how many of the newly identified coding exons were predicted by ab initio methods. Genomic sequences covering novel genes together with 2 kb upstream and downstream flanking sequences, were surveyed with GENSCAN programs [16]. Out of the 167 experimental protein-coding exons, 96 (57%) were correctly predicted by GENSCAN and 28 (17%) were partially predicted, while the remaining 43 (26%) exons were not predicted at all. It is well known that the specificity of ab initio prediction is less successful, and some of our novel genes put together several exons of different ab-initio-predicted genes. For example, QflA-11332 covered exons derived from five different genes predicted by GENSCAN and three predicted by another ab initio prediction, GENEMARK.hmm [17] (Figure 3). The putative function of the proteins was annotated using InterPro search [18] and protein homology search by BLAST. Some putative functions were deduced for about half of the 29 proteins. The results are summarized in the functional annotation column in Table 1.
Figure 3.
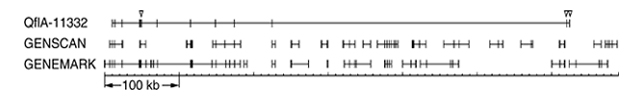
Genomic structure of QflA-11332 and ab-initio-predicted exons. QflA-11332 is only 2.1 kb of cDNA but spans more than 600 kb in the human genome. Vertical lines indicate the exons of clones or exons predicted by each computer program, and are connected by a horizontal line (intron). The exons under open triangles represent the exons not predicted by either computer program. Other exons were correctly predicted but segmented into several genes by ab initio prediction.
Conclusions
We have constructed 29 hypothetical human cDNAs on the basis of sequence similarity to novel cynomolgus monkey cDNAs, and comparisons between human and monkey sequences allowed us to select the cDNAs carrying protein-coding ORFs with high accuracy. These novel genes had not been discovered by recent explorations for novel genes in humans, and the ab initio method failed to predict all of their exons. They were also functionally annotated by computer analysis. Thus, the cDNA of a closely related monkey is a valuable resource for preparing a complete human gene catalog, which will facilitate future post-genomic studies, such as DNA microarray or proteome analysis.
Materials and methods
Cynomolgus monkey tissues
Tissue was collected from a 21-year-old male cynomolgus monkey. This monkey was cared for and handled according to guidelines established by the Institutional Animal Care and Use Committee of the National Institute of Infectious Diseases (NIID) of Japan and the standard operating procedures for monkeys at the Tsukuba Primate Center, NIID, Tsukuba, Ibaraki, Japan. Extirpation of the tissues was conducted in accordance with all guidelines required in the Laboratory Biosafety Manual, World Health Organization, and was carried out in the P3 facility for monkeys at the Tsukuba Primate Center, NIID.
Construction of oligo-capped cDNA libraries
Total RNA was isolated using a commercially available RNA isolation kit (Isogen, Nippon Gene;Rneasy, QIAGEN). Poly(A)+ RNA was purified using oligo-dT cellulose (Collaborative Biomedical Products; Roche). Oligo-capping was carried out as previously described [19]. After PCR amplification, the separated products longer than 2 kb were cloned into DraIII-digested pME18S-FL3, and the plasmid vectors containing cDNA were used to transform competent cells byelectroporation.
Reverse transcription-coupled polymerase chain reaction (RT-PCR)
The templates of the human brain total mRNA were purchased from Clontech. Total RNA of cynomolgus monkey brain was isolated from the cerebrum of the cynomolgus monkey described above with Trizol (Life Technologies). A 1 μl volume of total mRNA was amplified with a One Step RNA PCR Kit (Takara). The temperature and time schedule were 40 cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 90 sec. PCR products were separated on 1.5% agarose gel with a 100 bp ladder DNA marker (Gibco BRL).
DNA sequencing
The entire sequence of clones was determined on an ABI 3700 and 310 automated sequencers (Perkin-Elmer) by the primer walking method. Cycle sequencing was carried out using an ABI PRISM BigDye Terminator Sequencing kit (Perkin-Elmer) according to the manufacturer's instructions.
Acknowledgments
Acknowledgements
This study was supported in part by the Health Science Research Grant for the Human Genome Program from the Ministry of Health and Welfare of Japan.
References
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1086/172716. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Reese MG, Kulp D, Tammana H, Haussler D. Genie - gene finding in Drosophila melanogaster. Genome Res. 2000;10:529–538. doi: 10.1101/gr.10.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Guigo R, Agarwal P, Abril JF, Burset M, Fickett JW. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 2000;10:1631–642. doi: 10.1101/gr.122800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann S, Weil B, Wellenreuther R, Gassenhuber J, Glassl S, Ansorge W, Bocher M, Blocker H, Bauersachs S, Blum H, et al. Toward a catalog of human genes and proteins: sequencing and analysis of 500 novel complete protein coding human cDNAs. Genome Res. 2001;11:422–435. doi: 10.1101/gr.154701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG, Landsman D. A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res. 1997;25:1626–1632. doi: 10.1093/nar/25.8.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N, Hida M, Kusuda J, Tanuma R, Iseki K, Hirata M, Suto Y, Hirai M, Terao K, Suzuki Y, et al. Assignment of 118 novel cDNAs of cynomolgus monkey brain to human chromosomes. Gene. 2001;275:31–37. doi: 10.1016/S0378-1119(01)00665-5. [DOI] [PubMed] [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Goodman M, Tagle DA, Fitch DHA, Bailey W, Czelusniak J, Koop BF, Benson P, Slightom JL. Primate evolution at the DNA level and a classification of hominoids. J Mol Evol. 1990;30:260–266. doi: 10.1007/BF02099995. [DOI] [PubMed] [Google Scholar]
- Prediction of unidentified human genes based on sequence similarity to novel cDNAs of cynomolgus monkey brain http://www.nih.go.jp/yoken/genebank/Supplementary_data/prediction/index.html [DOI] [PMC free article] [PubMed]
- Human Genome Project Working Draft at UCSC http://genome.ucsc.edu/
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JV, Yan XH, Diamond J, Lin F, Cross NCP, Goldman JM. Reverse transcription/polymerase chain reaction (RT/PCR) amplification of very small numbers of transcripts: the risk in misinterpreting negative result. Leukemia. 1996;10:1217–1221. [PubMed] [Google Scholar]
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- GeneMark http://opal.biology.gatech.edu/GeneMark/
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Ishihara D, Sasaki M, Nakagawa H, Hata H, Tsunoda T, Watanabe M, Komatsu T, Ota T, Isogai T, et al. Statistical analysis of the 5' untranslated region of human mRNA using "Oligo-Capped" cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]


