Abstract
A recent proteomic analysis of germinating Arabidopsis thaliana seeds demonstrates the effectiveness of functional genomics for investigating the complexity of developmental regulatory networks, such as the development of the embryo into a young plant.
In flowering plant development, seed germination is the transition of the quiescent embryo, which has developed from the fertilized ovule, into a new photosynthetically active plant. The visible sign that germination has been completed is the protrusion of the radicle, the precursor of the root, through the seed coat; germination sensu stricto begins, however, with water uptake by the seed (imbibition) and ends with the start of elongation the embryonic axis inside the seed [1]. Germination results from a combination of many cellular and metabolic events, coordinated by a complex regulatory network that includes seed dormancy, an intrinsic ability to temporarily block radicle elongation in order to optimize the timing of germination [2]. In the field of seed biology, germination mechanisms and their control by dormancy have been investigated in a wide range of species. Nonetheless, how these processes are coordinated, how they contribute to germination, and the regulatory network leading to completion of germination remain poorly understood.
The availability of the complete genome sequence of the model plant Arabidopsis thaliana [3], together with the development of high-throughput procedures for global analyses of gene function, has launched the 'post-genomic' era of plant biology. Systematic analyses of RNA and protein expression patterns, and of post-translational modifications, are now feasible for a large set of genes [4]. These can provide important clues about protein-protein interactions and gene functions in a complex developmental context. A recent proteome study of germinating Arabidopsis seeds [5] highlights the effectiveness of using this model organism to provide information on germination that may prove general to all plants.
The physiology of seed germination
In temperate climates, most mature seeds, consisting of an embryo surrounded by endosperm and a seed coat (testa), are dispersed from the mother plant in a state of low moisture content (5-15%) and with metabolic activity at a standstill. Some physiologists have hypothesized that with regard to their structure, genetic information and macromolecular content, dry seeds are in a state of readiness to resume metabolism [6]. For germination to occur, quiescent seeds need only be hydrated under conditions that encourage metabolism, such as a suitable temperature and the presence of oxygen (Figure 1). The uptake of water by the seed, which is considered to be a trigger for germination, and the metabolic processes that take place as a result, are described in Figure 1. Seed germination can be delayed by dormancy, a process that involves interactions between two plant growth factors, gibberellins (GAs) and abscisic acid (ABA) [7]. This overall picture results from fragmentary physiological and biochemical studies in numerous species and remains to be refined with information from model species.
Figure 1.
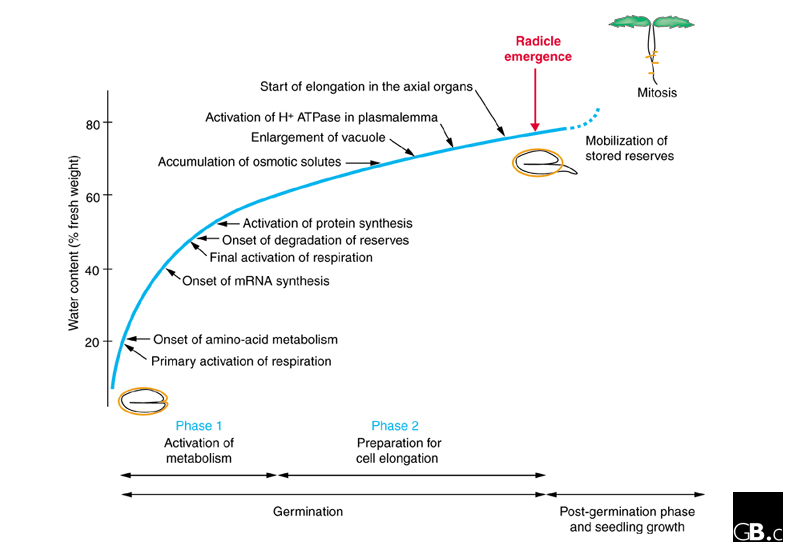
The cellular and metabolic events triggered by water uptake during seed germination. Germination is affected by both environmental factors (the availability of water, oxygen and light as well as the temperature) and intrinsic factors (dormancy, permeability of the testa to water and oxygen, and obstruction of radicle emergence by the endosperm). A rapid imbibition phase (phase 1) launches the resumption of basic metabolism. During this phase, known as 'physical' imbibition, a step-by-step activation of metabolic pathways results from the gradual increase in hydration (arrows). When the level of hydration exceeds 60%, the rate of hydration slows (phase 2) and new physiological mechanisms prepare cell expansion in the embryonic axes, culminating in the start of cell elongation. Osmotically active substances (solutes, such as sugars, amino acids, and potassium ions) are accumulated and acidification of the cell wall leads to a loosening of the bonds between cell-wall polymers. These events coincide with the activation of the H+ ATPase in the plasmalemma, which results in a further increase in water uptake that may coincide with weakening of the surrounding tissues (the endosperm) as the embryonic axes elongate and germination is completed. Completion of seed germination can be temporarily blocked by dormancy, which is in turn released by antagonistic interactions between the endogenous plant growth factors abscisic acid (ABA) and gibberellins (GAs) [7]. Storage nutrients (lipids, proteins or starch) accumulated in the embryo's cotyledon and/or endosperm start to be mobilized before completion of germination and are used in the post-germination steps to sustain the young plant in its early growth stages, before it becomes autotrophic. If the cell cycle resumes during germination, the first cell division (mitosis) occurs in the postgerminative phase. The arrows indicate the particular hydration levels that are known to correlate with individual metabolic events. The sequence of events shown in this model results from studies in various species. Modified from [6].
The genetic and molecular determinants of seed germination and dormancy has been intensively investigated in oat, tomato, Nicotiana plumbaginifolia and in Arabidopsis [8]. Several studies have led to the identification of many proteins or mRNAs, the accumulation of which is correlated with dormancy or seed germination. Many of these products have sequence homology with proteins involved in desiccation tolerance or protection against various injuries and with storage proteins [9,10,11,12,13,14]. Identifying these functions is important for understanding the differences between the dormant and non-dormant states, but to date no 'switch' function that could be involved in the choice between maintenance and release of dormancy and germinating has been discovered by these approaches. Curiously, screens for non-germinating Arabidopsis mutants have identified mutations in only three of the five loci involved in GA biosynthesis [15,16]. Screens for early germination (low dormancy) have led to the isolation of mutants that are deficient in the synthesis of, and sensitivity to, ABA [17], in testa color (as seen in the transparent testa mutants), testa structure (seed shape mutants), and in unknown downstream dormancy-inducing processes (rdo mutants) [18]. These mutants provide useful support to the results of physiological analyses, and they genetically confirm the roles of GA, ABA and the seed coat in the regulatory network of germination, including dormancy. But the lack of more sensitive screens and the high level of gene redundancy in plant genomes may explain why such 'forward genetic' approaches have not been very effective in identifying the genes involved in germination and dormancy and why they have, up to now, elucidated only a limited number of gene-function relationships.
Understanding of germination mechanisms and their regulatory networks is limited by the complexity of seed architecture and of the various spatio-temporally regulated processes involved. Fundamental questions about germination, such as how the embryo emerges from the seed to complete germination and how embryo emergence is blocked so that the seed can be maintained in a dormant state are still unanswered [2]. Using high-throughput procedures developed from having sequenced the genome of Arabidopsis therefore provides an opportunity to tackle these key questions and establish an integrated model of seed germination.
Proteomics of seed germination
Gallardo and coworkers [5] have initiated a proteomic analysis of the Arabidopsis seed-germination process using the ecotype Landsberg erecta. Two-dimensional gel electrophoresis was used to resolve and analyze seed proteins and the changes in their abundance during germination. Several of these proteins were identified by matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) mass spectroscopy. Protein extracts from mature dry seeds and from seeds that had been given water (imbibed) for 1 or 2 days were compared in order to follow the pattern of changes in protein expression in quiescent mature seeds, in imbibed germinating seeds and after radicle protrusion, respectively. About 1,300 proteins were resolved on the gels, and these were classified according to their specific accumulation patterns.
Quiescent seeds are ready to resume metabolism
Most of the proteins found by Gallardo et al. [5] (1,251) were present in dry seeds, and their abundance remained constant throughout the germination process. The germination process therefore appeared to be associated with modifications in the abundance of only a limited number of proteins, supporting the idea that dry seeds are essentially ready to germinate. Germination sensu stricto was characterized by changes in the abundance of 39 proteins in seeds imbibed for 1 day. Of these, none are respiratory enzymes, but an actin 7, tubulin subunits, and a WD-40-repeat protein resembling receptors of activated protein kinase C (RACKs) accumulated, correlating with the resumption of cell elongation and cell-cycle activity [5]. It appeared that metabolic activation during the first phase of hydration is predetermined in dry non-dormant seeds, and that the newly synthesized proteins contribute to the completion of processes that occur in the second hydration phase involved in the completion of germination (see Figure 1). Thus, resumption of metabolic activity during germination may rely mainly upon proteins that are stored during seed maturation. We should consider the possible involvement of proteins that are synthesized de novo but are not detected by the technique used here, however.
When are storage reserves mobilized?
Gallardo et al. [5] found that the accumulation of 12S seed-storage-protein subunits, and of some enzymes involved in triacylglycerol catabolism (catalase, aconitase, phosphoenolpyruvate carboxykinase, and others), correlated with initial events in the mobilization of protein and lipid reserves. Curiously, both precursor forms and proteolyzed forms of the 12S subunit were identified in dry mature seeds. Thus, protein reserves may be mobilized not only during germination and seedling growth but also during the maturation phase. This new finding is contrary to the well-known assumption that protein reserves are synthesized before germination and that this is developmentally separated from the catabolic processes that normally occur during germination [1,19].
Post-germination events
The second day of imbibition was characterized by changes in the level of 35 proteins [5]. Some of these were also linked to the mobilization of reserves and their levels may reflect the continuation of the processes started earlier. Indeed, the level of an isocitrate lyase that may be involved in triacylglycerol catabolism increased, whereas a β subunit of the 12S seed-storage protein was completely degraded. Some aspects of metabolic control in quiescent seeds and in young seedling are also highlighted by this study. The absence of S-adenosyl-methionine synthase in the dry seeds, compared with its high level after germination, may explain the metabolic repression that occurs during quiescence. A putative seed-maturation protein (SMP), which was absent during the second day of germination, may modulate the level of biotin, a cofactor of several housekeeping carboxylases, and thus contribute to the metabolic control of seed maturation and germination. The development of defense mechanisms that protect the seedling against herbivores and pathogens was also indirectly observed; a strong increase in levels of a myrosinase and of two jasmonate-inducible myrosinase-binding proteins was detected during radicle emergence. These proteins are involved in hydrolysis of glucosinolates into products that are toxic to herbivores and microorganisms [20]. This work [5] also shows that the germinated seeds prepare for photosynthesis, as illustrated by the accumulation of the chloroplast translation-elongation factor EF-Tu, which reflects the establishment of the plastid translational apparatus needed to build up the photosynthetic system.
These results [5] are in excellent agreement with many previous results obtained over more than a decade from several species, and also reveal new proteins associated with the hydration state of the seeds. Systematic identification of the 1,300 proteins in total that were detected in two-dimensional gel electrophoresis should also yield other important molecular clues to describe germination. The Arabidopsis ecotype used by Gallardo and coworkers [5] is not subject to seed dormancy, which is an important part of the germination control. We are therefore following up their work by analyzing the Arabidopsis ecotype Cap Verde Island (cvi), which has strongly dormant seeds, by analyzing mutants deficient in hormone synthesis or hormone perception, and by treating seeds with hormones during imbibition to identify the proteins involved in germination control (M.J., unpublished observations).
New methods and prospects for the near future
One limit of the proteomic analysis described here [5] is that classic two-dimensional gel electrophoresis does not give access to hydrophobic, highly insoluble or very basic proteins [21]. Moreover, proteins of very low abundance tend to be masked by proteins that are present at several orders of magnitude higher levels; they are thus invisible because of sensitivity limits [22]. Complementary information on the functions involved in germination could therefore be provided by microarray analysis (see, for example, [23]), and the combined use of proteomic and transcriptomic analyses may be powerful for unraveling the complex mechanisms involved in germination. No broad transcriptome analysis of germination has yet been published, however. We are using N. plumbaginifolia, a good physiological model of seed dormancy [7] to characterize by cDNA array several hundred genes that were previously identified by cDNA amplified fragment length polymorphism analysis (cDNA-AFLP) for their differential accumulation between dormant and non-dormant seeds during hydration (our unpublished data). It is interesting to note that 30% of these cDNA sequences can be assigned as homologs of Arabidopsis open reading frames available in public databasesand that among the functions of these, several were also found by Gallardo and coworkers [5]. We will perform functional analysis of these genes in Arabidopsis.
Comparison of proteins and mRNAs identified by proteomics and microarrays with sequence databases may give some working hypotheses about the mechanisms of seed germination, but their biological function in germination remains to be proved. Identifying knockout mutants in the gene of interest, either by PCR screening of collections of Arabidopsis mutagenized by an insertion element (T-DNA or transposon) or by interrogating databases of flanking sequence tags, is a promising approach for meeting this challenge. As most Arabidopsis knockout lines do not look different from the wild type in standard culture conditions [24], a wide range of physiological studies and the introgression of the mutation into a dormant ecotype of Arabidopsis may be needed to find mutants with altered germination phenotypes and to obtain functional clues. The sequencing of the Arabidopsis genome and the development of a large range of high-throughput technologies for assigning a function to a gene make this species the organism of choice for the molecular-genetic dissection of seed germination. The combinations of transcriptome and proteome analysis with reverse genetics will soon provide the means to characterize the regulatory genes in their developmental context.
Acknowledgments
Acknowledgements
We thank Michel Caboche for stimulating discussion and Beatrice Godin, Larissa Neves and Samantha Vernhettes for critical reading of the manuscript.
References
- Bewley JD, Black M. Seeds, germination, structure, and composition. In Seeds: Physiology of Development and Germination New York: Plenum Press; 1994. pp. 1–33.
- Bewley JD. Seed germination and dormancy. Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Bouchez D, Höfte H. Functional genomics in plants. Plant Physiol. 1998;118:725–732. doi: 10.1104/pp.118.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo K, Job C, Groot SPC, Puype M, Demol H, Vandekerckhove J, Job D. Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 2001;126:835–848. doi: 10.1104/pp.126.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obroucheva NV, Antipova OV. Physiology of the initiation of seed germination. Russ J Plant Physiol. 1997;44:250–264. [Google Scholar]
- Grappin P, Bouinot D, Sotta B, Miginiac E, Jullien M. Control of seed dormancy in Nicotiana plumbaginifolia : a post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta. 2000;210:279–285. doi: 10.1007/PL00008135. [DOI] [PubMed] [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO J. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Ried JL, Walker Simmons MK. Synthesis of abscisic acid-responsive, heat-stable proteins in embryonic axes of dormant wheat grain. Plant Physiol. 1990;93:662–667. doi: 10.1104/pp.93.2.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey PC, Lycett GW, Roberts JA. A molecular study of dormancy breaking and germination in seeds of Trollius ledebouri. Plant Mol Biol. 1996;32:559–564. doi: 10.1007/BF00019110. [DOI] [PubMed] [Google Scholar]
- Li B, Foley ME. Cloning and characterization of differentially expressed genes in imbibed dormand and afterripened Avena fatua embryos. Plant Mol Biol. 1995;29:823–831. doi: 10.1007/BF00041171. [DOI] [PubMed] [Google Scholar]
- Goldmark PJ, Curry C, Morris CF, Walker-Simmons MK. Cloning and expression of an embryo-specific mRNA up-regulated in hydrated dormant seeds. Plant Mol Biol. 1992;19:433–441. doi: 10.1007/BF00023391. [DOI] [PubMed] [Google Scholar]
- Stacy RAP, Munthe E, Steinum T, Sharma B, Aalen R. A peroxiredoxin antioxidant is encoded by a dormancy-related gene, Per1, expressed during late development in the aleurone and embryo of barley grains. Plant Mol Biol. 1996;31:1205–1216. doi: 10.1007/BF00040837. [DOI] [PubMed] [Google Scholar]
- Johnson RR, Cranston HJ, Chaverra ME, Dyer WE. Characterization of cDNA clones for differentially expressed genes in embryos of domant and nondormant Avena fatua L. caryopses. Plant Mol Biol. 1995;28:113–122. doi: 10.1007/BF00042043. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Karssen CM. Seed dormancy and germination. In Arabidopsis Edited by Meyerowitz EM, Sommerville CR Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. pp. 313–334.
- Dubreucq B, Grappin P, Caboche M. A new method for the identification and isolation of genes essential for Arabidopsis thaliana seed germination. Mol Gen Genet. 1996;252:42–50. doi: 10.1007/s004389670005. [DOI] [PubMed] [Google Scholar]
- Leung J, Giraudat J. Abscisic acid signal transduction. Ann Rev Plant Physiol Plant Mol Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, van de Bunt GA, Zeevaart JAD, Koornneef M. Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 1996;110:233–240. doi: 10.1104/pp.110.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS. Seed storage proteins: structures and biosynthesis. Plant Cell. 1995;7:945–956. doi: 10.1105/tpc.7.7.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J. Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol. 2000;42:93–113. [PubMed] [Google Scholar]
- Seigneurin-Berny D, Rolland N, Garin J, Jouyard J. Technical advance: Differential extraction of hydrophobic proteins from chloroplast envelope membranes: a subcellular-specific proteomic approach to identify rare intrinsic membrane proteins. Plant J. 1999;19:217–228. doi: 10.1046/j.1365-313X.1999.00511.x. [DOI] [PubMed] [Google Scholar]
- Naaby-Hansen S, Waterfield MD, Cramer R. Proteomics - post-genomic cartography to understand gene function. Trends Pharmacol Sci. 2001;22:376–383. doi: 10.1016/s0165-6147(00)01663-1. [DOI] [PubMed] [Google Scholar]
- CATMA: A Complete Arabidopsis Transcriptome Microarray http://jic-bioinfo.bbsrc.ac.uk/CATMA/
- Bouché N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]


