Abstract
FKBP65 (65-kDa FK506-binding protein) is a member of the highly conserved family of intracellular receptors called immunophilins. All have the property of peptidyl-prolyl cis-trans isomerization, and most have been implicated in folding and trafficking events. In an earlier study, we identified that FKBP65 associates with the extracellular matrix protein tropoelastin during its transport through the cell. In the present study, we have carried out a detailed investigation of the subcellular localization of FKBP65 and its relationship to tropoelastin. Using subcellular fractionation, Triton X-114 phase separation, protease protection assays, and immunofluorescence microscopy (IF), we have identified that FKBP65 is contained within the lumen of the endoplasmic reticulum (ER). Subsequent IF studies colocalized FKBP65 with tropoelastin and showed that the two proteins dissociate before reaching the Golgi apparatus. Immunohistochemical localization of FKBP65 in developing lung showed strong staining of vascular and airway smooth muscle cells. Similar areas stained positive for the presence of elastic fibers in the extracellular matrix. The expression of FKBP65 was investigated during development as tropoelastin is not expressed in adult tissues. Tissue-specific expression of FKBP65 was observed in 12-d old mouse tissues; however, the pattern of expression of FKBP65 was not restricted to those tissues expressing tropoelastin. This suggests that additional ligands for FKBP65 likely exist within the ER. Remarkably, in the adult tissues examined, FKBP65 expression was absent or barely detectable. Taken together, these results support an ER-localized FKBP65-tropoelastin interaction that occurs specifically during growth and development of tissues.
INTRODUCTION
FKBP65 is a member of a group of proteins termed immunophilins. Immunophilins were initially identified by their high binding affinity for the immunosuppressant drugs cyclosporin A (CsA) and FK506, the macrolide antibiotic produced by Streptomyces tsukubaensis. Based on their ability to bind either one of these two drugs, two structurally distinct subfamilies of immunophilins emerged–the cyclophilins (CyPs) and the FK506 binding proteins (FKBPs). The immunosuppressive effects of CsA and FK506 have been studied in detail and attributed to the formation of drug-ligand complexes involving CyPA and FKBP12, respectively. When complexed, these immunophilins bind the phosphatase calcineurin and ultimately prevent its function in T-cell activation (reviewed in Hamilton and Steiner, 1998; Göthel and Marahiel, 1999). Most immunophilins, however, do not appear to be involved in immunosuppressive activities, and relatively little is known concerning their natural ligands.
Members of the immunophilin family have been found in virtually every tissue and every cellular compartment where they exist as membrane-anchored, soluble, and even secreted species (Galat, 1993). Accordingly, although not well defined, immunophilins appear to function in a wide range of cellular activities including such events as folding, assembly, and trafficking of proteins; coregulation of molecular complexes including heat shock proteins, steroid receptors, and ion channels; cell-to-cell interactions; and transcription and translation of genes (reviewed in Galat and Rivière, 1998; Hamilton and Steiner, 1998).
All immunophilins possess the protein folding property of peptidyl-prolyl cis-trans isomerization. Because the endoplasmic reticulum (ER) is a principal site of protein synthesis, folding, and assembly, the presence of immunophilins in the ER is not unexpected. The first two immunophilins to be identified in the ER were CyPB (Price et al., 1991) and FKBP13 (Nigam et al., 1993). In contrast to CyPB, which has been shown to facilitate the folding of procollagen (Smith et al., 1995), no ligand for FKBP13 in the ER has been discovered. More recently, two additional FKBPs, FKBP23 (Nakamura et al., 1998) and FKBP60 (Shadidy et al., 1999), have been identified and localized to the ER. Both of these proteins possess a C-terminal HDEL sequence, which is characteristic of proteins retained in the ER (Pelham, 1990). Similar to FKBP13, no ligands or specific functions for these two FKBPs have been elucidated. Since PPIases catalyze the otherwise slow conversion between cis- and trans-isomers of Xaa-Pro bonds, however, and evidence has been reported that FKBP13 is up-regulated by misfolded proteins in the ER (Bush et al., 1994), it has been presumed that these ER-localized FKBPs function as general molecular chaperones or foldases.
FKBP65 was cloned and sequenced from a JB6 murine epidermal cell cDNA expression library (Coss et al., 1995). Sequence analysis predicted the protein to have four PPIase domains, a highly hydrophobic region at the N-terminus, and a putative ER retention sequence at the C-terminus. Using chemical cross-linking in intact cultured chondroblasts, we previously identified that FKBP65 associates with the extracellular matrix protein tropoelastin during its transport through the cell (Davis et al., 1998). Tropoelastin is a 70-kDa soluble protein that is cross-linked in the presence of extracellular microfibrils to form insoluble elastic fibers. These fibers are an abundant component of the extracellular matrix, where they provide the critical function of elasticity to tissues such as blood vessels and lung (Mecham and Davis, 1994). The tropoelastin monomer remains relatively unchanged as it traverses the secretory pathway en route to the cell surface, with no glycosylation or proteolytic processing. As a result, intracellular folding or maturation of the protein is difficult to determine. Although tropoelastin is not glycosylated, the protein does traverse the Golgi apparatus and appears to exit the cell after transport through an acidic compartment, a rapid process that takes ∼ 30 min from synthesis to secretion (Davis and Mecham, 1996, 1998). Ultrastructural studies have shown an intimate association of newly forming elastic fibers with the cell surface, suggesting that the tropoelastin monomers may be targeted to specific sites for assembly (reviewed in Mecham and Davis, 1994). Despite considerable research, however, the details concerning such a process, the involvement of trafficking molecules, and the final assembly of the monomers into elastic fibers remains poorly understood.
Because tropoelastin is a soluble secreted protein, our data showing an association of FKBP65 with tropoelastin was consistent with a localization of FKBP65 to the secretory pathway. However, Coss and colleagues (1998) reported that FKBP65 forms a heterocomplex with the cytosolic proteins Hsp90 and c-Raf-1 in NIH3T3 cells, and they suggested that FKBP65 may play a role in signal transduction processes. Clearly, the function of FKBP65 and the interpretation of experimental data concerning this protein will not be fully understood without better characterization of its subcellular localization.
In the present study, we have used biochemical assays and immunofluorescence microscopy to demonstrate that FKBP65 is indeed a resident protein of the ER lumen. Additionally, we have shown that FKBP65 is localized to smooth muscle cells (SMCs) in the developing lung–cells that synthesize and assemble elastic fibers in the extracellular matrix. Because tropoelastin is produced only in developing tissues, the expression of FKBP65 during development was also investigated. Remarkably, Northern analysis of FKBP65 showed a pattern of developmental regulation similar to that of tropoelastin, with virtually no expression seen in adult tissues.
MATERIALS AND METHODS
Cells and Reagents
Fetal bovine chondroblasts (FBCs) were obtained from 150–160-d fetal bovine auricular cartilage by collagenase digestion as previously described (Mecham, 1987). Cells were grown in DMEM supplemented with L-glutamine, nonessential amino acids, antibiotics, and 10% fortified fetal calf serum (Hyclone Laboratories, Inc., Logan, UT). All experiments in this report were conducted with first- or second-passage cells. Protease inhibitors included ε-amino-n-caproic acid, phenylmethylsulfonyl fluoride (PMSF), ethylenediamine-tetraacetic acid, and N-ethylmaleimide purchased from Sigma Chemical Company (St. Louis, MO) and used in lysis buffers at final concentrations of 10 mM, 2.5 mM, 5 mM, and 5 mM, respectively. Secretion-disrupting agents used before immunofluorescence labeling included brefeldin A (BFA) and monensin (Sigma). BFA was stored at −20°C as 10 mg/ml stocks in DMSO and used at a final concentration of 10 μg/ml. Monensin was prepared fresh as a 10 mM stock in ethanol and used at a final concentration of 10 μM. To inhibit ER-associated degradation, N-acetyl-leu-leu-norleucinal (ALLN; Sigma) was used at final concentration of 10 μg/ml from a 10 mg/ml stock in ethanol stored at −20°C.
Antibodies and cDNA Probes
Primary antibodies used for Western analysis and immunofluorescence included the following: a monoclonal tropoelastin antibody, BA-4, raised to bovine α-elastin (Wrenn et al., 1986); a polyclonal FKBP65 antipeptide antibody raised against a synthetic peptide to the C-terminus of mouse FKBP65 (Davis et al., 1998); a commercial monoclonal FKBP65 antibody (Transduction Laboratories, Lexington, KY); a commercial anti-BiP antibody (Affinity Bioreagents, Golden, CO); and polyclonal antibodies to calnexin (C-terminus), ribophorin I, Grp94, and Hsp90 (generous gifts from Dr. Williams, University of Toronto; Dr. David Meyer, UCLA; Dr. Michael Green, St. Louis University; and Dr. David Toft, Mayo Clinic, respectively). Secondary antibodies used for ECL detection were sheep antimouse and donkey antirabbit HRP-conjugated F(ab′)2 (Amersham Life Science, Arlington Heights, IL). For immunofluorescence, goat antimouse fluorescein-conjugated IgG (ICN, Cappel Research Reagents, Costa Mesa, CA) was used to detect tropoelastin and goat antirabbit fluorescein-conjugated IgG was used to detect FKBP65 and Grp94. For double labeling, goat antimouse fluorescein-conjugated IgG and goat antirabbit rhodamine-conjugated IgG were used concurrently. Probes used for Northern analysis included a 2.3-kb bovine tropoelastin cDNA probe (Parks et al., 1988), a 2.8-kb Grp94 cDNA probe (generous gift from Dr. Michael Green, Saint Louis University), and a 1.7-kb full-length FKBP65 cDNA probe. The FKBP65 probe was constructed using a mouse EST clone (GenBank Acc: W89347) and RT-PCR to obtain the 5′ end of the coding region.
Subcellular Fractionation
Confluent cultures of FBCs or NIH3T3 cells (six 300-mm dishes) were washed with ice cold PBS and scraped as sheets of cells into a total volume of 6 ml PBS. After pelleting the cells at 1000 rpm for 10 min, the PBS was removed, and the cells were gently resuspended in 3 ml of ice-cold homogenization buffer [HB: 250 mM sucrose, 50 mM triethanolamine, 50 mM potassium acetate, 6 mM magnesium acetate, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF]. The cells were then placed in a nitrogen cavitation bomb (Knotes Glass Company, Millville, NJ), pressurized to 60 psi with N2, and held under pressure for 15 min on ice. The cavitated cells were collected, and unbroken cells and nuclei were pelleted by centrifugation at 1000 rpm for 10 min at 4°C. The supernatant from this spin was centrifuged at 68,000 rpm for 2 h at 4°C to obtain cytosol and a pellet of undisrupted intracellular membranes. For additional experiments where only intracellular membranes were needed, the unbroken cavitated cells (1000 rpm pellet) were resuspended in 5 ml of ice cold HB and homogenized in a glass dounce homogenizer. The resulting cell homogenate was centrifuged at 1000 rpm, the pellet discarded, and the supernatant centrifuged at 68,000 rpm as described above to obtain a final pellet of intracellular membranes.
To analyze proteins in the cytosol versus intracellular membrane fractions, proteins in the membrane fraction were solubilized in lysis buffer [25 mM Tris-HCl (pH 7.5), 5 mM EDTA (pH 7.5), 250 mM NaCl, 1% Triton X-100] with protease inhibitors, and both fractions were precipitated with −20°C acetone. Precipitated proteins were resuspended in 100 μl Laemmli sample buffer containing 100 mM DTT, incubated at 100°C for 6 min, and 20 μl of each sample were electrophoresed on SDS-polyacrylamide gels. Following electrophoresis, samples were transferred to nitrocellulose, and nonspecific binding sites were blocked with 5% dry milk in TBS [20 mM Tris base, 137 mM NaCl (pH 7.6)] containing 0.1% Tween-20 overnight at 4°C. All remaining rinses, washes, and antibody dilutions were in 2% dry milk, 0.05% Tween-20 in TBS. The next day, the blot was rinsed and incubated in primary antibody for 1 h, washed several times, and then incubated with HRP-conjugated secondary antibody for 1 h. After washing, and final rinse in TBS, the blot was incubated with ECL detection reagents (Pierce, Rockford, IL) according to manufacturers directions.
Triton X-114 Phase Separation
Triton X-114 extraction was carried out on proteins contained in the intracellular membrane pellet as described (Bordier, 1981). Before phase separation, one third of the lysate was removed for a control (aqueous + detergent). Specific proteins in the control sample and the separated aqueous and detergent phases were immunoprecipitated as previously described (Davis et al., 1998), or total proteins were precipitated with −20°C acetone. After resuspension in Laemmli sample buffer containing 100 mM DTT and incubation at 100°C for 6 min, equal volumes of each sample were electrophoresed on SDS-polyacrylamide gels, transferred to nitrocellulose, and immunoblotted as described above.
Protease Protection Assay
Membranes obtained by subcellular fractionation, as described above, were resuspended in 1 ml reaction buffer [10 mM Tris-HCl (pH 7.8), 150 mM KCl, 2 mM MgCl2, 2 mMCaCl2, and 200 mM sucrose]. Three 300 μl aliquots of suspended membranes were incubated with either nothing added, with 100 μg/ml proteinase K (Sigma), or with 100 μg/ml proteinase K + 1% Triton X-100. After 1 h at 4°C, the reaction was stopped by the addition of 10 mM PMSF. Triton X-100 was added to each sample to give a final concentration of 1% before acetone precipitation, SDS-PAGE, and immunoblotting as described above.
Immunofluorescence
Subconfluent cultures of FBCs in 4-well LabTek Chamber slides (Nunc no. 177437, Fisher) were left untreated, treated with 10 μg/ml BFA + 10 μM ALLN, or treated with 10 μM monensin for 3 h. Cell layers were then washed with PBS and fixed with 2% paraformaldehyde in PBS for 30 min. Following several washes in PBS and quenching in 50 mM NH4Cl for 30 min, cell layers were rinsed in three changes of PBS containing 1% BSA and 0.1% saponin, then further permeabilized for 30 min in the same buffer. All antibody dilutions and subsequent washes were with permeabilization buffer. The permeabilized cells were incubated with primary antibody for 1 h at room temperature. For double labeling, cells were incubated concurrently with the two primary antibodies. After four washings of 5 min each, the cells were incubated with fluorescent-labeled secondary antibody diluted 1:200 for 1 h. Cell layers were then washed 4 times, for 5 min each, then rinsed once with PBS, before mounting in 80% glycerol in PBS containing 1 mg/ml p-phenylenediamine and visualized with a Leica confocal microscope.
RNA Isolation and Northern Analysis
Mouse tissues were dissected from 14.5-d embryos, 12-d postnatal and adult animals, and directly frozen in liquid nitrogen. Frozen tissues were homogenized in Trizol reagent (GIBCO BRL, Gaitherburg, MD), total RNA isolated according to manufacturers directions, and 10 μg separated by electrophoresis through a 1% agarose gel containing 1 M formaldehyde. RNA was transferred overnight to Hybond-N+ nylon membrane (Amersham, Arlington Heights, IL) and UV cross-linked using a Spectrolinker XL-100 UV Cross-linker (Spectronics Corporation, Lincoln, NE). cDNA probes were labeled with [32P]dCTP using the Rediprime II random prime labeling system (Amersham), and unincorporated nucleotides were removed using the QIAquick Nucleotide Removal kit (Qiagen, Valencia, CA). Membranes were hybridized overnight at 65°C using 250 mM Na2PO4 and 7% SDS, or they were hybridized for 3 h at 65°C using Rapid-hyb buffer (Amersham). Membranes were then washed once in 2X SSPE, 0.1% SDS for 15 min at room temperature, followed by 2 washes in 1X SSPE, 0.1% SDS for 15 min at the hybridization temperature. Hybridized complexes were detected by exposure of the membrane to X-OMAT AR film (Kodak, Rochester, NY). For reprobing, membranes were stripped using boiling 0.5% SDS.
Immunohistochemistry
Formalin-fixed sections of 200-d gestation fetal bovine lung were deparaffinized and hydrated through xylene and a graded series of ethanol. Sections were then incubated in 0.3% Triton X-100 in TBS [10 mM Tris, 150 mM NaCl (pH 7.4)] for 15 min, rinsed in TBS followed by absolute MeOH for 2 min each, and then quenched of endogenous peroxidase activity in 0.6% H2O2 in absolute MeOH for 1 h. After rinsing in absolute MeOH for 2 min, the sections were washed three times in 0.1% BSA in TBS for 5 min each time. Sections were then incubated in a blocking solution consisting of 0.5% BSA in TBS, with normal blocking serum from the Vectastain Elite ABC Kit (Vector Laboratories, Inc., Burlingame, CA) added according to kit instructions. After 30 min, excess blocking solution was drained from the sections, and the sections were incubated in primary antibody diluted in blocking solution overnight in a humidity chamber at 4°C. The following day, the sections were washed in 0.1% BSA in TBS, incubated in diluted biotinylated secondary antibody for 1 h at room temperature, washed again, and incubated in ABC reagent for 1 h. The biotinylated secondary antibody and ABC reagent were contained in the Vectastain kit and were prepared according to kit instructions. After washing, the sections were incubated in Vector NovaRED substrate, counterstained with Vector hematoxylin, and mounted using VectaMount (Vector Laboratories, Inc.). Sections were viewed with a Zeiss Axioskop 2 microscope and images captured with an AxioCam digital camera using AxioVision software (Carl Zeiss, Inc., Thornwood, NY).
RESULTS
FKBP65 Is Found Exclusively in the Membrane Fraction
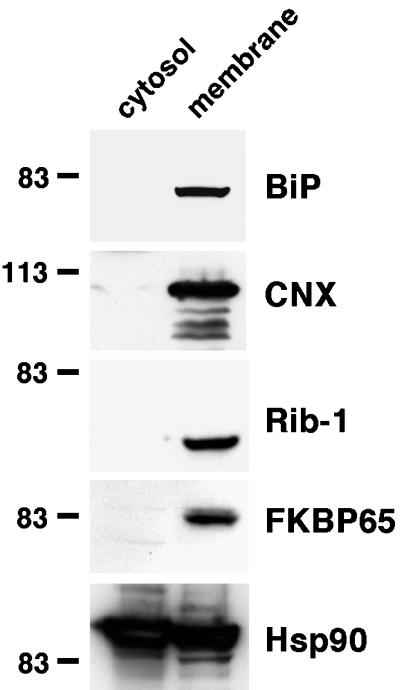
To determine the intracellular distribution of FKBP65, it was important to maintain complete integrity of the intracellular membranous compartments. NIH3T3 cells disrupted by nitrogen cavitation were therefore subjected to differential centrifugation without any prior homogenization. Following a low-speed spin to remove nuclei and unbroken cells, the supernatant was fractionated into cytosolic (200,000g supernatant) and membranous (200,000g pellet) fractions. Western blot analysis of the proteins in each fraction showed the presence of FKBP65 only in the membranous fraction (Figure 1). The presence of intracellular membranes in this fraction was demonstrated by positive immunoreactivity for the ER membrane proteins, calnexin and ribophorin-I. Localization of BiP, a soluble ER lumenal protein, entirely in the membrane fraction indicated the lack of any protein leakage from intravesicular compartments during cell disruption. Hsp90, a cytosolic protein that is known to associate with intracellular membranes, was found in both fractions.
Figure 1.
Cytosol/membrane fractionation. FKBP65 is contained entirely within the membrane fraction. NIH3T3 cells were disrupted by nitrogen cavitation, unbroken cells were pelleted, and the resulting supernatant separated into cytosol and membrane fractions by a 200,000g spin. The membrane fraction was solubilized in Triton X-100, then proteins in both fractions were acetone precipitated. Pellets were dissolved in sample buffer, resolved by SDS-PAGE, and Western blotted. CNX, calnexin; Rib-1, ribophorin-1.
FKBP65 Is Contained Within the Lumen of an Intracellular Membranous Compartment
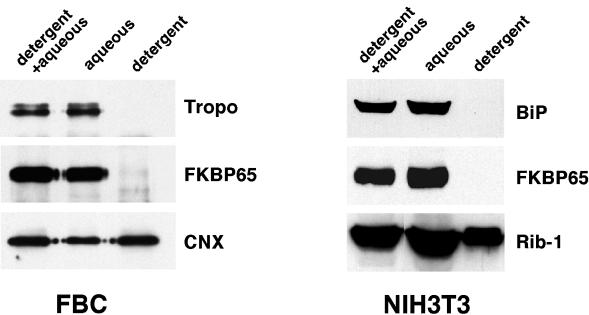
Molecular cloning of FKBP65 identified an N-terminal hydrophobic region indicative of a signal sequence and a short stretch of hydrophobic residues (VIPPQASLVFYVLLL) in the second PPIase domain that was suggested to be a potential membrane-spanning domain (Coss et al., 1995). To investigate the hydrophobic nature of FKBP65, Triton X-114 was used to solubilize FBCs and NIH3T3 cells, and the solubilized material was subjected to phase separation. In both FBCs and NIH3T3 cells, FKBP65 partitioned entirely into the aqueous phase indicating the hydrophilicity of this protein (Figure 2). This result was consistent with the partitioning of tropoelastin and BiP, both of which are soluble proteins contained within membranous compartments of the secretory pathway. In contrast, considerable proportions of the integral membrane proteins, calnexin and ribophorin-I, were found in the detergent phase. Partitioning of an integral membrane protein into the aqueous phase, as seen here, has been shown to be caused by residual Triton X-114 remaining in the aqueous phase (Pryde, 1986).
Figure 2.
Triton X-114 phase separation. FKBP65 does not have a hydrophobic domain indicative of a transmembrane protein. NIH3T3 cells and FBCs were solubilized in Triton X-114, warmed to 30°C, and separated into detergent and aqueous phases by centrifugation. The two phases were collected and either immunoprecipitated (FBCs) or acetone precipitated (NIH3T3 cells). Samples were resolved by SDS-PAGE and Western blotted. A sample from the original solubilization (detergent + aqueous) was also analyzed. Tropo, tropoelastin; CNX, calnexin; Rib-1, ribophorin-1.
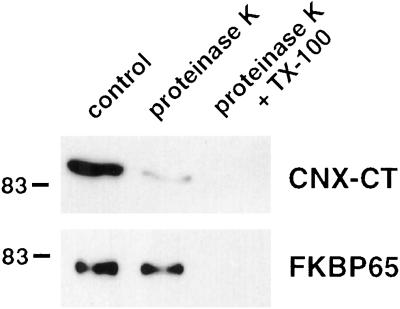
To confirm that FKBP65 is not a membrane-spanning protein, membranes obtained from NIH3T3 cells after nitrogen cavitation and differential centrifugation (200,000g pellet) were subjected to a protease protection assay. Figure 3 shows that the cytosolically exposed C-terminus of calnexin was almost completely digested by proteinase K, whereas FKBP65 was unaffected by the enzyme. Because the putative membrane-spanning domain in FKBP65 was reported to be centrally located in the molecule, it is unlikely that our antibody recognized a nearly full-length, protected fragment. Taken together, the Triton X-114 and proteinase K experiments demonstrate that FKBP65 is an intravesicular protein.
Figure 3.
Protease protection assay. FKBP65 is entirely intravesicular. The 200,000g microsomal pellet from NIH3T3 cells was treated with buffer alone (control), 100 μg/ml proteinase K alone, or 100 μg/ml proteinase K + 1% Triton X-100. After 30 min at room temperature, excess PMSF was added to each sample to inhibit the enzyme, and the membranes in all samples were solubilized with Triton X-100. Proteins were acetone precipitated and Western blotted. CNX-CT = calnexin C-terminal antibody.
FKBP65 is an ER Resident Protein that Dissociates from its Cargo before the Cis-Face of the Golgi Apparatus
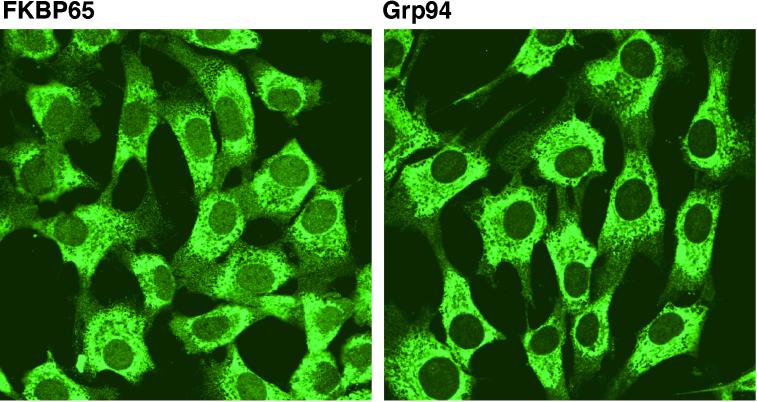
Results from the cell fractionation studies showed that FKBP65 is located within the vesicular compartments of the cell. To obtain more precise information concerning the subcellular location of this protein, immunofluorescence staining was carried out on permeabilized FBCs. In all cells, a perinuclear reticular pattern was observed with little or no staining in the cell periphery (Figure 4). In many cells, a region devoid of staining could be observed at one pole of the nucleus typical of the site occupied by the Golgi apparatus (see Figure 5B). This pattern of staining is characteristic of an ER-localized protein as demonstrated by the identical immunofluorescence pattern for Grp94, a resident protein of the ER (Figure 4).
Figure 4.
Indirect immunofluorescence labeling of FKBP65 is identical to that of Grp94. FBCs were fixed with 2% paraformaldehyde, permeabilized with 0.1% saponin in PBS containing 1% BSA, and immunolabeled for either FKBP65 or Grp94. Both antibodies show the same ER pattern of staining, and in many cells a negatively stained area representing the perinuclear Golgi cap can be seen.
Figure 5.
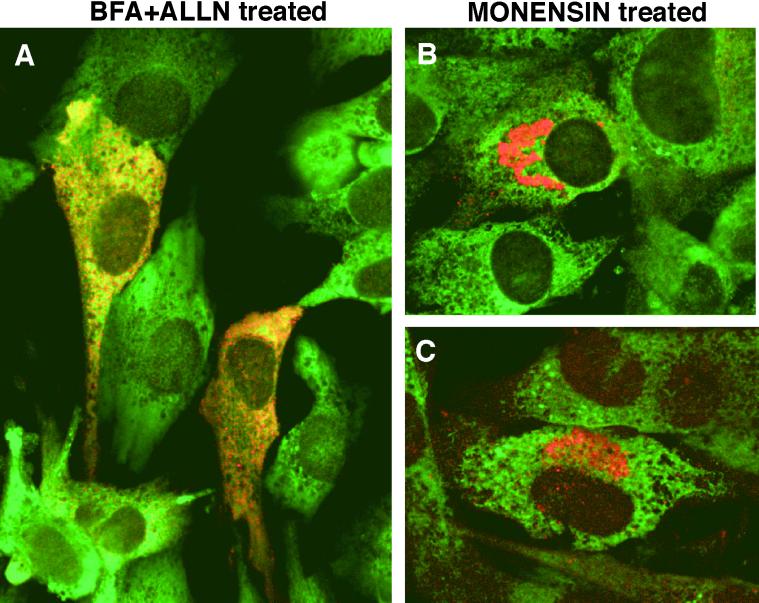
FKBP65 is a resident protein of the ER. Subconfluent cultures of FBCs were treated for 3 h with either (A) 10 μg/ml brefeldin A + 10 μg/ml ALLN to “hold” the tropoelastin in the fused ER/Golgi compartment (ALLN is needed to prevent tropoelastin degradation), or (B, C) 10 μM monensin to “hold” the tropoelastin in the cis-Golgi cisternae. Cells were then fixed, permeabilized, and double labeled with a monoclonal antibody to tropoelastin (red) and a polyclonal antibody to either (A, B) FKBP65 (green) or (C) Grp94 (green). After treatment with BFA+ALLN, tropoelastin and FKBP65 were colocalized in the fused ER/Golgi apparatus. When tropoelastin was accumulated in the Golgi, however, the two proteins showed distinct patterns of immunolabeling with tropoelastin in the peri-nuclear Golgi region and FKBP65 remaining in the ER. The same distribution of labeling was seen when Grp94 was colocalized with tropoelastin after monensin treatment. Note that the production of tropoelastin by FBCs in culture is density-dependent, thus not all the cells are synthesizing tropoelastin at subconfluency.
Although the amino acid sequence of FKBP65 predicts a putative ER-retention sequence (HEEL) at the C-terminus (Pelham, 1990), proteins with such sequences have been reported to exit the ER and traverse the secretory pathway to ultimately gain access to the cell surface (Colley et al., 1991; Krause and Michalak, 1997). To investigate the extent to which FKBP65 may traffic through the secretory pathway with tropoelastin, colocalization of FKBP65 and tropoelastin was conducted on FBCs treated with secretion disrupting drugs. When FBCs were treated with BFA to keep newly synthesized tropoelastin in the fused ER/Golgi compartment, colocalization of FKBP65 and tropoelastin was observed (Figure 5A). The cysteine protease inhibitor, ALLN, was included in the treatment because we have previously shown that tropoelastin undergoes ER-associated degradation if retained in the ER under these conditions (Davis and Mecham, 1996). In contrast, cells treated with monensin to block the transport of tropoelastin at the cis-side of the Golgi apparatus showed distinct patterns of localization for the two proteins (Figure 5B). As expected, strong immunofluorescence staining for tropoelastin was seen in a juxtanuclear Golgi cap, whereas the reticular ER pattern of FKBP65 staining remained undisturbed. An identical immunofluorescence pattern was obtained with monensin-treated cells double-labeled for tropoelastin and Grp94 (Figure 5C). These results indicate that FKBP65 dissociates from tropoelastin before reaching the cis-face of the Golgi apparatus and remains ER-localized.
Localization of FKBP65 in Developing Lung Correlates with Sites of Tropoelastin Deposition
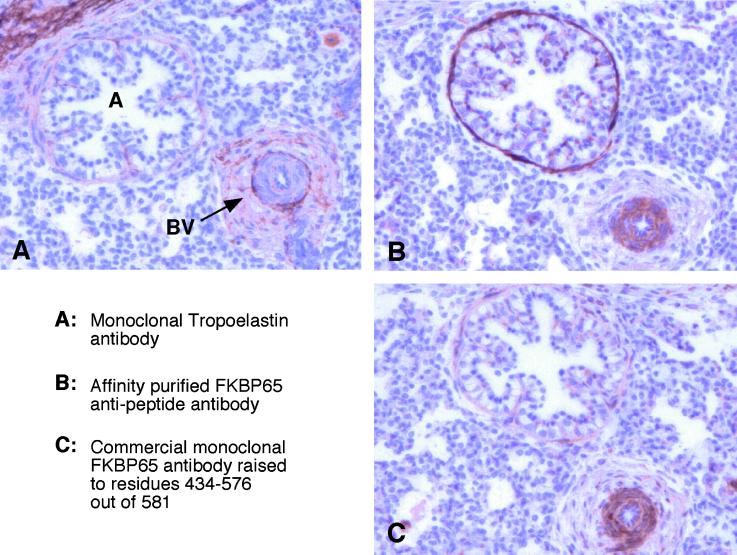
In the developing lung, tropoelastin is synthesized by SMCs that underlie airways and by endothelial cells, medial SMCs, and adventitial fibroblasts of the pulmonary vessels. Because FKBP65 associates with tropoelastin in the ER, FKBP65 should be present in those cells that produce tropoelastin. To investigate the localization of FKBP65 with respect to tropoelastin, immunostaining of the two proteins was carried out on serial sections of paraffin-embedded fetal bovine lung. Tropoelastin staining was found in the regions of the lung containing airways and blood vessels (Figure 6A). The extracellular elastic fibers that are assembled from the secreted monomer react strongly with the antielastin antibody, while the cells synthesizing tropoelastin stain only weakly, as they contain relatively little tropoelastin at any one time. Consistent with the data that FKBP65 associates with tropoelastin, strong staining of airway SMCs and cells of the pulmonary vessels was found (Figure 6B). Immunoreactivity for FKBP65 could also be seen in the epithelial cells of the airway. The localization of FKBP65 to these cells was confirmed using a commercial FKBP65 antibody raised to a different region of the protein (Figure 6C).
Figure 6.
FKBP65 and tropoelastin colocalize in developing lung. Immunostaining for FKBP65 and tropoelastin on serial sections of paraffin-embedded fetal bovine lung. (A) Tropoelastin staining, seen surrounding an airway and small blood vessel, represents elastic fibers in the extracellular matrix. (B, C) The SMCs that synthesize the tropoelastin to form these fibers stain strongly for FKBP65. Note that the blood vessel stains with equal intensity with both FKBP65 antibodies, but stronger staining of the airway SMCs is seen with the antipeptide FKBP65 antibody than with the commercial antibody. A, airway; BV, blood vessel.
Northern Analysis of FKBP65 Indicates Tissue Specificity and Developmental Regulation
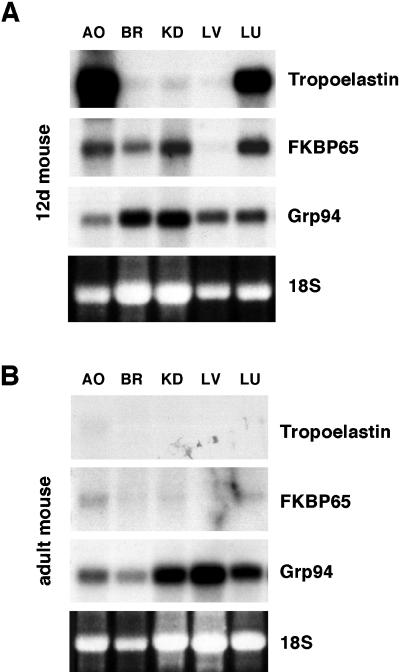
To date, tropoelastin has been the only ligand identified for an FKBP in the secretory pathway. Tropoelastin expression is tissue-specific, being present at very high levels only in those tissues that undergo stretch and recoil, such as the great vessels and the lungs. Because of this distinct tissue expression, the degree of specificity of FKBP65 as a molecular chaperone and/or foldase for tropoelastin was investigated by Northern analysis. Five tissues were removed from 12-d postnatal mice and probed for tropoelastin and FKBP65. As expected, tropoelastin and FKBP65 were both expressed in the developing aorta and lungs (Figure 7A). However, FKBP65 was also expressed in brain and kidney, two tissues that do not express tropoelastin. This result suggests that additional ligands for FKBP65 likely exist within the ER. A second observation was that FKBP65 was not ubiquitously expressed because no expression was seen in liver. In contrast, Grp94 a well-characterized ER chaperone, was expressed at various levels in all five developing tissues (Figure 7A).
Figure 7.
FKBP65 is developmentally regulated. (A) Northern analysis of tissues from 12-d postnatal mice shows no expression of FKBP65 in liver, whereas elevated-to-moderate levels of expression are seen in aorta, brain, kidney, and lung. Expression of Grp94 is seen in all five tissues. (B) Northern analysis of the same tissues from adult mice shows developmental down-regulation of tropoelastin. FKBP65 expression was also found to be absent, or barely detectable, in all tissues examined. In contrast, Grp94 is constituitively expressed. Adult blots were exposed relative to a control lane of 12-d postnatal mouse lung RNA (control lane not shown). AO, aorta; BR, brain; KD, kidney; LV, liver; LU, lung.
Unlike most other components of the extracellular matrix, elastic fibers are formed only during development, with little or no synthesis in adult tissues (Davis, 1993). For this reason, it was of interest to investigate expression levels for FKBP65 in adult tissues as tropoelastin expression would be absent at this time. Consistent with published data, no tropoelastin expression was detected in the adult tissues (Figure 7B). Remarkably, the expression of FKBP65 was also found to be absent, or barely detectable, in all five tissues. Unlike FKBP65, Grp94 continued to be expressed and was found in all of the adult tissues examined (Figure 7B).
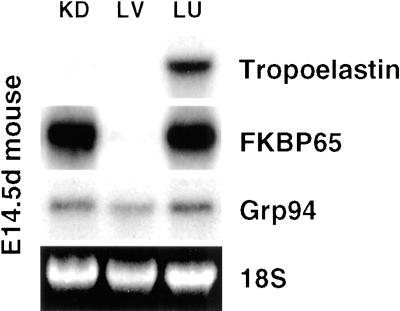
The expression of FKBP65 in developing tissues and its absence in adult tissues suggests that FKBP65 may play a role in organogenesis. It is possible, therefore, that FKBP65 may be expressed at an earlier time point in the liver, since this organ is one of the earliest to develop. FKBP65 expression was investigated in liver from mouse embryos at 14.5 d of gestation. In contrast to embryonic kidney and lung, where message levels for FKBP65 were very high, FKBP65 expression in the liver was still absent (Figure 8).
Figure 8.
FKBP65 is not expressed in liver. Northern analysis of kidney, liver, and lung from 14.5-d mouse embryos shows strong expression FKBP65 in kidney and lung but no expression in liver. KD, kidney; LV, liver; LU, lung.
DISCUSSION
In the present study, we have identified FKBP65 as an ER-localized PPIase. This result is consistent with the molecular characteristics of the protein, which include an N-terminal signal sequence, tunicamycin and Endo-F sensitive glycosylation, and a C-terminal ER-retention motif (Coss et al., 1995). However, our ER-localization of FKBP65 conflicts with data showing an association of FKBP65 with c-Raf-1 and Hsp90 in the cytosol of NIH3T3 cells (Coss et al., 1998). Such a discrepancy has also been reported for FKBP13. Immunolocalization of FKBP13 following subcellular fractionation and immunofluorescence labeling of cells overexpressing FKBP13 have both shown the protein to be contained in the ER (Nigam et al., 1993). FKBP13 was also reported to be up-regulated by conditions that cause an ER accumulation of misfolded proteins (Bush et al., 1994), a response typical of resident ER proteins like BiP and Grp94 (Chapman et al., 1998; Kaufman, 1999). More recently, however, FKBP13 was shown to be a component of the red blood cell membrane cytoskeleton (Walensky et al., 1998). Although red blood cells are specialized cells that are devoid of organelles, this result demonstrated that FKBP13 can reside outside the ER. The possibility exists, therefore, that alternative processing of the N-terminal signal sequences of FKBP13 and FKBP65 may allow for a small percentage of these proteins to reside in the cytosol.
As a site of protein synthesis, the ER is loaded with unfolded proteins, all with the potential to aggregate and/or misfold. To alleviate this problem, the ER contains a multitude of molecular chaperones, which reduce the risk of misfolding and aggregation by associating with proteins in their unfolded state, and foldases, which catalyze folding reactions (Gilbert, 1994). PPIases are foldases in that they catalyze the otherwise slow isomerization of peptidyl-prolyl bonds. Although such an activity has yet to be shown for FKBP65 on tropoelastin in the ER, the amino acid sequence of tropoelastin strongly predicts that proline isomerization would be of great importance for the final conformation of the monomer. As illustrated in Figure 9, 12% of the residues in tropoelastin are proline. These prolines are located in both repeating domains, such as the pentapeptide PGVGV, which repeats 11 times in bovine tropoelastin (Indik et al., 1990), and in cross-linking domains, which are critical for linking the tropoelastin monomers together to form insoluble elastic fibers (Mecham and Davis, 1994). Interestingly, all of the cross-linking domains either contain proline residues or are immediately or closely preceded by a proline (Figure 9). Based on the location of these residues, it is clear that cis-trans isomerization of the Xaa-Pro bonds would have considerable influence over the ultimate structure of this molecule.
Figure 9.
Amino acid sequence of bovine tropoelastin (Indik et al., 1990). Proline residues make up ∼12% of the tropoelastin molecule. Prolines appear in repeating sequences (see line 11, dotted underline) and preceding cross-linking domains (double underlines). The sequence in this figure is arranged with the cross-linking domains and preceding proline aligned to the left.
In addition to acting as a foldase, FKBP65 may also act as a molecular chaperone by preventing tropoelastin aggregation before transport of the monomer from the ER to the Golgi apparatus. Under physiological conditions, tropoelastin has an interesting property in that it can undergo a concentration-dependant phase transition to form a coacervate (Cox et al., 1974). Although this property has been suggested to be important for the interaction of tropoelastin monomers during the assembly of elastic fibers in the extracellular matrix (Wu et al., 1999), such a property would be potentially lethal if it occurred inside the cell. Previously, we have shown the tropoelastin undergoes selective degradation if retained in the ER (Davis and Mecham, 1996). Based on this result and the physical properties of tropoelastin, the necessity for timely folding and transport of tropoelastin out of the ER in cells that produce massive amounts of this protein is clear.
Although little is known concerning the biological function and substrate specificity of the FKBPs in the ER, precedence for chaperone and/or foldase activity of these proteins has come from work on two other ER-localized immunophilins; ninaA and CyPB. The Drosophila ninaA gene encodes a CyP homologue that is specific for photoreceptor cells (Schneuwly et al., 1989; Shieh et al., 1989). Despite an ER-retention sequence, ninaA colocalizes with its substrate, Rh1 rhodopsin, throughout the secretory pathway, and mutations in the ninaA gene cause an ER accumulation of Rh1 and reduced transport of the protein to the cell surface (Colley et al., 1991). These results support an intracellular transport or chaperone function for ninaA. In contrast, CyPB remains within the ER. Together with the molecular chaperone Hsp47, CyPB facilitates procollagen I folding and exit from the ER (Smith et al., 1995). The amino acid sequence of collagen is extremely regular, with every third residue being glycine, and an abundance of prolines, frequently in the triplet Gly-Pro-Xaa. Remarkably, the triple helical structure of procollagen can accommodate only peptide bonds that are in trans-conformation. Experimental evidence that cis-trans isomerization of peptidyl-prolyl bonds in procollagen is a significant factor in the secretion and assembly of collagen has been shown by treating cells with CsA. As would be predicted, the addition of CsA to cells in culture caused a reduced rate of procollagen folding, delayed secretion, increased intracellular degradation, and a decrease in the rate of Hsp47 release from the molecule (Steinmann et al., 1991; Smith et al., 1995). CsA has also been shown to slow the initial steps of transferrin folding within the ER, thereby delaying its maturation and secretion from the cell (Lodish and Kong, 1991). This observation suggests that the folding of transferrin is also catalyzed by a CyP. With similar reasoning, it will now be of interest to examine the effects of FK506 on the synthesis and secretion of tropoelastin.
Molecular chaperones and foldases that reside in the ER are generally thought to be ubiquitously expressed and present at all stages of development. It was surprising, therefore, to find virtually no expression of FKBP65 in the adult tissues examined. This result suggests that the function of FKBP65 is not necessary in the adult and is specific to developing tissues. Although such a feature is not typical of resident ER proteins, increasing evidence suggests that this may be the case for the immunophilin family of proteins. Perhaps the strongest support against a generalized function for these proteins has come from work on yeast immunophilins. To investigate the functions of CyPs and FKBPs, Dolinski and colleagues (1997) produced null mutant strains lacking each of the eight CyPs and four FKBPs. When none of the 12 yeast immunophilins were individually found to be necessary for viability, various yeast strains were engineered with multiple immunophilins deleted to test for functional overlap. Remarkably, all of the multiple mutant strains were viable, including strains lacking all of the CyPs, all of the FKBPs, and even all 12 of the yeast immunophilins (Dolinski et al., 1997). Results from this work allowed for the conclusion that each immunophilin likely performs a specialized function through interactions with a unique and restricted set of target proteins.
Another observation from the present study is that FKBP65 is expressed at high levels in some developing tissues that do not significantly express tropoelastin. This result suggests that additional ligands for FKBP65 exist in the ER apart from tropoelastin. In an investigation of collagen-binding proteins, Zeng and coworkers (1998) found that FKBP65 eluded from a gelatin-sepharose column following incubation of the column with a chick embryo protein extract. The chicken FKBP65 did not bind to the column once purified, however, indicating that FKBP65 was likely bound indirectly as part of a protein complex. The proteins linking FKBP65 to the column were not identified. FKBP65 can also catalyze the folding of type III collagen in vitro (Zeng et al., 1998). Evidence supporting the activity of FKBPs on the folding of collagen in vivo, however, is not strong. In cell culture studies, only a slight effect of FK506 on the rate of collagen folding was found, whereas CsA greatly inhibited the process (Bächinger et al., 1993; Ikeda and Fujiwara, 1995). Interestingly, in the present study, immunohistochemical localization of FKBP65 in developing lung showed comparable staining of vascular SMCs using two different FKBP65 antibodies, but weaker staining of the airway SMCs with the commercial antibody compared with our antipeptide antibody (see Figure 6B and C). This observation suggests that the C-terminal epitope recognized by our polyclonal antibody is equally accessible in both smooth muscle cell types, whereas the epitope recognized by the commercial antibody, which includes the majority of the fourth PPIase domain, is not as readily accessible in airway SMCs as it is in vascular SMCs. With the evidence presented in this study that additional ligands for FKBP65 likely exist, it is possible that the accessibility of the PPIase epitope could differ based on the specific protein ligands produced in the different cell types. Furthermore, temporal expression of a single ligand may differ in different regions of the tissue and thus account for the altered accessibility of the epitope. Further studies to identify the specific set of ligands recognized by FKBP65 will aid in the understanding of this observation.
In summary, FKBPs are becoming a significant subset of resident ER proteins. Unlike many of the molecular chaperones and foldases in the ER, these proteins appear unlikely to function in a generalized global manner. In the present study, we have identified FKBP65 as one of the ER-localized PPIases and have provided evidence for developmental regulation of this protein. The localization and expression of FKBP65 in lung is consistent with that of tropoelastin, thus providing additional evidence for a functional link between these two proteins.
ACKNOWLEDGMENTS
The authors thank Drs. David Williams (University of Toronto), David Meyer (UCLA), Michael Green (St. Louis University), and David Toft (Mayo Clinic) for their generous gifts of antibodies, Dr. William Snell for his critical reading of this manuscript, and Kevin Kennon for administrative assistance. This work was supported by National Institutes of Health grant HL-60394 to E.C. Davis.
REFERENCES
- Bächinger HP, Morris NP, Davis JM. Thermal stability and folding of the collagen triple helix and the effects of mutations in osteogenesis imperfecta on the triple helix of type I collagen. Am J Med Genet. 1993;45:152–162. doi: 10.1002/ajmg.1320450204. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Bush KT, Hendrickson BA, Nigam SK. Induction of the FK506-binding protein, FKBP13, under conditions which misfold proteins in the endoplasmic reticulum. Biochem J. 1994;303:705–708. doi: 10.1042/bj3030705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Colley NJ, Baker EK, Stamnes MA, Zuker CS. The cyclophilin homolog ninaA is required in the secretory pathway. Cell. 1991;67:255–263. doi: 10.1016/0092-8674(91)90177-z. [DOI] [PubMed] [Google Scholar]
- Coss MC, Stephens RM, Morrison DK, Winterstein D, Smith LM, Simek SL. The immunophilin FKBP65 forms an association with the serine/threonine kinase c-Raf-1. Cell Growth Differ. 1998;9:41–48. [PubMed] [Google Scholar]
- Coss MC, Winterstein D, Sowder RC, Simek SL. Molecular cloning, DNA sequence analysis, and Biochemical characterization of a novel 65-kDa FK506-binding protein (FKBP65) J Biol Chem. 1995;270:29336–29341. doi: 10.1074/jbc.270.49.29336. [DOI] [PubMed] [Google Scholar]
- Cox BA, Starcher BC, Urry DW. Coacervation of tropoelastin results in fiber formation. J Biol Chem. 1974;249:997–998. [PubMed] [Google Scholar]
- Davis EC. Stability of elastin in the developing mouse aorta: a quantitative radioautographic study. Histochem. 1993;100:17–26. doi: 10.1007/BF00268874. [DOI] [PubMed] [Google Scholar]
- Davis EC, Mecham RP. Intracellular trafficking of tropoelastin. Matrix Biol. 1998;17:245–254. doi: 10.1016/s0945-053x(98)90078-6. [DOI] [PubMed] [Google Scholar]
- Davis EC, Broekelmann TJ, Ozawa Y, Mecham RP. Identification of tropoelastin as a ligand for the 65-kD FK506-binding protein, FKBP65, in the secretory pathway. J Cell Biol. 1998;140:295–303. doi: 10.1083/jcb.140.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EC, Mecham RP. Selective degradation of accumulated secretory proteins in the endoplasmic reticulum: a possible clearance pathway for abnormal tropoelastin. J Biol Chem. 1996;271:3787–3794. [PubMed] [Google Scholar]
- Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galat A. Peptidylproline cis-trans-isomerases: immunophilins. Eur J Biochem. 1993;216:689–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- Galat A, Rivière S, Sheterline P, editors. Peptidyl-Prolyl Cis/Trans Isomerases. Oxford, UK: Oxford University Press; 1998. , 117. [Google Scholar]
- Gilbert HF. Protein chaperones and protein folding. Curr Opin Biotech. 1994;5:534–539. doi: 10.1016/0958-1669(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Göthel SF, Marahiel MA. Peptidyl-prolyl cis-trans-isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton GS, Steiner JP. Immunophilins. Beyond Immunosuppression. J Med Chem. 1998;41:5119–5143. doi: 10.1021/jm980307x. [DOI] [PubMed] [Google Scholar]
- Ikeda H, Fujiwara K. Cyclosporin A and FK-506 in inhibition of rat Ito cell activation in vitro. Hepatology. 1995;21:1161–1166. [PubMed] [Google Scholar]
- Indik Z, Yeh H, Ornstein-Goldstein N, Rosenbloom J. Structure of the elastin gene and alternative splicing of elastin mRNA. In: Sandell L, Boyd C, editors. Genes for Extracellular Matrix Proteins. New York, NY: Academic Press; 1990. pp. 221–250. [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Krause K-H, Michalak M. Calreticulin. Cell. 1997;88:439–443. doi: 10.1016/s0092-8674(00)81884-x. [DOI] [PubMed] [Google Scholar]
- Lodish HF, Kong N. Cyclosporin A inhibits an initial step in folding of transferrin within the endoplasmic reticulum. J Biol Chem. 1991;266:14835–14838. [PubMed] [Google Scholar]
- Mecham RP. Modulation of elastin synthesis: in vitro models. Methods Enzymol. 1987;144(D):232–246. doi: 10.1016/0076-6879(87)44181-5. [DOI] [PubMed] [Google Scholar]
- Mecham RP, Davis EC. Elastic fiber structure and assembly. In: Yurchenko P D, Birk D E, Mecham R P, editors. Extracellular Matrix Assembly and Structure. San Diego, CA: Academic Press.; 1994. pp. 281–314. [Google Scholar]
- Nakamura T, Yabe D, Kanazawa N, Tashiro K, Sasayama S, Honjo T. Molecular cloning, characterization, and chromosomal localization of FKBP23, a novel FK506-binding protein with Ca2+-binding activity. Genomics. 1998;54:89–98. doi: 10.1006/geno.1998.5571. [DOI] [PubMed] [Google Scholar]
- Nigam SK, Jin Y-J, Jin M-J, Bush KT, Bierer BE, Burakoff SJ. Localization of the FK506-binding protein, FKBP 13, to the lumen of the endoplasmic reticulum. Biochem J. 1993;294:511–515. doi: 10.1042/bj2940511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks WC, Secrist H, Wu LC, Mecham RP. Developmental regulation of tropoelastin isoforms. J Biol Chem. 1988;263:4416–4423. [PubMed] [Google Scholar]
- Pelham HRB. The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem Sci. 1990;15:483–486. doi: 10.1016/0968-0004(90)90303-s. [DOI] [PubMed] [Google Scholar]
- Price ER, Zydowsky LD, Jin MJ, Baker CH, McKeon FD, Walsh CT. Human cyclophilin B. A second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc Natl Acad Sci USA. 1991;88:1903–1907. doi: 10.1073/pnas.88.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde JG. Triton X-114: a detergent that has come in from the cold. Trends Biochem Sci. 1986;11:160–163. [Google Scholar]
- Schneuwly S, Shortridge RD, Larrivee DC, Ono T, Ozaki M, Pak WL. Drosophila ninaA gene encodes an eye-specific cyclophilin (cyclosporin A binding protein) Proc Natl Acad Sci USA. 1989;86:5390–5394. doi: 10.1073/pnas.86.14.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadidy M, Caubit X, Olsen R, Seternes O, Moens U, Krauss S. Biochemical analysis of mouse FKBP60, a novel member of the FKBP family. Biochim Biophys Acta. 1999;1446:295–307. doi: 10.1016/s0167-4781(99)00080-9. [DOI] [PubMed] [Google Scholar]
- Shieh B-H, Stamnes MA, Seavello S, Harris GL, Zuker CS. The ninaA gene required for visual transduction in Drosophila encodes a homologue of cyclosporin A-binding protein. Nature. 1989;338:67–70. doi: 10.1038/338067a0. [DOI] [PubMed] [Google Scholar]
- Smith T, Ferreira LR, Hebert C, Norris K, Sauk JJ. Hsp47 and cyclophilin B traverse the endoplasmic reticulum with procollagen into pre-Golgi intermediate vesicles. J Biol Chem. 1995;270:18323–18328. doi: 10.1074/jbc.270.31.18323. [DOI] [PubMed] [Google Scholar]
- Steinmann B, Bruckner P, Superti-Furga A. Cyclosporin A slows collagen triple-helix formation in vivo: indirect evidence for a physiologic role of peptidyl-prolyl cis-trans-isomerase J. Biol Chem. 1991;266:1299–1303. [PubMed] [Google Scholar]
- Walensky LD, Gascard P, Fields ME, Blackshaw S, Conboy JG, Mohandas N, Snyder SH. The 13-kD FK506 binding protein, FKBP13, interacts with a novel homologue of the erythrocyte membrane cytoskeletal protein 4.1. J Cell Biol. 1998;141:143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn DS, Griffin GL, Senior RM, Mecham RP. Characterization of biologically active domains on elastin: identification of a monoclonal antibody to a cell recognition site. Biochem. 1986;25:5172–5176. doi: 10.1021/bi00366a028. [DOI] [PubMed] [Google Scholar]
- Wu WJ, Vrhovski B, Weiss AS. Glycosaminoglycans mediate the coacervation of human tropoelastin through dominant charge interactions involving lysine side chains. J Biol Chem. 1999;274:21719–21724. doi: 10.1074/jbc.274.31.21719. [DOI] [PubMed] [Google Scholar]
- Zeng B, MacDonald JR, Bann JG, Beck K, Gambee JE, Boswell BA, Bächinger HP. Chicken FK506-binding protein, FKBP65, a member of the FKBP family of peptidylprolyl cis-trans isomerases, is only partially inhibited by FK506. Biochem J. 1998;330:109–114. doi: 10.1042/bj3300109. [DOI] [PMC free article] [PubMed] [Google Scholar]