Abstract
In the budding yeast Saccharomyces cerevisiae, movement of the mitotic spindle to a predetermined cleavage plane at the bud neck is essential for partitioning chromosomes into the mother and daughter cells. Astral microtubule dynamics are critical to the mechanism that ensures nuclear migration to the bud neck. The nucleus moves in the opposite direction of astral microtubule growth in the mother cell, apparently being “pushed” by microtubule contacts at the cortex. In contrast, microtubules growing toward the neck and within the bud promote nuclear movement in the same direction of microtubule growth, thus “pulling” the nucleus toward the bud neck. Failure of “pulling” is evident in cells lacking Bud6p, Bni1p, Kar9p, or the kinesin homolog, Kip3p. As a consequence, there is a loss of asymmetry in spindle pole body segregation into the bud. The cytoplasmic motor protein, dynein, is not required for nuclear movement to the neck; rather, it has been postulated to contribute to spindle elongation through the neck. In the absence of KAR9, dynein-dependent spindle oscillations are evident before anaphase onset, as are postanaphase dynein-dependent pulling forces that exceed the velocity of wild-type spindle elongation threefold. In addition, dynein-mediated forces on astral microtubules are sufficient to segregate a 2N chromosome set through the neck in the absence of spindle elongation, but cytoplasmic kinesins are not. These observations support a model in which spindle polarity determinants (BUD6, BNI1, KAR9) and cytoplasmic kinesin (KIP3) provide directional cues for spindle orientation to the bud while restraining the spindle to the neck. Cytoplasmic dynein is attenuated by these spindle polarity determinants and kinesin until anaphase onset, when dynein directs spindle elongation to distal points in the mother and bud.
INTRODUCTION
The regulated orientation of cell divisions in response to positional cues is critical for morphogenesis and cell-fate specification during development in many organisms (Drubin and Nelson, 1996; Gonczy and Hyman, 1996). Positional information, either cell-intrinsic or derived from extrinsic cues, directs the alignment of the mitotic spindle along a specified axis, leading to oriented cell division. In the budding yeast Saccharomyces cerevisiae, the site of bud emergence becomes the mother-bud neck, where cytokinesis takes place (Chant and Pringle, 1995; Pringle et al., 1995). Thus, the mitotic spindle must orient perpendicular to this prespecified cleavage plane to accurately partition chromosomes into mother and daughter cells. Spindle orientation is achieved through interactions between astral microtubules, microtubule-based motor proteins, and cortical determinants, as well as the actin cytoskeleton (Palmer et al., 1992; Carminati and Stearns, 1997; Shaw et al., 1997b; Theesfeld et al., 1999; Heil-Chapdelaine et al., 2000).
Cytoplasmic astral microtubules in yeast emanate from the spindle pole body (SPB; functionally equivalent to the centrosome in animal cells), which is embedded in the nuclear envelope (Peterson and Ris, 1976; Byers, 1981). The SPB is duplicated near the time of the G1/S transition, and in G2 the duplicated SPBs separate to opposite sides of the nucleus as the mitotic spindle is formed. Astral microtubules extend from each SPB toward the cell cortex, while intranuclear microtubules extending between the SPBs and from SPBs to kinetochores form the spindle (as with other fungi, the nuclear envelope does not break down during mitosis). Before anaphase, the spindle is positioned close to the neck such that one SPB directs one or more astral microtubule toward the bud cortex and the other SPB nucleates microtubules predominantly within the mother cell. This feature results in spindle orientation along the mother-bud axis; anaphase spindle elongation then proceeds until the spindle poles reach the distal ends of the budded cell, resulting in accurate segregation of chromosomes at cell division.
Our understanding of the molecular basis for spindle orientation has benefited greatly from genetic and cell biological approaches, in which specific microtubule motors (primarily dynein and the kinesin homologue Kip3p), a putative microtubule plus-end capturing protein (Kar9p)(Miller and Rose, 1998), and asymmetrically distributed cortical proteins (Kar9p, Num1p, Bni1p, and Bud6p) were all shown to be important for spindle positioning. Cells lacking any one of these proteins often contain misaligned spindles, but in all cases this defect is rectified before cytokinesis. In contrast, cells lacking dynein in combination with one of several other proteins (e.g., Kip3p, Kar9p, Bud6p, Bni1p) exhibit conditional phenotypes (this study and Lee et al., 1999; Miller et al., 1999). Two partially redundant “pathways” for spindle orientation have been proposed, one, involving Kip3p, Kar9p, Bni1p, and Bud6p, that acts early in the cell cycle to position the preanaphase spindle, and a second involving dynein, that acts late in the cell cycle to promote spindle segregation (Cottingham and Hoyt, 1997; Heil-Chapdelaine et al., 1999; Lee et al., 1999; Miller et al., 1999).
Accurate spindle positioning also depends upon an intact actin cytoskeleton (Palmer et al., 1992). Actin's role in spindle positioning versus its essential role in bud growth was determined through perturbations of F-actin and use of mutations that selectively disrupt actin cables (Pruyne et al., 1998; Theesfeld et al., 1999). Actin is required early in the cell cycle, temporally coincident with the Kip3p-dependent phase, to establish spindle orientation. Two proteins required for the Kip3p-dependent phase (Bni1p, Bud6p) are themselves actin-interacting proteins. Bni1p/She5p contains a formin homology domain, is required for integrity of the actin cytoskeleton, and is itself related to a protein, Cdc12p, in S. pombe that is transported on microtubules and actin filaments (Chang, 1999). Bud6p/Aip3p (Amberg et al., 1997) is delivered to cortical sites via the secretory machinery, where it contributes to polarized cell growth (Jin and Amberg, 2000). Bud6p/Aip3p and Bni1p/She5p are required for the integrity of the actin cytoskeleton, localized distribution of Kar9p (Miller et al., 1999), and regulating mating competency in the mother versus the bud (ASH1 mRNA; Beach and Bloom 1999). The pleiotropic phenotype of bud6 and bni1 mutants point to a more general role for these proteins in polarized events. Bud6p and Bni1p have been proposed to comprise a cortical scaffold for docking ASH1 mRNA as well as for providing a target for microtubule capture via Kar9p. The role of the actin cytoskeleton is therefore critical for timely delivery and/or deposition of these cortical components (e.g., Bni1p, Bud6p, Kar9p) that in turn facilitate microtubule–cortical interactions.
Microtubule interactions with cortical sites are mediated in part by Bim1p. Bim1p, a member of the EB1 family of proteins that interact with the adenomatous polyposis coli tumor suppressor, binds the plus ends of microtubules (Tirnauer et al., 1999) and physically interacts with Kar9p (Korinek et al., 2000; Lee et al., 2000). The capture of microtubule ends at the cell cortex (Adames and Cooper, 2000), together with findings that shortening astral microtubules lose tubulin subunits from the plus ends (Maddox et al., 2000) indicate that cortical sites of Kar9p promote plus-end disassembly through Bim1p, while maintaining contact with the shortening end. Loss of the cortical interactions can be corrected through dynein-dependent microtubule sliding (Adames and Cooper (2000)), consistent with previous findings that the early pathway is dispensable (Cottingham and Hoyt, 1997; DeZwaan et al., 1997). The proposed functional overlap of cortical capture mechanisms with dynein-dependent sliding processes (Adames and Cooper, 2000) was not predicted from the temporal separation of these pathways in genetic studies.
A remarkably small number (3–5) of dynamic astral microtubules provide the basis for nuclear movement throughout the cell cycle. The astral microtubules have an unexpectedly short average lifetime (< 3.65 min; Maddox et al., 1999), suggesting that spindle positioning may result from multiple transient interactions. In addition, preexisting microtubules are partitioned unequally upon SPB duplication and separation. One SPB retains all of the dynein-GFP labeled astral microtubules. The other SPB acquires dynein-GFP after a delay of ∼10 min, during which time intranuclear microtubules push the SPBs apart (Shaw et al., 1997b). This “delayed acquisition” of dynein-GFP to one SPB is regulated by the Cdc28p cyclin-dependent kinase and appears to be important for preventing the late-acquiring SPB from nucleating bud-directed microtubules (Segal et al., 2000). These observations suggest a simple model in which the stable orientation of one SPB toward the bud and the other SPB toward the mother comes about through the preferential association of one SPB at the neck with bud-directed astral microtubules. The rigid intranuclear spindle predisposes the second SPB away from the neck, steering astral microtubules from that SPB away from the bud.
In this study we have examined microtubule and spindle behavior in living cells that lack individual proteins (or pairs of proteins) involved in spindle orientation. Our results suggest that Kip3p, Kar9p, Bni1p, and Bud6p provide directional cues for spindle orientation, as well as oppose the action of dynein until anaphase onset. Cytoplasmic dynein is the dominant cytoplasmic force and drives spindle elongation to distal reaches in the mother and bud. The hierarchy of motors, their interaction with each other and transient interaction with astral microtubules reflect distinct but coordinated pathways toward faithful nuclear migration.
MATERIALS AND METHODS
Strains, Media, Growth Conditions, and Genetic Techniques
S. cerevisiae strains used in this study are described in Table 1 and where appropriate. YEF473, 1345, 1358, AM102, AM16, AM29, and AM8 were kindly provided by J. Pringle (University of North Carolina at Chapel Hill). dhc1::URA3 and dhc1::LEU2 deletions were constructed as described previously (Li et al., 1993). kar9::LEU2 deletions were constructed by transforming strains with SmaI cut pRM386 (kindly provided by R. Miller and M. Rose). A kip3::KANMX cassette was designed that results in a deletion of the entire KIP3 coding region (kip3::KANMX 5′ catattccctgattgcatcgtttatctgattatcatttcattgcattcttcagctgaagcttcgtacgc (69mer), kip3::KANMX3′ gcgtttaatgctggcggaaagaagttatattcgatagtttacgtaggatagcataggccact-agtggatctg (72mer)). The deletion cassettes were introduced into DLY2 (DLY3650) and W303 (KBY58 and KBY61) strain backgrounds. All three strains displayed similar numbers of binucleate cells in logarithmically growing populations. The aip3/bud6 phenotype has been reported to vary in different genetic backgrounds (Amberg et al., 1997). The aip3-Δ3 mutant used herein (1345) and described in Amberg et al. (1997) is resistant to 37°C and contains very few binucleated cells. Strains derived from YEF473 were used in the real-time analysis. Microtubules were visualized in live cells either by transformation with the plasmid pKB701 containing a DHC1-GFP fusion protein on the GAL1 promoter or by integration of pASF125 containing GFP::TUB1 at URA3 (kindly provided by A. Straight). To visualize DHC1-GFP, cells were grown overnight in SD-URA medium containing 2% glucose and 2% galactose. Yeast cultures were grown at 32°C, except where otherwise noted. Yeast rich medium (YPD) contained 2% glucose, 2% peptone, and 1% yeast extract. Synthetic Dropout (SD) medium lacking particular nutrients (SD-Leu, SD-His) and synthetic sporulation medium supplemented with amino acids were made as described (Rose et al., 1991).
Table 1.
Yeast strains and plasmids
| Yeast strains | Genotype |
|---|---|
| YEF473 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200 |
| YEF1345 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, aip3Δ3(bud6)∷TRP1 |
| YEF1358 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, bni1∷HIS3 |
| CYY118 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, dhc1Δ∷LEU2 |
| KBY474 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, kip3Δ∷KANMX |
| KBY475 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, kar9Δ∷LEU2 |
| AM16 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, bud7∷HIS3 |
| AM29 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, bud3∷TRP1 |
| AM8 | MATa ura3-52, trp1Δ63, leu2Δ1, lys2-801, his3Δ200, bud8∷TRP1 |
| KBY61 | MATα leu2-3,112, ura3-52, trp1, ade2, his3Δ200, dhc1Δ7∷URA3 |
| KBY58 | MATα leu2-3,112, ura3-52, trp1, ade2, his3Δ200, dhc1Δ∷LEU2 |
| DLY2 | MATα ura3Δns, leu2-3,112, trp1-1a, his2, ade1 |
| DLY3650 | MATα ura3Δns, leu2-3,112, trp1-1a, his2, ade1, dhc1Δ∷LEU2 |
| DLY2719 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, bem2∷URA3 |
| JMY1011 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, bem1∷URA3 |
| JMY1020 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, tpm1∷URA3 |
| JMY1023 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, cap2∷URA3 |
| JMY1030 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, spa2∷URA3 |
| JMY1080 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, bed1∷URA3 |
| JMY2-26 | MATa ura3Δns, leu2-3,112, trp1-1a, his2, ade1, myo2-66 |
| Plasmids | |
| pKB701 | GALpDHC1-GFP, 2μ, URA3 |
| pAFS125 | TUB1-GFP integrated at URA3 |
| pRM386 | kar9ΔLEU2 disruption plasmid |
Genetic Interactions.
dhc1 kar9 and dhc1 bni1 double-mutant strains were not viable when grown at temperatures outside a narrow permissive range (DHC1 encodes the dynein heavy chain) as reported by (Lee et al., 1999; Miller et al., 1999). In addition to these interactions dhc1,bud6 double mutants exhibited a conditional growth phenotype (>75% of double-mutant spore colonies from a dhc1 BUD6 X DHC1 bud6 cross were not viable at 37°C, compared with < 15% lack of viability for wild-type or single-mutant spore colonies from the cross). Mutations in other genes affecting actin organization did not show synthetic growth defects when crossed to dhc1 mutants (this group included bem1 [Chenevert et al., 1992], bem2 [Peterson et al., 1994], tpm1 [Liu and Bretscher, 1992], cap2 [Amatruda and Cooper, 1992; Karpova et al., 1995], bed1 [Mondesert and Reed, 1996], and spa2 [Snyder, 1989; Zahner et al., 1996]).
Microscopy and Quantitation
Cells expressing the Dhc1p-GFP fusion protein or GFP-Tub1p were observed using time-lapse video microscopy. Strains 1345 and 1358 containing plasmid pKB701 were grown overnight to midlogarithmic phase in glucose/galactose-containing medium at 24°C. Double mutants (dhc1/bni1 from 1358 x KBY61; dhc1/bud6 from 1345 x KBY58) containing GFP-Tub1 were grown overnight to midlogarithmic phase in glucose-containing medium at 24°C. Cells were applied to microscope slides coated with minimal medium containing 25% gelatin. Coverslips were applied and sealed with valap (1:1:1 vaseline:lanolin:paraffin), and cells were observed with a microscope (Microphot FXA, Nikon, Garden City, NY) using a 100X/1.4NA Plan Apo objective. Metamorph software (Universal Imaging Corp., West Chester, PA) was used to control a Biopoint 99B100 filter wheel (Ludl Electronic Products, Hawthorne, NY) (Salmon et al., 1998). A shuttered mercury lamb was used for fluorescence excitation (490 nm) at 0.3-s exposures. Images were collected with a cooled CCD camera (C4880; Hamamatsu, Photonics, Bridgewater, NJ) at either 30-s or 1-min intervals throughout the course of cell growth. A total of five fluorescence images were acquired at 0.75 μm intervals through the cell and projected into a single reconstruction. A single differential interference contrast (DIC) image was made at the middle z-step by rotating the analyzer into the light path and taking a 0.6-s exposure (Shaw et al., 1997a).
RESULTS
Spindle Pole Asymmetry Is Retained in bni1, bud6, and kar9 Mutants
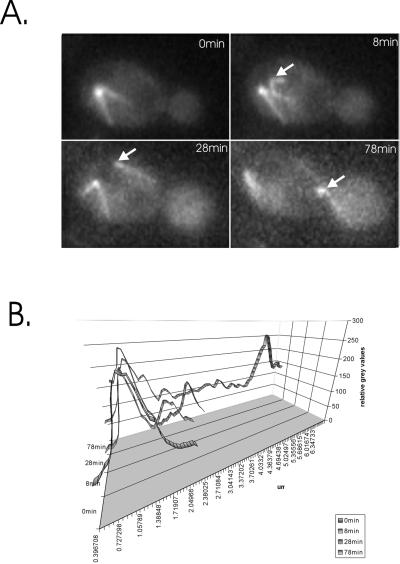
Observation of bni1, bud6, or kar9 single-mutant cells containing GFP-tagged tubulin or dynein (see MATERIALS AND METHODS) confirmed previous findings that these cells frequently misorient the spindle (Miller and Rose, 1998; Lee et al., 1999). Failure to regulate the asymmetric behavior of spindle pole bodies during spindle assembly also results in a spindle position defect in cdc28–4 clb5Δmutants (Segal et al., 2000). To assess spindle pole asymmetry in bni1, bud6, or kar9 cells, we monitored dynein-GFP accumulation during SPB separation. In every case (>50 examples for each mutant) there was a delay after visible separation of the SPBs before the second spindle pole body accumulated cytoplasmic dynein (and astral microtubules), similar in timing to that seen in wild-type cells (e.g., kar9 mutant, Figure 1A; quantitative fluorescence accumulation in Figure 1B). Like wild-type cells these mutants are not defective in the asymmetric partitioning and/or acquisition of dynein-GFP during SPB separation.
Figure 1.
Segregation of the “late acquiring” SPB into the bud in kar9 mutants. kar9 mutants expressing dynein-GFP were grown to midlogarithmic phase and were examined by fluorescence microscopy. (A) Images from a time-lapse series are displayed (times in min indicated in the upper right of each panel). The nucleus lies distal to the neck of a budded cell before spindle elongation. Microtubules emanate from a single spindle pole body (0 min). The poles have duplicated by 8 min, and the 2nd pole (indicated with white arrowhead) acquires cytoplasmic dynein. At 28 min, late acquiring pole nucleates an astral microtubule(s) directed toward the bud. At 78 min, the spindle has aligned along the mother-bud axis, and a single microtubule from the late-acquiring pole penetrates the bud. The late-acquiring SPB migrates into the bud. (B) Quantitative fluorescence accumulation as a function of time and distance between the spindle pole bodies. The time points in (A) are plotted. Note spindle pole separation as increased distance from peak to peak (one pole at 0 min, two at 8, 28, and 78,min). The delayed acquisition of dynein-GFP is evident in the fluorescent intensity of the two peaks (8 and 28 min) until spindle elongation, whereupon the two poles are equally fluorescent (78 min).
Defective Spindle Pole Orientation Toward the Bud in bni1, bud6, and kar9 Mutants
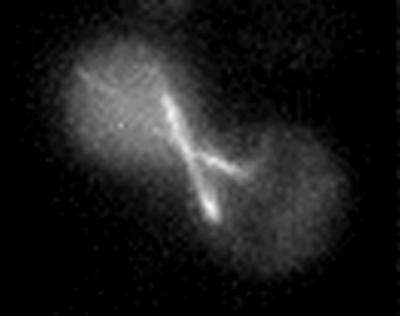
The pole that initially acquires astral microtubules in G1 orients the nucleus toward the neck and leads spindle migration into the bud upon anaphase onset (Shaw et al., 1997b). While spindle pole asymmetry was maintained in kar9, bud6, and bni1 mutants, neither SPB was predisposed to lead spindle migration into the bud (e.g., migration of late acquiring SPB into the bud in kar9 mutant, Figure 1A). In some cases (bni1 mutant, Figure 2), defects in SPB orientation toward the bud occurred through depolymerization of the bud-directed microtubule and subsequent penetration of an astral microtubule from the other SPB into the bud. In other instances, microtubules from the second pole penetrated into the bud even though the first pole retained bud-directed microtubules, generating cells with astral microtubules from both spindle poles extending into the bud (bud6 mutant, Figure 3). Loss of SPB orientation and/or dual microtubule penetration was observed in 25–35% of the mutant cells (7/28 for bud6, 11/35 for bni1, and 5/14 for kar9). Loss of spindle orientation along the mother-bud axis was often accompanied by cycles of microtubule polymerization/depolymerization in the bud, alternating between each SPB (shown in Figure 2). These transition periods persisted for ∼ 1–8% of the time before anaphase onset (3.7% in bud6, 8.5/230 total minutes; 8.1% in bni1, 50/617 total min; 1.6% in kar9, 7.5/464 total min). Neither loss of SPB orientation nor penetration of astral microtubules from both poles into the bud were observed in > 6000 min of image analysis in wild-type or dynein mutant cells (Shaw et al., 1997b).
Figure 2.
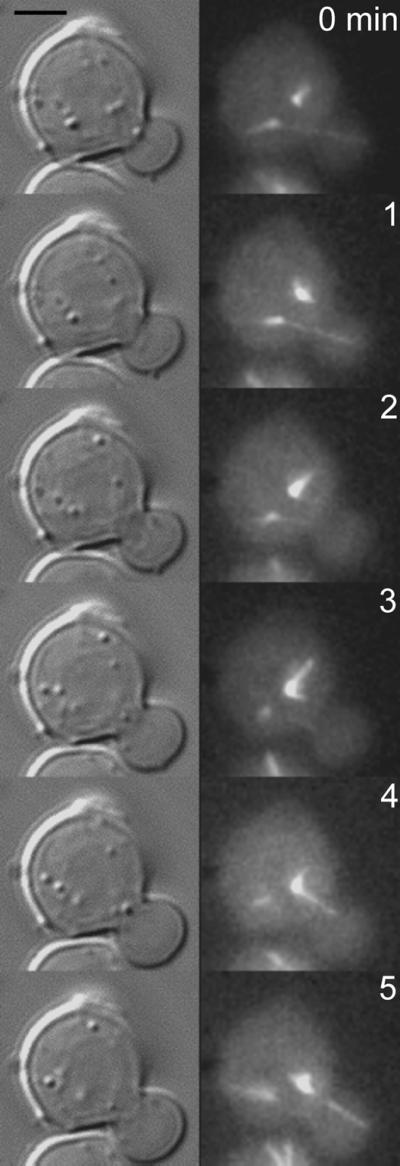
SPB nucleating microtubules that first penetrate the bud are not the SPB destined for the bud in bni1 mutants. bni1 mutants expressing dynein-GFP were grown to midlogarithmic phase and examined by fluorescence microscopy. Images from a time-lapse series are displayed (right, times in min indicated in the upper right of each panel). Corresponding DIC images are shown to the left. Two spindle pole bodies can be seen in the upper cell (4 o'clock and 6 o'clock, respectively). Astral microtubules from the pole at 6 o'clock penetrate the bud (0,1 min). At 2 and 3 min, the bud is devoid of astral microtubules. At 4 and 5 min, the pole at 4 o'clock nucleates an astral microtubule that penetrates the bud. This pole leads the nucleus into the bud at later time points.
Figure 3.
Simultaneous penetration of astral microtubules in the bud in bud6 mutants. Bud6 mutants expressing dynein-GFP were visualized by fluorescence microscopy. The mother cell is in the bottom right quadrant, the daughter to the top left. Astral microtubules emanating from spindle pole bodies in the mother (8 and 10 o'clock respectively) cross over the neck and penetrate the bud simultaneously. These defects were observed in 3/18 bud6 mutants for a total of 3.7 percentage of the time before anaphase onset (8 min of alternating cycles of microtubule polymerization/depolymerization in the bud emanating from each pole, 30 s where microtubules from both poles penetrate the bud; 230 total minutes). 5/20 bni1 mutants exhibited dual penetration from both spindle poles for 8.1% of the time (50/617 total min) and 2/11 kar9 mutants for a total duration of 1.6% of the time before anaphase onset (7.5/464 total min).
Despite the frequent appearance of spindles with microtubules from both poles penetrating the bud in bni1, bud6, or kar9 mutants, there was not a commensurate migration of the entire spindle into the bud (few transient events, <5 min, observed over the course of 20–30 h of filming) as observed in clb5-cdc28–4 mutants (Segal et al., 2000).
Defective Retention of the Spindle Near the Neck in bni1, bud6, dhc1, kar9, and kip3 Mutants
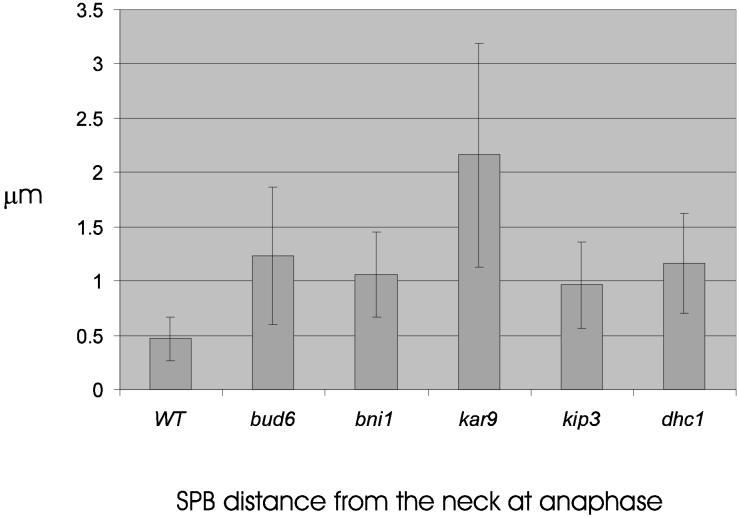
If the failure to predispose one SPB to the bud reflects loss of spindle positioning determinants in the bud, then not only will the spindle be misaligned before anaphase, but it may not migrate to the bud neck. To test this hypothesis, we examined the distance between the bud-proximal SPB and the neck at 1 min before anaphase B, defined as the onset of 2 μ spindle elongation (stage III, (Yeh et al., 1995)). The spindle was significantly closer to the neck in wild-type cells (95% confidence) relative to all 5 mutant strains examined (Figures 4,5). The spindle was further displaced in kar9 mutants relative to bud6, dhc1, bni1, and kip3. In addition, spindle position was highly variable from cell to cell in kar9 mutants (noted by large SD for kar9 in Figure 4). These data indicate a defect in the mechanisms responsible for juxtaposition of the SPB to the neck before anaphase onset.
Figure 4.
Distance from SPB proximal to neck 1 min before spindle pole elongation. Wild-type, bud6, bni1, kar9, kip3, and dhc1 mutants containing Tub1-GFP were grown to midlogarithmic growth and examined by fluorescence microscopy. Cells containing a 1–1.5 μm spindle were examined and followed by time-lapse microscopy through spindle elongation. At 1 min before anaphase spindle elongation, a point at the center of the neck was determined in DIC and was used as the origin when measuring the distance to the center of SPB fluorescence. Track Points software in Metamorph (Universal Imaging Corp.) recorded the SPB position at each time point. The distance was determined by converting pixels to microns using the image of the stage micrometer. Values are the average of ten to twenty cells. Wild-type spindles are statistically closer to the neck than spindles in bud6, bni1, kar9, and kip3 mutants as determined by 2-tailed unequal variance Students t test (95% confidence). kar9 spindles are significantly further from the neck relative to bud6, bni1, kip3, and wild-type. Spindle position in bud6, bni1, kip3, and dhc1 are statistically indistinguishable.
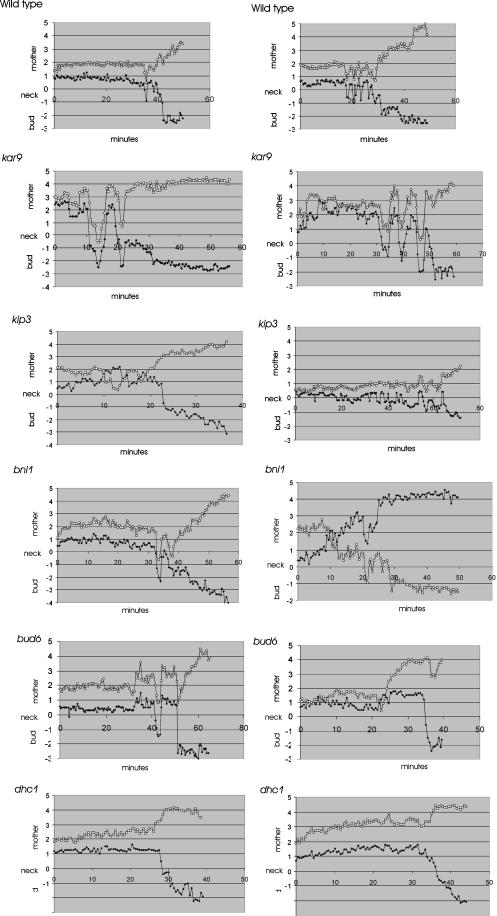
Figure 5.
Metaphase and anaphase spindle movements in wild-type and mutant cells. Spindle movements in wild-type, bud6, bni1, kar9, kip3, and dhc1 mutants. The distances between each spindle pole body and neck were measured (μm) at 30-s intervals using Track Points software in Metamorph (Universal Imaging Inc.). The position of the SPB relative to the mother is plotted as positive numbers, the distance between the neck and the bud is plotted as a negative number. The position of the neck is set as zero. Two examples of each cell type are shown. The spindle pole that is initially distal to the neck at t = 0 is indicated by a circle, the pole proximal to the neck by a diamond. Preanaphase spindle oscillations were observed in 5/7 wild-type cells, 5/5 kar9, 10/13 kip3, 9/16 bni1, 8/15 bud6, and 0/8 dhc1 mutants, respectively.
Spindle Alignment and Oscillations Before Anaphase in bni1, bud6, dhc1, kar9, and kip3 Mutants
Cytoplasmic forces acting on the preanaphase spindle are manifested as oscillations of the spindle along the mother/bud axis at constant spindle length (Yeh et al., 1995). The oscillations begin ∼ 5–15 min before anaphase onset and average between 2 and 3 per cell (range 1–5, Figure 5 and (Yeh et al., 1995)). Cells lacking cytoplasmic dynein did not display spindle oscillations: the spindle migrated to within 1.0 μm of the neck (as in wild-type cells) and remained static (Figure 5, dhc1 panels), suggesting that the preanaphase spindle oscillations are indeed powered by dynein. Displacement of the spindle from the neck in spindle position mutants shown in Figure 4 could reflect a general loss of cytoplasmic forces, or specific loss of a bud-directed force. To distinguish between these mechanisms we examined spindle oscillations before and through the initial stages of anaphase B.
Spindle oscillations were quantitated using a semiautomated tracking system to determine the position of both SPBs relative to the neck at 30-s intervals. The neck provided a reference point for comparing spindle movement from cell to cell. Loss of spindle alignment along the mother-bud axis was observed as converging or overlapping points, indicating that the SPBs have rotated perpendicular to the image plane (e.g., bni1 right panel 10min, kip3 left panel 5, 14min, bud6 right panel 1–5min, 22min). Spindles in wild-type, bud6, bni1, and kip3 cells exhibited a similar number of oscillation, however the average distance from the neck was slightly higher in mutant versus wild-type (wild-type 1.04 ± 0.57 μm, bni1 1.24 ± 0.60 μm, bud6 1.57 ± 0.89 μm, kip3 1.63 ± 0.68 μm). Spindles in bni1 and bud6 were observed to transit into and out of the bud (Figure 5, bni1 left panel 35min, right panel 20min; complete spindle transit left panel bud6 42min)(Lee et al., 1999). In kar9 mutants, the few cells (n = 5) that contain spindles aligned along the mother/bud axis also displayed frequent oscillations that spanned an even greater distance (3.7 ± 0.89 μm; Figure 5), as well as complete transits into the bud (left panel 13 min, right panel 35, 40 and 48 min). These spindle oscillations are consistent with the high variability found in spindle position relative to the neck (kar9, Figure 4). The increased spindle motility indicates that spindle displacement from the neck does not reflect a general loss of cytoplasmic forces. KIP3, which has been proposed to act in the same pathway as BNI1 and KAR9, is likewise not required for spindle oscillations before anaphase. Spindle oscillations in kip3 precede anaphase by 30 min (similar to kar9) and are characterized by loss of pole orientation where the initially distal pole (circles) came to lie proximal to the neck (left panel 10–14 min). However the amplitude of oscillations in kip3 was not as great as kar9. This finding is consistent with results above demonstrating decreased displacement from the neck in kip3 relative to kar9 mutants, and indicates kip3 and kar9 have distinguishable spindle position and motility phenotypes before anaphase onset. The dependency of these preanaphase spindle oscillations on dynein is indicative of early dynein function. Kar9p, Bni1p, and Bud6p are not required for dynein-dependent spindle oscillations. Rather, they appear to restrain the spindle in the vicinity of the neck. This might be achieved either through a role of these proteins in down-regulating dynein activity, or through a role in anchoring the spindle and/or astral microtubules near the neck in the face of dynein-dependent pulling forces.
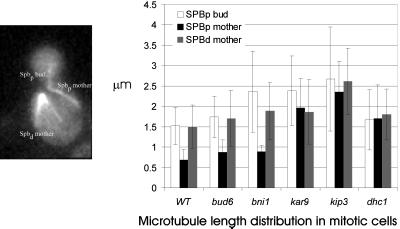
Astral Microtubule Length and Orientation in bni1, bud6, kar9, kip3, and dhc1 Mutants
One possible cause of spindle orientation and motility defects are alterations in astral microtubule dynamics. To characterize the preanaphase astral microtubules we examined the length distributions of three classes of astral microtubules in preanaphase mitotic cells. The three classes include microtubules extending into the mother cell and nucleated from the spindle pole distal to the neck (SPBd mother), those extending into the mother cell and nucleated from the spindle pole proximal to the neck (SPBp mother), and those extending into the bud and nucleated from the spindle pole proximal to the neck (SPBp bud)(Figure 6). All three classes of microtubules were significantly longer in kip3 mutants (>95% confidence interval), extending previous reports and consistent with the interpretation that Kip3p affects microtubule dynamics. Interestingly, bud6, bni1, kar9, and dhc1 SPBd mother microtubules were only slightly longer than their wild-type counterpart (20% on average) but not statistically different from each other. The increased microtubule lengths observed in SPBp mother (kip3 = kar9 = dhc1 > bud6 = bni1 = wt) and SPBp bud (kip3 = kar9= bni1 > dhc1 = bud6 = wt) may reflect spindle position relative to the neck (see Figure 4) and, to a lesser extent, alterations in microtubule dynamics. kar9 exhibits the most severe spindle position defect, which may account for the increase in microtubule length of the SPBp bud class due to the increased distance from that SPB to the tip of the bud. In contrast, dhc1 affects primarily SPBp mother microtubules. Spindle oscillations are significantly decreased in dhc1 (Figure 5), which may indirectly influence the dynamics and length distribution of these microtubules. The total number of astral microtubules (average) was similar in all mutant and wild-type cells.
Figure 6.
Average microtubule lengths during mitosis. Microtubule lengths were determined in wild-type and the indicated mutants expressing Tub1-GFP. The three classes include those extending into the mother cell and nucleated from the spindle pole distal to the neck (SPBd mother), those extending into the mother cell and nucleated from the spindle pole proximal to the neck (SPBp mother), and those extending into the bud and nucleated from the spindle pole proximal to the neck (SPBp bud). Lengths were determined by measuring the distance from the center of SPB fluorescence to the microtubule end. Pixels were converted to microns using the image of the stage micrometer. Time-lapsed image sequences from five to ten metaphase and anaphase cells were used. The average lengths, plus or minus the SD, are indicated. Microtubule numbers are for wild-type (Spbpb = 39, Spbpm = 8, Spbd = 25); bud6 (Spbpb = 36, Spbpm = 7, Spbd = 23); bni1 (Spbpb = 30, Spbpm = 3, Spbd = 25), kar9 (Spbpb = 25, Spbpm = 24, Spbd = 30), kip3 (Spbpb = 32, Spbpm = 6, Spbd = 42), and dhc1 (Spbpb = 57, Spbpm = 10, Spbd = 43). All three classes of microtubules were significantly longer in kip3 mutants versus other mutants and wild-type (>95% confidence). Statistical analysis (2-tailed unequal variance Students t test) indicate the following significant differences in microtubule length relationships: SPBd mother kip3> kar9 = bud6 = bni1= dhc1>wild-type; SPBp mother kip3 = kar9 = dhc1 > bud6 = bni1 = wild-type; SPBp bud kip3 = kar9= bni1 > dhc1 = bud6 = wild-type.
In the course of these measurements we observed that in kar9 mutants the spindle pole nearest the bud was less likely to be linked to a bud-directed microtubule compared with wild-type or other mutant cells. In particular, astral microtubules emanating from this pole were equally likely to be bud-directed (30% of cells) or mother-directed (30% of cells). For comparison, wild-type and other mutant cells were approximately five times more likely to have bud-directed (40–54% of cells) than mother-directed (5–11% of cells) microtubules emanating from that pole. These results are consistent with previously reported data on fixed cells (Miller and Rose, 1998), suggesting that Kar9p promotes microtubule entry into the bud, perhaps in conjunction with the actin cytoskeleton (Theesfeld et al., 1999).
Dynein-dependent Pulling During Anaphase
Spindle movements parallel to the mother-bud axis may arise from microtubule-cortical interactions, or (during anaphase) from sliding of intranuclear microtubules. By analyzing kar9 mutant cells in which spindle elongation took place along an eccentric axis, we are able to separate pole movements directed by astral microtubules from those involving the central spindle.
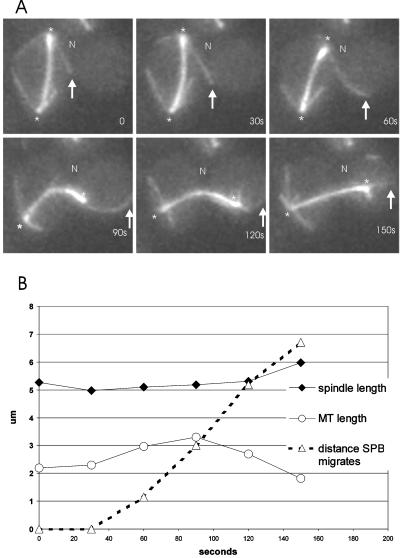
We analyzed the movements of anaphase spindles that initially failed to enter the bud at the onset of spindle elongation (e.g., Figure 7). These spindles elongate in the mother cell to ∼ 5 μm and are present in ∼ 5% of cells in a kar9 mutant population. In the cell shown in Figure 7, a microtubule from one pole (12 o'clock) grows toward the neck and penetrates the bud by 30 s. In the following 30 s, this microtubule does not change in length but moves toward the bud tip, concomitant with movement of the SPB toward the neck and into the bud. The spindle itself bends toward the neck, indicating that the pole is being pulled into the bud by the astral microtubule, rather than being pushed by the central spindle or microtubules from the opposite spindle pole. SPB movement into the bud proceeds at ∼ 3.58 ± 0.8 μm/min in these situations (n = 6), a speed 3.6-fold faster than the fast phase (1 μm/min) of anaphase spindle elongation (Kahana et al., 1995; Yeh et al., 1995; Straight et al., 1998). These data suggest that SPB movements during anaphase are driven by microtubule sliding along cortically anchored motor proteins (Adames and Cooper, 2000). In contrast to fast migration of maloriented spindles in kar9 (Figure 7), similarly positioned spindles were virtually static in dhc1 mutants (Figure 5) or migrate away from the bud in dhc1, bud6 or dhc1, kar9 mutants (Figure 8). No pulling forces were observed in the absence of dynein.
Figure 7.
Spindle translocation in the absence of spindle elongation. Astral microtubules pull the nucleus into the bud in the absence of Kar9p (n = 9). (A). The spindle has elongated in the mother perpendicular to the mother-bud axis. N denotes the position of the neck, an arrow marks the end of the microtubule, and the spindle pole indicated by an asterisk. An astral microtubule from the SPB at 12 o'clock grows into the bud in the top three panels (0–60 s). Once the astral microtubule interacts with the bud cortex, there is net movement of the spindle toward the bud (90 s). (B). Graph of the microtubule length (open circles), spindle length (closed diamonds), and SPB movement (open triangles) as a function of time (sec). Movement of the SPBp into the bud is concomitant with astral microtubule interaction with the bud cortex. SPB movement is independent of spindle elongation.
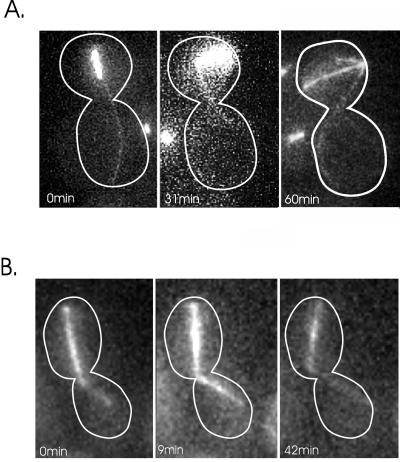
Figure 8.
Spindle movement in dhc1,bud6, and dhc1, kar9 mutants. dhc1, bud6 (A), and dhc1, kar9 (B) double mutants containing GFP-Tub1 were grown at 24°C. GFP-Tub1 was used to visualize the spindle and astral microtubules. Dynein-GFP could not be used to image astral microtubules in the double mutants because dynein-GFP complements the heavy chain deletion. The spindle in bud6, dhc1 (A) is visible as the bright bar and remains in the body of the mother cell. The astral microtubules originate from the spindle pole body and extend into the bud. The spindle is aligned along the mother/bud axis at t = 0. The spindle is 1.91 μm in length and lies 1.34 μm from the neck of the budded cell. At t = 31 min, the spindle elongates slightly (2.82 μm) and migrates away from the neck (2.01 μm from the neck). By 60 min, spindle length = 5.74 μm and the spindle lies 2.11 μm from the neck, and at 81 min the spindle has disassembled. There is no evidence for nuclear migration toward the bud even though an astral microtubule penetrates the bud throughout 81 min of continuous observation. The spindle in kar9, dhc1 (B) is aligned along the mother-bud axis at t = 0. An astral microtubule extends into the bud at t = 0 min. The spindle does not migrate into the bud; in contrast, spindle movement away from the neck is apparent at 42 min.
Ten seconds after the pulling event described above, the astral microtubule begins shortening as the spindle continues to migrate to the bud tip (120s Figure 7). We have frequently observed that astral microtubules begin depolymerizing (from the plus-end; Maddox et al., 2000) during movements that begin as sliding interactions, indicative of mechanisms that may couple spindle translocation with microtubule depolymerization in the bud.
Astral Microtubules Exert a Pushing Force in bud6, dhc1 or kar9, dhc1
If cytoplasmic dynein is responsible for pulling the spindle into the bud in the absence of spindle elongation, and BNI1/BUD6/KAR9/KIP3 provide early directional cues, then spindles should be largely confined to the mother cell in the double mutants. The only mechanism for nuclear migration into the bud would be from anaphase spindle elongation. dhc1, bud6 and dhc1,kar9 double mutants are viable at room temperature, providing a unique opportunity to examine spindle dynamics in cells lacking both these gene products. Spindle and astral microtubules were visualized with GFP-Tub1. The rate of anaphase spindle elongation and morphology of the mitotic spindle in the dhc1, bud6 or dhc1, kar9 was typical of wild-type cells (data not shown). Late anaphase spindles were readily observed in the mother cell in these mutants and exhibited erratic movement and rotation (Figure 8A and B). Microtubules penetrated the bud, and spindle poles migrated opposite to the direction of astral microtubule growth (dhc1, bud6 Figure 8A, 0 -31 min). There was no evidence for pulling forces toward the bud in bud6, dhc1 mutants. In contrast, the spindle migrated away from the neck from t = 0 (1.34 μm) to t = 31 min (2.01 μm) despite the penetration of microtubules into the bud. Movement away from the microtubule plus-end was reminiscent of nuclear rotations in G1, in which the nucleus is propelled opposite to the direction of microtubule growth (Shaw et al., 1997b). Similarly, in dhc1, kar9 double mutants, astral microtubules penetrated the bud, yet there was no net movement of the spindle toward the bud (Figure 8B). In addition, preanaphase spindle oscillations (shown in Figure 5) were completely absent in the dhc1, kar9 double mutant (data not shown). Elongation of astral microtubule in the bud and contact with the bud cortex lead to spindle movement away from the neck (Figure 8B, 42 min). Thus cells lacking Kar9p or Bud6p and dynein are devoid of all positional cues for directing nuclear movement via astral microtubules.
Dynein Exerts the Dominant Cytoplasmic Pulling Force during Anaphase
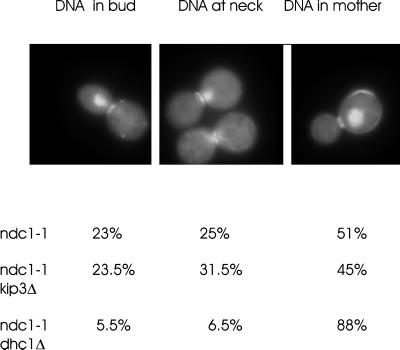
In a separate approach, we examined segregation of the SPB containing attached chromosomes in ndc1–1 cells lacking a bipolar spindle. SPB duplication is aberrant in ndc1–1 mutants at the restrictive temperature, and the new SPB is not correctly assembled into the nuclear envelope, preventing it from nucleating intranuclear microtubules and precluding bipolar spindle assembly. Astral microtubules are nucleated from both SPBs, and the SPBs eventually segregate away from each other, resulting in one pole in the bud and one pole in the mother. There is no bias in otherwise wild-type cells as to which pole (chromosome-associated, or defective) enters the bud (Thomas and Botstein, 1986; Yeh et al., 1995). In our studies, we found that the chromosome-associated pole (visualized by DAPI staining) was segregated to the bud in 23% of ndc1–1 cells (Figure 9, 51% of the chromosome-associated pole remained in the mother, while 25% of the cells contained chromatin at the neck). This pattern was virtually unchanged in ndc1–1 kip3Δ double-mutants (23.5% to the bud, n = 200) but was dramatically altered in ndc1–1, dhc1Δ double mutants (5.5% to the bud, n = 200; Figure 9) (Yeh et al., 1995). This suggests that, while either dynein or kinesin (kip3p) forces suffice to pull the defective SPB into the bud, only cytoplasmic dynein provides enough force to translocate the SPB with associated chromosomes into the bud in the absence of an intranuclear spindle. Dynein therefore exerts the dominant cytoplasmic pulling force for spindle translocation into the bud postanaphase.
Figure 9.
Chromatin segregation in the absence of a bipolar spindle and cytoplasmic motor proteins. Ndc1; ndc1, kip3 Δ and ndc1, dhc1Δ mutants were grown to early logarithmic phase at 25°C before shifting to 14°C, the nonpermissive growth temperature for ndc1–1, for 24 h. Cells arrest as large budded cells with a single nucleus. The cells were stained with Calcofluor to visualize the birth scar and with DAPI to visualize the chromatin DNA. An example of DNA in the bud is displayed to the left, DNA at the neck in the middle panel, and DNA in the mother to the right. Percentages of each phenotype in the different mutants are indicated below. Greater then 100 cells were counted for each determination.
DISCUSSION
The sequential action of nuclear movement toward the bud, alignment and retention at the neck, and translocation through the aperture of the budded cell at the time of anaphase onset, is required for the fidelity of nuclear segregation. The mechanisms that contribute to this process include establishment of spindle pole asymmetry (Vallen et al., 1992; Shaw et al., 1997b), directional cues to orient one spindle pole toward the neck (Heil-Chapdelaine et al., 1999; Lee et al., 1999; Miller et al., 1999), and pulling forces to guide the separated spindle pole bodies and associated chromatin into mother and bud (Yeh et al., 1995). Cytoplasmic motor proteins play a key role in this process via regulation of microtubule dynamics (dynein, Yeh et al., 1995; Kip2p and Kip3p, Cottingham and Hoyt, 1997; DeZwaan et al., 1997; Miller et al., 1998; and Kar3p, Cottingham et al., 1999), and generation of pulling or pushing forces along astral microtubule. It has been difficult to determine the role of any single motor and/or anchor as they have redundant functions in this highly dynamic process. Using a combination of genetics and dynamic imaging of intracellular structures, we have been able to distinguish temporal and functional roles for microtubule-based motor proteins, microtubule dynamics, and key spindle positioning determinants.
Several spindle polarity determinants including Kar9p, Bud6p and Bni1p provide positional cues for directing the nucleus to the bud (this study, (Lee et al., 1999; Miller et al., 1999)). Their role in initial spindle orientation is to promote nuclear orientation to the bud via astral microtubules. Stable microtubule attachments to cortical sites in the bud (persistence > 3 min.) have not been observed in vegetative cell growth (Carminati and Stearns, 1997; Shaw et al., 1997b; Maddox et al., 1999), and therefore a simple tethering mechanism for astral microtubules is unlikely. An alternative mechanism wherein these proteins couple microtubule dynamics to force generation (Lombillo et al., 1995) is more consistent with the dynamic aspects of microtubules observed in live cells, as well as Bud6p protein dynamics (Beach et al., 1999). Transiently tethering Kar9p to cortical sites in the neck or bud via Bud6p and Bni1p may result in plus-end microtubule disassembly accompanied by net movement of the nucleus toward the dynamic plus end. In support of this hypothesis Kar9p has recently been shown to bind microtubule-plus ends via Bim1p (Korinek et al., 2000; Lee et al., 2000), and in live cells, microtubule growth and shortening while tethered to the shmoo tip in mating cells is responsible for nuclear movement toward and away from the shmoo tip (Maddox et al., 1999).
Redundancies in spindle positioning are evident from examining the mitotic spindle before anaphase onset. Mitotic spindles are indeed directed toward the bud in bud6, bni1, or kar9 mutants but are slightly displaced from the neck at the time of anaphase onset (Figure 4). In addition, spindles in these mutants are not aligned along the mother-bud axis and exhibit heightened oscillations (increased frequency, length, and duration) relative to wild-type cells (Figure 5). Thus bud6, bni1, kip3, or kar9 are required to attenuate or antagonize mechanisms of spindle movement before anaphase onset. Cytoplasmic dynein provides the primary force for spindle oscillations, as dhc1 spindles are virtually static at the bud neck and remain aligned along the mother-bud axis (dhc1, Figure 5). In vitro motility studies indicate that, when conventional kinesin and axonemal dynein are loaded at similar concentrations on the same microtubule, microtubule translocation proceeds in a kinesin-directed manner, i.e. conventional kinesin is dominant to dynein (Vale et al., 1992). If Kip3p is a conventional kinesin-like protein, it could mask dynein function in vivo as well. Upon deletion of Kip3p or potential effectors of Kip3p (Bud6p, Bni1p, Kar9p), dynein function is revealed in increased frequency and duration of spindle oscillations (Figures 5 and 7). Analysis of spindle oscillations revealed two important functional aspects of motor and spindle positioning determinants. First, cytoplasmic dynein functions before anaphase onset, and second, spindle positioning determinants (bud6, bni1, and kar9) act to attenuate spindle oscillations, perhaps through regulating cytoplasmic dynein before anaphase onset.
The live cell assay provides an opportunity to measure the velocity of the dynein-generated pulling force. Translocation into the bud in the absence of spindle elongation proceeds at 3.6 μm/min, considerably greater than the 1 μm/min spindle elongation characteristic of the fast phase of anaphase B (Kahana et al., 1995; Yeh et al., 1995). Spindle translocation into the bud is governed by the rate of spindle elongation, and/or astral microtubule assembly/disassembly at the cell cortex. Nuclear maintenance at the neck is therefore not a “static” phase of the nuclear/spindle cycle. Rather, we propose that early Kip3p function, in addition to providing directional cues for initial spindle orientation, antagonizes cytoplasmic dynein. Preanaphase spindle oscillations at and through the neck reflect this balance between Kip3p and dynein. Upon anaphase onset, forces from spindle elongation and limitations in microtubule dynamics prevent dynein from pulling the entire nucleus into the bud.
We have attempted to determine the hierarchy of cytoplasmic motor function in yeast using mutants that fail to assemble a bipolar spindle but contain two spindle pole bodies, each capable of nucleating astral microtubules. The newly formed spindle pole body in ndc1–1 mutants is not correctly assembled into the nuclear envelope, and thus the bipolar spindle fails to assemble. In ndc1–1 cells at the restrictive temperature, the nucleus and associated chromatin DNA is either deposited into the bud or remains in the mother cell (∼ 50:50 bud:mother)(Thomas and Botstein, 1986; Yeh et al., 1995). In the absence of dynein, the nuclear mass is not translocated to the bud (<3% in bud) (Yeh et al., 1995). In contrast, the nuclear mass is translocated into the bud (23.5%) in the presence or absence of kip3 (Figure 9). Thus dynein is dominant to Kip3p in its ability to pull the nucleus into the bud in the absence of forces from spindle elongation. Spindle retention at the neck may reflect this hierarchy of motor function. The early force directing spindle migration to the neck may be weak and lack sufficient power to pull the spindle through the aperture of budded cells. Dynein provides the power for getting the nucleus through the neck upon anaphase onset.
The pattern of distribution of the asymmetric determinants, Bud6p (Amberg et al., 1997; Beach et al., 1999) and Bni1p make it unlikely that these proteins interact exclusively with elements of the microtubule cytoskeleton. Bud6p and Bni1p more likely recruit or anchor other key components (e.g., Kar9p) that provide cortical linkage to microtubules (Bloom and Beach, 1999). Bni1p was identified as a She mutant (she5, symmetric HO expression (Bobola et al., 1996), and she mutants are defective in either the formation of an Ash1 mRNA particle or the polar distribution of ASH1 mRNA. Beach et al. (1999), using a live cell assay for mRNA dynamics, demonstrated that ASH1 mRNA is distributed to the bud of large budded cells in bni1 mutants, but ASH1 mRNA in the bud is delocalized. Thus bni1 mutants lack an asymmetric determinant that keeps mRNA distributed to the tip of budded cells (Bloom and Beach, 1999). Bud6 has a slight she phenotype (Beach and Bloom, unpublished), and ASH1 mRNA is also delocalized in bud6 mutants (Beach et al., 1999). Bud6p and Bni1p may therefore comprise a multipurpose cortical scaffold that directs several elements (microtubules, mRNA) to sites of polarized growth. Interaction between cytoskeletal elements and cortical sites is likely to proceed via individual effector molecules, with Kar9p as the primary candidate for directing the microtubule cytoskeleton. In contrast to bni1 and bud6, kar9 mutants show no defect in ASH1 mRNA localization (Beach and Bloom, unpublished results).
The spindle polarity determinants (Bud6p, Bni1p, and Kar9p) must recognize sites of polarized growth or other asymmetries in cell patterning. In yeast, polarized cell growth is initialized by the selection of bud-site (Chant and Pringle, 1995; Pringle et al., 1995), followed by polarization of the actin cytoskeleton, spindle orientation, nuclear migration, and other events including ASH1 mRNA deposition. How various cytoskeletal elements interact with a specific local at a specific time is central to both cell and developmental processes. We posit that construction of a multipurpose scaffold (defined by Bud6p and Bni1p) that in turn binds different effectors (Kar9p for microtubules, She proteins for mRNA) is one mechanism for attaining specificity. These effectors, at least for nuclear migration, are likely to interact with the cytoplasmic microtubule-based motor proteins that can generate force from transient interactions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dale Beach, Doug Thrower, Chad Pearson, and members of the Bloom and Salmon laboratories for helpful discussions. C. Yang was supported in part by a fellowship for Undergraduate Research from the University of North Carolina at Chapel Hill.
This work was supported by research grants from the National Institutes of Health (K. Bloom, GM-32238; D.J. Lew, GM-53050; and E.D. Salmon, GM-24364).
Footnotes
Online version of this article contains video material.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Cooper JA. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J Cell Biol. 1992;117:1067–1076. doi: 10.1083/jcb.117.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a Yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Bloom K, Beach DL. mRNA localization: Motile mRNA, asymmetric anchors. Curr. Opin. Microbiol: Genet. Dev. 1999;2:604–609. doi: 10.1016/s1369-5274(99)00029-6. [DOI] [PubMed] [Google Scholar]
- Bobola N, Jansen R-P, Shin TH, Nasmyth K. Asymmetric accumulation of ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. Movement of a cytokinesis factor Cdc12p to the site of cell division. Curr Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Gheber L, Miller DL, Hoyt MA. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–349. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Gonczy P, Hyman AA. Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 1996;6:382–387. doi: 10.1016/0962-8924(96)10035-0. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Adames NR, Cooper JA. Formin' the connection between microtubules and the cell cortex. J Cell Biol. 1999;144:809–811. doi: 10.1083/jcb.144.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Tran NK, Cooper JA. Dynein-dependent movements of the mitotic spindle in Saccharomyces cerevisiae do not require filamentous actin. Mol Biol Cell. 2000;11:863–872. doi: 10.1091/mbc.11.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Amberg DC. The secretory pathway mediates localization of the cell polarity regulator Aip3p/Bud6p. Mol Biol Cell. 2000;11:647–661. doi: 10.1091/mbc.11.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova TS, Tatchell K, Cooper JA. Actin filaments in yeast are unstable in the absence of capping protein or fimbrin. J Cell Biol. 1995;131:1483–1493. doi: 10.1083/jcb.131.6.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek WS, Copeland MJ, Chaudhuri A, Chant J. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 2000;287:2257–2259. doi: 10.1126/science.287.5461.2257. [DOI] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Bonne C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Tirnauer JS, Li J, Schuyler SC, Liu JY, Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Li YY, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bretscher A. Characterization of TPM1 disrupted yeast cells indicates an involvement of tropomyosin in directed vesicular transport. J Cell Biol. 1992;118:285–299. doi: 10.1083/jcb.118.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombillo VA, Stewart RJ, McIntosh JR. Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature. 1995;373:161–164. doi: 10.1038/373161a0. [DOI] [PubMed] [Google Scholar]
- Maddox P, Bloom K, Salmon ED. Polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P, Chin E, Mallavarapu A, Yeh E, Salmon ED, Bloom K. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1999;144:977–987. doi: 10.1083/jcb.144.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Matheos D, Rose M. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Rose MD. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondesert G, Reed RI. BED1, a gene encoding a galactosyltransferase homologue, is required for polarized growth and efficient bud emergence in Saccharomyces cerevisiae. J Cell Biol. 1996;132:137–143. doi: 10.1083/jcb.132.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JB, Ris H. Electron-microscopic study of the spindle and chromosome movement in the yeast Saccharomyces cerevisiae. J Cell Sci. 1976;22:219–242. doi: 10.1242/jcs.22.2.219. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De VC, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. (Review) (111 refs) Cold Spring Harb Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1991. [Google Scholar]
- Salmon ED, Shaw SL, Waters J, Waterman-Storer CM, Maddox PS, Yeh E, Bloom K. A high-resolution multimode digital microscope system. Methods Cell Biol. 1998;56:185–215. [PMC free article] [PubMed] [Google Scholar]
- Segal M, Clarke DJ, Maddox P, Salmon ED, Bloom K, Reed SJ. Coordinated spindle assembly and orientation requires clb5p-dependent kinase in budding yeast. J Cell Biol. 2000;148:441–451. doi: 10.1083/jcb.148.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Bloom K, Salmon ED. Imaging green fluorescent protein fusion proteins in Saccharomyces cerevisiae. Curr Biol. 1997a;7:701–704. doi: 10.1016/s0960-9822(06)00299-5. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: A microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. JCell Biol. 1997b;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The SPA2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theesfeld CL, Irazoqui JE, Bloom K, Lew DJ. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1999;146:1019–1032. doi: 10.1083/jcb.146.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Botstein D. A gene required for the separation of chromosomes on the spindle apparatus in yeast. Cell. 1986;44:65–76. doi: 10.1016/0092-8674(86)90485-x. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, O'Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bimlp promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Malik F, Brown D. Directional instability of microtubule transport in the presence of kinesin and dynein, two opposite polarity motor proteins. J Cell Biol. 1992;119:1589–1596. doi: 10.1083/jcb.119.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.