Abstract
A number of peroxisome-associated proteins have been described that are involved in the import of proteins into peroxisomes, among which is the receptor for peroxisomal targeting signal 1 (PTS1) proteins Pex5p, the integral membrane protein Pex13p, which contains an Src homology 3 (SH3) domain, and the peripheral membrane protein Pex14p. In the yeast Saccharomyces cerevisiae, both Pex5p and Pex14p are able to bind Pex13p via its SH3 domain. Pex14p contains the classical SH3 binding motif PXXP, whereas this sequence is absent in Pex5p. Mutation of the conserved tryptophan in the PXXP binding pocket of Pex13-SH3 abolished interaction with Pex14p, but did not affect interaction with Pex5p, suggesting that Pex14p is the classical SH3 domain ligand and that Pex5p binds the SH3 domain in an alternative way. To identify the SH3 binding site in Pex5p, we screened a randomly mutagenized PEX5 library for loss of interaction with Pex13-SH3. Such mutations were all located in a small region in the N-terminal half of Pex5p. One of the altered residues (F208) was part of the sequence W204XXQF208, that is conserved between Pex5 proteins of different species. Site-directed mutagenesis of Trp204 confirmed the essential role of this motif in recognition of the SH3 domain. The Pex5p mutants could only partially restore PTS1-protein import in pex5Δ cells in vivo. In vitro binding studies showed that these Pex5p mutants failed to interact with Pex13-SH3 in the absence of Pex14p, but regained their ability to bind in the presence of Pex14p, suggesting the formation of a heterotrimeric complex consisting of Pex5p, Pex14p, and Pex13-SH3. In vivo, these Pex5p mutants, like wild-type Pex5p, were still found to be associated with peroxisomes. Taken together, this indicates that in the absence of Pex13-SH3 interaction, other protein(s) is able to bind Pex5p at the peroxisome; Pex14p is a likely candidate for this function.
INTRODUCTION
Peroxisomes are ubiquitous organelles bound by a single membrane that are present in almost all eukaryotic cells. Genetic screens in yeasts and in Chinese hamster ovary cell lines, and analysis of cells from patients with peroxisomal diseases have resulted in the identification of at least 23 genes encoding Pex proteins (peroxins) that play a role in the biogenesis of the peroxisome (a recent update can be viewed on the Web site www.mips.biochem.mpg.de/proj/yeast/reviews/pex_table.html). Most peroxins function in the import of matrix proteins into the peroxisome (reviewed in Erdmann et al., 1997; Subramani, 1998; Hettema et al., 1999; Tabak et al., 1999). Exceptions are Pex3p, Pex16p, and Pex19p, which are required for the proper localization of peroxisomal membrane proteins (Honsho et al., 1998; Kinoshita et al., 1998; Matsuzono et al., 1999; Snyder et al., 1999; South and Gould, 1999; Hettema et al., 2000). Proteins that reside in the peroxisomal matrix are synthesized on free polyribosomes in the cytosol and are posttranslationally imported into the peroxisome (Lazarow and Fujiki, 1985). The majority of these matrix proteins contain the peroxisomal targeting signal type I (PTS1) that consists of the carboxyl-terminal tripeptide SKL or a derivative thereof (Gould et al., 1989; Purdue and Lazarow, 1994; Elgersma et al., 1996b). Only a few proteins contain a PTS2, which is located in the N-terminal part of the protein (Osumi et al., 1991; Swinkels et al., 1991; Purdue and Lazarow, 1994). The PTSs are specifically recognized by their matching soluble receptors Pex5p (for PTS1 proteins) (Dodt et al., 1995; Wiemer et al., 1995; Elgersma et al., 1996a; Gould et al., 1996) or Pex7p (for PTS2 proteins) (Marzioch et al., 1994; Elgersma et al., 1998). In yeast, both receptors are able to function independently of each other, establishing separate cytosolic PTS1 and PTS2 protein-import routes (Subramani, 1996; reviewed in Erdmann et al., 1997; Hettema et al., 1999). Receptors with bound PTS proteins converge on a common translocation machinery. Two proteins of this machinery, Pex13p and Pex14p, have been shown to interact with Pex5p and Pex7p, implying a role for Pex13p and Pex14p in docking of the receptors (Elgersma et al., 1996a; Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997; Brocard et al., 1997; Fransen et al., 1998; Girzalsky et al., 1999; Schliebs et al., 1999). Pex13p and Pex14p form a complex with a third peroxin, Pex17p, which was characterized as a peripheral peroxisomal membrane protein (Huhse et al., 1998). Furthermore, three other peroxins have been suggested to play a role in the PTS import pathway downstream of the membrane-docking event. These are Pex10p, Pex12p, and Pex4p (Van der Klei et al., 1998; Chang et al., 1999).
Pex13p is an integral peroxisomal membrane protein possessing a C-terminal SH3 domain exposed to the cytosol. Src homology 3 (SH3) domains constitute a family of protein–protein interaction modules that participate in diverse signaling pathways (Pawson and Scott, 1997). X-ray crystallography, and nuclear magnetic resonance techniques have now resolved the three-dimensional structure of various SH3 domains and their contact sites with peptide ligands. Highly conserved aromatic amino acid residues form a hydrophobic binding pocket for typical polyproline helix structures, usually composed of two prolines spaced by two amino acids (PXXP motif) (Ren et al., 1993; Lim et al., 1994; Yu et al., 1994). Motifs containing a single proline have also been reported. For instance, binding of the SH3 domains of Hck and Src to an intramolecular peptide sequence in the protein requires only one proline residue (Sicheri et al., 1997; Xu et al., 1997). Recently, a novel ligand site has been identified for the Eps8-SH3 domain that conforms to the consensus sequence proline-X-X-aspartate-tyrosine (PXXDY) (Mongiovi et al., 1999). Cocrystallization of the Fyn SH3 domain and a high-affinity ligand peptide of Nef also showed that the (highly variable) RT-loop of the SH3 domain contributes to a higher binding affinity and specificity for the ligand by creating additional contact sites outside the PXXP motif (Lee et al., 1996).
The SH3 domain of Pex13p was shown to interact with both Pex5p and Pex14p (Elgersma et al., 1996a; Erdmann and Blobel, 1996; Gould et al., 1996; Albertini et al., 1997; Brocard et al., 1997; Girzalsky et al., 1999). The interaction with Pex14p is dependent on a typical PXXP motif (PTLPHR) present in the N-terminal half of the protein (Girzalsky et al., 1999). The second SH3 domain-binding partner Pex5p, however, does not possess a recognizable PXXP motif. A key issue that remains to be resolved is how Pex5p contacts the SH3 domain of Pex13p.
Here we report the identification of the region in Pex5p that is responsible for interaction with Pex13-SH3, based on a two-hybrid screen with a pex5 mutant library. Mutations locate in or near a motif, W204XXQF208, that is conserved between Pex5p proteins of different species and does not resemble a canonical PXXP motif. Moreover, binding of Pex5p to Pex13-SH3 containing a mutation in either the RT-loop (E320K) or in one of the aromatic residues of the PXXP binding cleft (W349A) was not affected, whereas binding of Pex14p to these mutants was destroyed, suggesting that Pex5p contacts a nonclassical binding site on Pex13-SH3. In vivo, pex5 mutants that had lost SH3 domain binding displayed a partially disturbed PTS1 protein import and showed reduced ability to grow on oleate. Mutant Pex5p was still partially associated with peroxisomes like in wild-type cells, indicating that the interaction with Pex13-SH3 is not solely responsible for membrane association of Pex5p. Because we could show that Pex14p can form a bridge between Pex13-SH3 and mutant Pex5p in vitro, we suggest that Pex14p might function as an alternative docking site in vivo.
MATERIALS AND METHODS
Yeast Strains and Culture Conditions
The yeast strains used in this study were Saccharomyces cerevisiae BJ1991 (MATα, leu2, trp1, ura3-251, prb1-1122, pep4-3, gal2), pex5Δ (MATα pex5::LEU2, leu2, trp1, ura3-251, prb1-1122, pep4-3, gal2) (Van der Leij et al., 1993), PCY2 (MATα, Δgal4, Δgal80, URA3::GAL1-LacZ, lys2-801, his3-Δ200, trp1-Δ63, leu2, ade2-101), PCY2pex5Δ (as PCY2 plus pex5::LYS2, ura3::KanMX), HF7c (MATa, ura3-52, his3-200, ade2-101, lys2-801, trp1-901, leu2-3112, gal4-542, gal80-538, LYS2::GAL1UAS, GAL1TATA-HIS3 URA3::GAL417mers(x3)-CyC1TATA-LacZ). Yeast transformants were selected and grown on minimal medium containing 0.67% yeast nitrogen base without amino acids (YNB-WO; Difco, Detroit, MI), 2% glucose, and amino acids (20–30 μg/ml) as needed. For subcellular fractionations and Nycodenz gradients, log-phase cells grown on 0.3% glucose media were shifted to oleate media containing 0.5% potassium phosphate buffer pH 6.0, 0.1% oleate, 0.2% Tween 40, 0.67% YNB-WO, and amino acids (20–30 μg/ml) as needed. To follow growth on oleate, log-phase cells were grown on 0.3% glucose and shifted to oleate media containing 0.5% potassium phosphate buffer pH 6.0, 0.5% peptone, and 0.3% yeast extract at 2 × 104cells/ml (OD600 = 0.001). Oleate plates contained 0.5% potassium phosphate buffer pH 6.0, 0.1% oleate, 0.5% Tween 40, 0.67% YNB-WO, and amino acids as needed.
Plasmids and Cloning Procedures
Plasmids encoding GAL4 DB fusions of Pex13-SH3(284-386) and Pex13-SH3(284-358) were described previously (Elgersma et al., 1996a). To generate GAL4 DNA-binding domain (DB) fusions with Pex13-SH3(301-386) (pGB17) and Pex13-SH3(310-386) (pGB16), polymerase chain reaction (PCR) was performed with primers P257, P258, and P256 (Table 1) on GAL4 DB PEX13-SH3(284-386) as template. The PCR products were digested with EcoRI and SpeI and cloned between the EcoRI and SpeI sites of pPC97 (Chevray and Nathans, 1992). Pex13-SH3(304-377) (pGB15) was obtained by cutting MTP 429 (a kind gift from M.T. Pisabarro, Genentech, San Francisco, CA) with NcoI and making the ends blunt with Klenow polymerase. After digestion with BamHI, the fragment was cloned between the SmaI and BglII sites of pPC97 (pGB19). To introduce the E320K mutation in pGB17 the plasmid was cut with BstBI and SpeI and the obtained fragment was exchanged for the BstBI-SpeI fragment from plasmid 20.50 (Elgersma et al., 1996a). GAL4 activation domain (AD) fusion with PEX5 (pAN4) will be described in detail elsewhere (Klein, Barnett, Bottger, Konings, Tabak, Distel, unpublished data). The PEX14 open reading frame was generated by PCR on genomic DNA of S. cerevisiae with primers P243 and P244 (Table 1). The PCR fragment was cut with BamHI and PstI and ligated into the pUC19 vector creating pGB4. GAL4 DB or GAL4 AD fusions were generated by digestion of pGB4 with EcoRI and SpeI and ligation of the PEX14 fragment between the EcoRI and SpeI sites of pPC86 or pPC97 (Chevray and Nathans, 1992). GAL4 DB fused to MDH3 SKL was generated by cutting pEL102 (Elgersma et al., 1996b) with BamHI, making the ends blunt with Klenow polymerase. After digestion with SpeI, the fragment was cloned between the SmaI and BglII sites of pPC97. The two-hybrid plasmid encoding GAL4 DB Pex8p was a kind gift from Dr. W.H. Kunau (Bochum, Federal Republic of Germany). All PCR fragments were verified by sequencing.
Table 1.
Primer sequences used for PCR and site-directed mutagenesis
| Primer | DNA sequence (5′ → 3′) | Purpose |
|---|---|---|
| P257 | CGGAATTCTAGGATCCGAGCCTATTGATCCTTCG | Forward SH3(301) |
| P258 | CGGAATTCTAGGATCCTTTGCAAGAGCGTTATATGAT | Forward SH3(310) |
| P256 | TTTTCTGCAGACTAGTGTGTACGCGTTTC | Reverse SH3(386) |
| P243 | CGGGATCCATGAGTGACGTGGTCAGTAAAG | Forward Pex14-ATG |
| P244 | AACTGCAGCTATGGGATGGAGTCTTCGAC | Reverse Stop-Pex14 |
| SH3: | ||
| W349A5′ | GGGAGGGATTCTGACGCCTGGAAAGTGAGGA | Site-directed mutagenesis |
| W349A3′ | TCCTCACTTTCCAGGCGTCAGAATCCCTCCC | Site-directed mutagenesis |
| Pex5p: | ||
| W204A5′ | GAGCAAGAACAACAACCCGCGACAGATCAGTTTG | Site-directed mutagenesis |
| W204A3′ | CAAACTGATCTGTCGCCGGGTTGTTGTTCTTGCTC | Site-directed mutagenesis |
| F208L5′ | CCCTGGACAGATCAGTTAGAAAAGCTGGAAAAAG | Site-directed mutagenesis |
| F208L3′ | CTTTTTCCAGCTTTTCTAACTGATCTGTCCAGGG | Site-directed mutagenesis |
| E212V5′ | CAGATCAGTTTGAAAAGCTGGTAAAAGAAGTCTCAGAAAACTTG | Site-directed mutagenesis |
| E212V3′ | CAAGTTTTCTGAGACTTCTTTTACCAGCTTTTCAAACTGATCTG | Site-directed mutagenesis |
| TRP5′ | CAGTTAGAAAAGCTGGTAAAAGGAGTCTCAGAAAACTTGG | Site-directed mutagenesis |
| TRP3′ | CCAAGTTTTCTGAGACTCCTTTTACCAGCTTTTCTAACTG | Site-directed mutagenesis |
Point mutations in PEX5 were introduced using the Quick-change site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers were used as listed in Table 1. As template pAN4 was used. To introduce the triple mutation Pex5p(F208L, E212V, E214G), the yeast-expression plasmid encoding Pex5p(F208L) under the control of the PEX5 promoter was used as a template. The introduced base pair changes were verified by sequencing. To create plasmids for expression of Pex5p in yeast, the PEX5 promoter was obtained from the genomic library plasmid originally isolated by Van der Leij et al. (1992). The plasmid was digested with XbaI (located 488 nucleotides upstream of the PEX5 start codon) and the ends were made blunt with Klenow polymerase, and subsequently digested with BamHI. This fragment was ligated between the blunted SacI site and the BamHI sites of the yeast expression vector Ycplac33 (Gietz and Sugino, 1988), generating pEL91. PEX5 was obtained from pAN4 or mutant plasmids derived from pAN4, by digestion of the plasmid with BamHI and HindIII. PEX5 fragments were cloned between the BamHI and HindIII sites of pEL91. Wild-type PEX5 cloned this way was fully capable of complementing the growth defect on oleate of the pex5Δ strain.
To create glutathione S-transferase (GST) fusions of Pex5p for expression in Escherichia coli, PEX5 inserts were excised from pAN4 (wild-type) or from mutant plasmids derived from pAN4 (F208L and E212V, described above) with NcoI and HindIII. The fragments were ligated between the NcoI and HindIII restriction sites of pRP265nb (a kind gift from Dr. B. Werten, Utrecht, The Netherlands) resulting in in-frame fusions of GST with Pex5p. To generate maltose-binding-protein (MBP) fusions with the SH3 domain, the PCR product generated with primers 256 and 257 [SH3(301–386)] was cut with EcoRI and PstI and cloned between the EcoRI and PstI restriction sites of pUC19, creating pGB7. For introduction of the E320K mutation into pGB7, plasmid 20.50 was cut with BstBI and SpeI, and the SH3 fragment containing the mutation was exchanged for the BstBI-SpeI fragment of pGB7, generating pGB18. Wild-type and mutant (E320K) SH3 fragments were isolated by cutting plasmids pGB7 and pGB18 with BamHI and PstI, respectively. The obtained fragments were cloned into pMALc2 (New England Biolabs, Beverly, MA) digested with BamHI and PstI. MBP fusion of Pex14p was obtained by cutting pGB4 with BamHI and PstI, and ligation of the PEX14 fragment into pMALc2 (described above). Digestion of pGB4 with BamHI and PstI and by ligating the PEX14 fragment between the BamHI and PstI restriction of pQE9 (Qiagen, Chatsworth, CA) created a 6xHis fusion of Pex14p.
Plasmids for expression of green fluorescent protein fused to SKL (GFP-SKL) and N-terminal hemagglutinin-tagged (NH) Mdh3p in yeast are discribed elsewhere (Elgersma et al., 1996b; Hettema et al., 1998). To create plasmids for overexpression of Pex13p and Pex13p(E320K) in yeast, plasmids 20.46 and 20.50 (Elgersma et al., 1996a) were cut with SacI and HindIII and PEX13 fragments were cloned behind the catalase A (CTA1) promoter (pEL30, described in Elgersma et al., 1993) digested with SacI and HindIII. For overexpression of Pex14p, pGB4 was cut with BamHI and PstI and the PEX14 fragment was ligated between the BamHI and PstI sites of pEL30. For overexpression of Pex5p, pAN1 (Klein, Barnett, Bottger, Konings, Tabak, Distel, unpublished data) was digested with BamHI and HindIII and the PEX5 fragment was cloned behind the CTAI promoter in 2 μ plasmid (pEL26, Elgersma et al., 1993).
In Vitro Binding Assay
All in vitro assays were set up according to the following regimen. Cultures (250 ml) of E. coli BL21 cells expressing either MBP or GST fusion proteins were induced with 1 mM isopropyl β-d-thiogalactoside and centrifuged; cell pellets were resuspended in 5 ml of phosphate-buffered saline (PBS; 100 mM sodium phosphate buffer pH 7.4, 140 mM NaCl, 2 mM phenylmethylsulfonyl fluoride [PMSF]). Cell suspensions were subsequently lysed by sonication. All GST constructs used for binding assays with MBP fusions were purified on glutathione S-sepharose (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's recommendations. A 200-μl amylose resin column was equilibrated in PBS and subsequently loaded with 250 μl of a bacterial lysate containing the appropriate MBP fusion. The resin was then washed with 1 ml of PBS. The GST fusion (100 μg) was then run through the column at a flow rate of ∼100 μl/min. The column was then washed with 3 ml of PBS and subsequently eluted with 500 μl of 10 mM maltose in PBS. Fractions were collected and subjected to SDS-PAGE and Western blot analysis. In vitro experiments involving 6x His-tagged Pex14p were conducted similarly except that before loading of the GST fusion, 200 μl of a bacterial lysate containing 6xHis-fused Pex14p was loaded and the column washed with 1 ml of PBS. The protocol then continued with GST fusion loading as described above.
Pex5 Mutant Screen and Two-Hybrid Assays
Random mutations were introduced in the PEX5 gene by error-prone PCR on plasmid pAN1. pAN1 contains the complete PEX5 open reading frame with a unique XbaI site at position 1140, which was introduced by site-directed mutagenesis. PCR was carried under standard conditions with the nonproofreading Taq DNA polymerase. The PCR product was digested with XbaI and BamHI and ligated into pAN1 to create the N-terminal library composed of mutagenized nucleotides 1–1140 (amino acids 1–380) and the wild-type C terminus of the protein. To create the C-terminal library the PCR product was digested with XbaI and PstI and the mutagenized nucleotides 1441–1836 (amino acids 381–612) were ligated into XbaI-PstI–digested pAN1. Sequence analysis of 20 randomly picked clones revealed that approximately one nucleotide in every 550 nucleotides was mutated. Both libraries were cloned between the EcoRI and SpeI sites of the two-hybrid plasmid pPC86, generating GAL4 AD fusions. One microgram of each two-hybrid library was transformed to the yeast two-hybrid strain HF7c containing the GAL4 DB Pex13-SH3(284-386) plasmid, and double transformants were selected on glucose plates without leucine and tryptophan. Colonies were replica-plated onto glucose plates without leucine, tryptophan, and histidine; 15,000 colonies of the C-terminal PEX5 library and 1,500 colonies of the N-terminal PEX5 library were screened, yielding 130 and 75 clones, respectively, that failed to grow in the absence of histidine. These colonies were selected and pex5 mutant plasmids were rescued from these colonies for further analysis. β-Galactosidase filter assays were performed as described by Fields and Song (1989).
Quantification of β-galactosidase activity was performed with the Galacto-Light kit (Tropix, Bedford, MA). Double-transformed PCY2 cells (10 OD units) were harvested, washed with distilled H2O, and resuspended in 200 μl of breaking buffer (100 mM Tris pH 7.5, 20% vol/vol glycerol, 1 mM PMSF) plus 0.4 g of glass beads and lysed by mixing on a vortex for 30 min. The homogenates were centrifuged for 15 min at 13,000 × g and the cleared lysates were used to measure β-galactosidase activity. Protein concentrations were determined with the method described by Bradford (1976).
Subcellular Fractionation and Gradient Analysis
One liter of oleate-grown transformants was converted to spheroplasts by using Zymolyase 100T (1 mg/g cells). Spheroplasts were washed with 1.2 M sorbitol in 2 [N-morpholino]ethanesulfonic acid (MES) buffer (5 mM MES pH 5.5, 1 mM KCl, 1 mM EDTA) and lysed by osmotic shock in MES buffer containing 0.65 M sorbitol and 1 mM PMSF. Intact cells and nuclei were removed by centrifuging twice at 600 × g for 2 min. The obtained postnuclear supernatants were centrifuged for 30 min at 20,000 × g. The volumes of the pellet fractions were made equal to the volumes of the supernatant fractions. For Nycodenz gradient analysis, pellet fractions were resuspended in 1 ml of hypotonic lysis buffer and loaded on top of a continuous 15–35% Nycodenz gradient (12 ml) underlaid with a 1-ml cushion of 50% Nycodenz in MES buffer containing 8.5% sucrose. Gradients were spun in an MSE-Europe 24 M centrifuge equipped with a vertical rotor for 2.5 h at 19,000 rpm. Fractions with a volume of 0.5 ml were collected and analyzed by SDS-PAGE and Western blotting.
SDS-PAGE, Western Blotting, and Enzyme Assays
Proteins were separated on 10% SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were blocked in PBS (pH 7.4) supplemented with 0.1% Tween 20 and 2% skimmed milk powder (Protifar). Blots were incubated with rabbit antibodies diluted in PBS with 0.1% Tween 20. The antibodies used were anti-Pex13p, anti-3-ketoacyl CoA-thiolase, anti-Pex5p (Elgersma et al., 1996a), and anti-Pat1p (Hettema et al., 1996). Anti-NH was a generous gift from Dr. P. van der Sluys (Utrecht, The Netherlands); anti-Hsp60 was a generous gift from Dr. S. Rospert, Basel, Switzerland. Polyclonal antisera for Pex14p were raised against the full-length Pex14 protein isolated as a 6xHis fusion protein from E. coli. Antibody complexes were detected by incubation with goat anti-rabbit Ig-conjugated alkaline phosphatase. 3-Hydroxyacyl-CoA dehydrogenase (3HAD) activity was measured on a Cobas-Fara centrifugal analyzer by following the 3-keto-octanoyl-CoA-dependent rate of NADH consumption at 340 nm (Wanders et al., 1990). Catalase A activity was measured as described by Lucke (1963).
RESULTS
Pex5p and Pex14p Bind Directly to the SH3 Domain of Pex13p
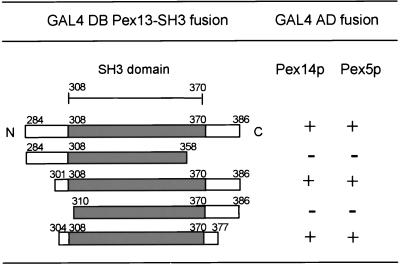
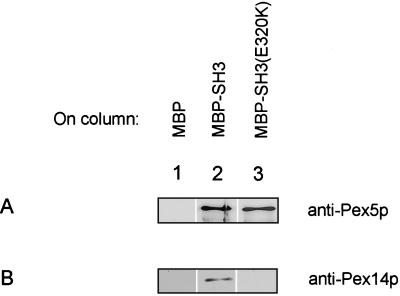
Based upon sequence alignment with other SH3 domains, the SH3 domain of Pex13p extends from amino acid 308 to 370. To determine the functional boundaries of this domain we constructed deleted versions of Pex13p (Figure 1). These constructs were tested in the two-hybrid system for interaction with Pex5p and Pex14p. Figure 1 shows that the SH3 domain flanked by four amino acids N-terminally and seven amino acids C-terminally was sufficient for interaction with Pex14p and Pex5p. Further deletion of either the N or C terminus disrupted the interactions. We performed in vitro reconstitution experiments to prove that these interactions are direct. A bacterial lysate containing MBP fused to the SH3 domain of Pex13p was loaded onto an amylose column. After washing, the column containing immobilized MBP-SH3 was incubated with extracts of bacteria expressing either a GST fusion of Pex5p or a 6xHis fusion of Pex14p. After washing, MBP-SH3 and bound proteins were eluted from the column with maltose. Proteins in the eluates were visualized by SDS-PAGE and Western blotting. Figure 2 shows that in separate binding experiments Pex5p (A) and Pex14p (B) were efficiently coeluted with MBP-SH3 (lanes 2) and did not bind to a column with MBP alone (lanes 1). These in vitro reconstitution assays indicate that Pex5p and Pex14p can bind to the Pex13-SH3 domain directly and independently of each other.
Figure 1.
Pex13-SH3 domain binds Pex5p and Pex14p in the two-hybrid assay. Truncated versions of Pex13-SH3 fused to GAL4 DB were cotransformed with GAL4 AD fusions of Pex5p or Pex14p into the yeast two-hybrid reporter strain PCY2. Interaction was monitored by determining β-galactosidase activity with a filter lift assay. “+” indicates blue staining of colonies within 1 h, “−” indicates that colonies remained white after incubation overnight.
Figure 2.
Pex13-SH3 domain interacts with Pex5p and with Pex14p in vitro. MBP, MBP-SH3(wild-type) and MBP-SH3(E320K) were expressed in E. coli and immobilized on amylose columns. E. coli lysates containing either GST-Pex5p (A) or 6xHisPex14p (B) were then passed over the columns. The columns were washed with five column volumes and bound proteins were eluted with maltose. Eluates were analyzed by SDS-PAGE and Western blotting by using antibodies specific for Pex14p and Pex5p.
pex5 Mutants Disturbed in Interaction with the Pex13-SH3 Domain
Pex14p contains a canonical SH3-binding motif, PXXP, and mutagenesis studies have shown that the two prolines within this motif are essential for its interaction with Pex13-SH3 (Girzalsky et al., 1999). Pex5p, however, does not contain a recognizable SH3 binding motif. To identify the region in Pex5p that contacts the SH3 domain, two libraries were constructed in which either the N-terminal or the C-terminal half of PEX5 was randomly mutagenized by error-prone PCR. These libraries were screened for mutants that had lost the interaction with Pex13-SH3 in the two-hybrid assay. Loss of binding was scored by the inability to grow on media lacking histidine. Such colonies were picked from the master plate and lysates were analyzed by Western blotting to verify that full-length Pex5p was expressed. The frequency of selected full-length pex5 mutants was much higher in the N-terminal library (5% of total) compared with the C-terminal library (0.9% of total). Moreover, all pex5 mutants isolated from the C-terminal library were either truncated or unstable and were not analyzed further. These findings suggest that the region in Pex5p involved in binding to the Pex13-SH3 domain is located in the N-terminal half of Pex5p. To exclude mutants with changes in overall structure, we tested two-hybrid interactions with other known partner proteins of Pex5p (Table 2). Five pex5 mutants were disturbed in binding to Pex13-SH3, but maintained interaction with Pex14p, a protein that binds the N-terminal half of Pex5p (Schliebs et al., 1999; our unpublished results), and Mdh3p, a PTS1 containing protein that binds to the C-terminal tetratricopeptide repeat (TPR) domains of Pex5p (Brocard et al., 1994; Klein, Barnett, Bottger, Konings, Tabak, Distel, unpublished data). Additionally, the interaction with Pex8p, a protein that contacts both the N-terminal and C-terminal half of Pex5p (Rehling et al., 2000), was also unaffected for these mutants. It is noteworthy that only mutant N19 had completely lost two-hybrid interaction with Pex13-SH3. Other mutants still displayed some growth in the absence of histidine, suggesting residual binding capacity with Pex13-SH3. We conclude that these pex5 mutants are specifically affected in binding the Pex13-SH3 domain and that the overall structure of these mutant proteins is still intact.
Table 2.
N-terminal pex5 mutants are selectively disturbed in two-hybrid interaction with Pex13-SH3 in HF7c
| GAL4 AD fusion
|
GAL4 DB fusion
|
|||
|---|---|---|---|---|
| Pex5 | Pex13-SH3 | Pex14p | PTS1 | Pex8p |
| Wild-type | ++ | ++ | ++ | ++ |
| N3 | +/− | ++ | ++ | ++ |
| N8 | +/- | ++ | ++ | ND |
| N19 | - | ++ | ++ | ++ |
| N84 | +/- | ++ | ++ | ND |
| N100 | +/- | ++ | ++ | ND |
Proteins were fused to GAL4 or GAL4 DB as indicated. Double transformants were grown on glucose plates lacking histidine. ++, growth after 3 d; +/-, growth after a minimum of 4 d; +/−, growth in small colonies after a minimum of 4 d; -, no growth; ND, not determined.
Pex5p Is a Non-PXXP Ligand for the Pex13-SH3 Domain
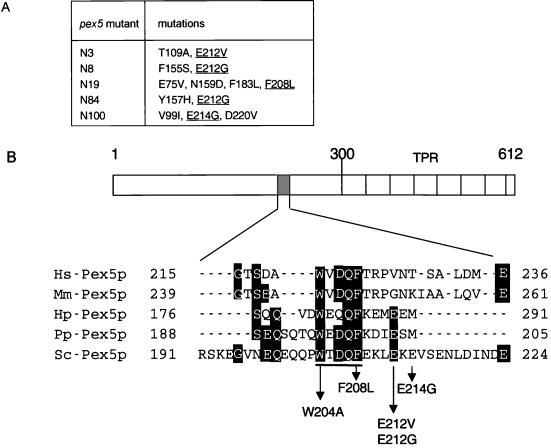
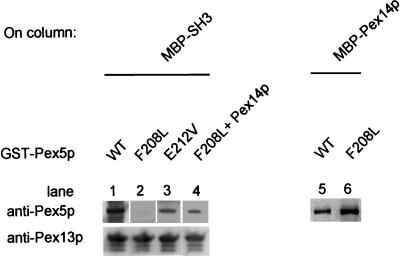
The five selected pex5 mutants were sequenced to determine the site of the mutations. All mutants contained multiple amino acid substitutions (Figure 3A). Three independent mutants were mutated in the same residue: glutamic acid 212 (E212). This residue was replaced by a valine (mutant N3), or a glycine (mutants N8 and N84). In addition, clones N19 and N100 had mutations in the same region (residues 208 and 214, respectively). These amino acid residues are in or near a block of amino acids, W204XXQF208 (where X stands for any amino acid), that is conserved between Pex5 proteins of yeast and higher eukaryotes (Figure 3B). To investigate which mutations were responsible for the loss of Pex13-SH3 domain binding, single amino acid substitutions were made using site-directed mutagenesis. Mutations were made at position 109(T109A) and position 212 (E212V) (both found to be mutated in mutant N3), and at position 208 (F208L) (found mutated in the quadruple mutant N19). These three single mutants were tested against Pex13-SH3 in the two-hybrid assay (Table 3). As a control, they were also tested for interaction with other Pex5p binding partners. Interactions were monitored by a quantitative β-galactosidase assay and by growth in the absence of histidine in the two-hybrid strains PCY2 and HF7c, respectively. The F208L mutation was sufficient to disrupt the two-hybrid interaction with Pex13-SH3. In addition, the E212V mutation disturbed the Pex13-SH3 interaction, although some growth in the absence of histidine could be detected. The T109A mutation showed a two-hybrid interaction with Pex13-SH3 comparable to wild-type Pex5p. The single mutants that had lost SH3-domain binding appeared not to be affected in their interaction with Pex14p and Mdh3p-SKL (Table 3; our unpublished results). These results indicate that E212 and F208, but not T109, are involved in Pex13-SH3 domain binding, but do not play a detectable role in the interaction with other Pex5p partners. The two-hybrid results were backed up by in vitro reconstitution experiments. Figure 4 shows that in contrast to wild-type GST-Pex5p (lane 1), GST-Pex5p(F208L) (lane 2) could not be coeluted with MBP-SH3, whereas a small amount of GST-Pex5p(E212V) (lane 3) was recovered from the elution. The F208L mutation did not affect in vitro binding to MBP-Pex14p. In separate binding experiments comparable amounts of wild-type GST-Pex5p (lane 5) and GST-Pex5p(F208L) (lane 6) could be coeluted with MBP-Pex14p from the column. In vitro binding of GST-Pex5p(E212V) to MBP-Pex14p appeared also not to be affected (our unpublished results). Together, these data indicate that residue F208 (and to a lesser extent residue E212) in Pex5p is essential for direct and specific contact of Pex5p with the SH3 domain.
Figure 3.
Amino acid substitutions in pex5 mutants selected for loss of two-hybrid interaction with Pex13-SH3 are clustered in the region between amino acids 208 and 214. (A) Amino acid substitutions in pex5 mutants selected for loss of interaction with Pex13-SH3. Mutations in or near the conserved W204XDQF208 motif are underlined. (B) Multiple sequence alignment (GeneInspector) of the region in Pex5p important for Pex13-SH3 interaction. Amino acid substitutions in pex5 mutants are indicated. The loss of interaction mutant W204A created by site-directed mutagenesis is also shown. The conserved WXDQF motif is underlined. Hs, Homo sapiens; Mm, Mus musculus; Hp, H. polymorpha; Pp, P. pastoris; Sc, S. cerevisiae.
Table 3.
Pex5p mutants are selectively disturbed in two-hybrid interaction with Pex13-SH3
| GAL4 AD fusion | GAL4 DB fusion | β-Galactosidase activity (RLU/mg protein ± SD) | Growth on his− |
|---|---|---|---|
| Pex5p(F208L) | — | 1.2 ± 0.1 | - |
| Pex5p | Pex13-SH3 | 4,800 ± 800 | + |
| Pex5p(T109A) | Pex13-SH3 | 3,300 ± 300 | + |
| Pex5p(E212V) | Pex13-SH3 | 2.0 ± 0.1 | +/− |
| Pex5p(F208L) | Pex13-SH3 | 1.7 ± 0.1 | - |
| Pex5p(W204A) | Pex13-SH3 | 2.3 ± 0.5 | +/- |
| Pex5p | Mdh3p | 6,900 ± 2,600 | + |
| Pex5p(F208L) | Mdh3p | 5,100 ± 500 | + |
| Pex5p | Pex14p | 10,300 ± 400 | + |
| Pex5p(F208L) | Pex14p | 3,200 ± 400 | + |
| Pex5p(W204A) | Pex14p | 12,000 ± 900 | ND |
Plasmids encoding GAL4 DB and GAL4 AD fusions as indicated were transformed to yeast two-hybrid strains PCY2 and HF7c. Two-hybrid interaction was quantitated in PCY2 by measuring β-galactosidase activity. Indicated is the mean of two measurements of duplo cultures ± SD. Interaction was also measured in HF7c by growth in the absence of histidine (see legend to Table 2). RLU, relative light units.
Figure 4.
Pex5p(F208L) and Pex5p(E212V) are disturbed in Pex13-SH3 binding, but maintain interaction with Pex14p in vitro. GST-Pex5p(wild-type) (WT) or mutant GST-Pex5p(F208L or E212V) were expressed in E. coli and passed over a column with immobilized MBP-SH3 (lanes 1–3) or MBP-Pex14p (lanes 5 and 6). Columns were washed, eluted, and analyzed as described below. Pex13-SH3, Pex14p, and Pex5p(F208L) can form a trimeric complex in vitro (lane 4): an E. coli lysate containing 6xHisPex14p was passed over an amylose column with immobilized MBP-SH3. After washing, an E. coli lysate containing GST-Pex5p(F208L) was passed over the column. Proteins were eluted with maltose and eluates were analyzed by SDS-PAGE and Western blotting by using antibodies specific for Pex13-SH3 and Pex5p.
To further investigate the role of the W204XXQF208 motif in Pex13-SH3 domain interaction, an additional pex5 mutant was created by site-directed mutagenesis. The strictly conserved tryptophan (W204) was mutated to alanine and tested in the two-hybrid assay. The W204A mutation disturbed interaction with Pex13-SH3, although some activation of the HIS3 reporter could be detected (Table 3). The binding of this mutant to Pex14p was completely unaffected. This data underscore the importance of the W204XXQF208 motif for Pex13-SH3 domain binding.
Pex5p and Pex14p Bind the Pex13-SH3 Domain in Different Ways
The presence of a nonclassical SH3 interaction motif in Pex5p raised the possibility that Pex5p may interact at a site on the Pex13-SH3 domain distinct from the PXXP binding pocket. To test this hypothesis we made use of two mutated forms of the Pex13p-SH3 domain. One mutation originates from a previously isolated mutant of Pex13p [Pex13p(E320K)] (Elgersma et al., 1996a). Pex13p(E320K) has a point mutation in the RT-loop of the SH3 domain. This loop has been shown to be important in determining the specificity of and affinity for SH3 ligands (Lee et al., 1995, 1996; Arold et al., 1998; Pisabarro et al., 1998). The second SH3 domain mutant was created by site-directed mutagenesis. This mutant contains an amino acid substitution in the conserved tryptophan that is part of the hydrophobic cleft, which forms the binding platform for polyproline ligands (Lim et al., 1994). The interaction of the wild-type and mutant SH3 domains with Pex14p and Pex5p was assayed in the two-hybrid system. β-Galactosidase activity was measured to quantitate the interaction strength. The results shown in Table 4 reveal that Pex14p is unable to interact with SH3(E320K) and SH3(W349A). However, Pex5p interaction with both SH3(E320K) and SH3(W349A) is largely unaffected. The controls included show that expression of either of the fusion proteins alone did not support the activation of the reporter genes. Similar results were obtained in an in vitro binding assay (Figure 2). E. coli-expressed 6xHis-Pex14p could be coeluted with MBP-SH3 (Figure 2B, lane 2), whereas in a parallel experiment 6xHis-Pex14p did not bind to MBP-SH3(E320K) because it did not appear in the eluate (Figure 2B, lane 3). Furthermore, GST-Pex5p could be coeluted with both wild-type MBP-SH3 (Figure 2A, lane 2) and MBP-SH3(E320K) (Figure 2A, lane 3), indicating that the direct interaction between Pex5p and mutant Pex13-SH3 is not affected. Taken together, these results show that the E320K and the W349A mutations affect Pex14p interaction, but do not interfere with Pex5p binding. They suggest, therefore, that Pex14p is the canonical SH3 domain ligand, whereas Pex5p binds the Pex13-SH3 domain in an alternative way.
Table 4.
Pex13-SH3 mutants have lost two-hybrid interaction with Pex14p but not with Pex5p
| GAL4 AD fusion | GAL4 DB fusion | β-Galactosidase activity (RLU/mg protein ± SD) | Growth on his− |
|---|---|---|---|
| — | — | 1.0 ± 0.1 | - |
| Pex5p | — | 0.5 ± 0.1 | - |
| Pex5p | Pex13-SH3 | 3,000 ± 900 | ++ |
| Pex5p | Pex13-SH3(E320K) | 3,500 ± 400 | ++ |
| Pex5p | Pex13-SH3(W349A) | 1,600 ± 800 | ND |
| — | Pex13-SH3 | 3.8 ± 1.0 | - |
| — | Pex13-SH3(E320K) | 3.1 ± 0.2 | - |
| — | Pex13-SH3(W349A) | 1.3 ± 0.1 | - |
| Pex14p | — | 0.8 ± 0.1 | - |
| Pex14p | Pex13-SH3 | 39 ± 12 | ++ |
| Pex14p | Pex13-SH3(E320K) | 2.7 ± 0.2 | - |
| Pex14p | Pex13-SH3(W349A) | 2.0 ± 0.5 | - |
Plasmids encoding GAL4 DB and GAL4 AD fusions as indicated were transformed to two-hybrid yeast strains PCY2 and HF7c. Two-hybrid interaction was quantitated in PCY2 by measuring the β-galactosidase activity as described in MATERIALS AND METHODS. Indicated numbers are the mean of two independent measurements in triplicate cultures ± SD. Two-hybrid interaction was measured in HF7c by growth in the absence of histidine (++, growth; -, no growth; ND, not determined). RLU, relative light units.
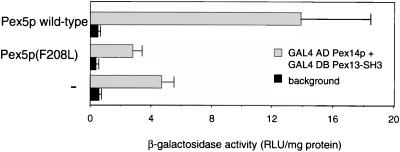
To obtain further support for this notion we investigated the effect of Pex5p expression on the two-hybrid interaction between Pex13-SH3 and Pex14p. A two-hybrid reporter strain isogenic to PCY2 was constructed in which the PEX5 gene was deleted (PCY2pex5Δ). This strain was transformed with plasmids encoding either wild-type or a mutant version of Pex5p under the control of the PEX5 promoter, or it was transformed with an empty expression vector. Figure 5 shows that deletion of endogenous Pex5p reduced the Pex13-SH3/Pex14p interaction about threefold, indicating that the strength of this interaction is dependent on the presence of Pex5p. Reexpression of the Pex5p(F208L) mutant that is specifically disturbed in SH3 interaction does not restore the SH3-Pex14p interaction to wild-type levels. Together, these results show that in vivo binding of Pex5p to Pex13-SH3 cooperatively stabilizes the SH3/Pex14p interaction, which suggests that Pex5p and Pex14p bind separate sites on the Pex13-SH3 domain.
Figure 5.
Two-hybrid interaction of Pex13-SH3 and Pex14p is reduced in the absence of wild-type Pex5p. A PCY2 strain harboring a deletion of the PEX5 gene (PCY2 pex5Δ) was transformed with expression constructs encoding either wild-type Pex5p or mutant Pex5p, or with an empty plasmid (−). The two-hybrid interaction of Pex13-SH3 and Pex14p was determined in these different strains by measuring the β-galactosidase activity. Indicated is the mean of two measurements in triplicate cultures ± SD. Background (▪) represents β-galactosidase activity in strains transformed with either GAL4DB-Pex13-SH3 or GAL4AD-Pex14p alone.
Pex13p and Pex14p Operate Stoichiometrically
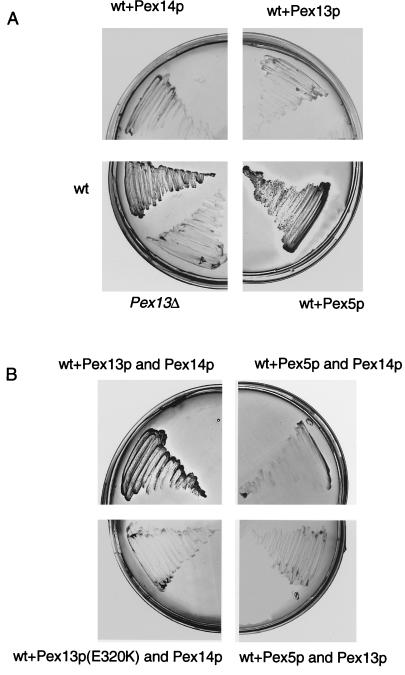
To further investigate complex formation in vivo we carried out experiments in which PEX13, PEX14, or PEX5 alone or in combination were overexpressed in wild-type cells. The transformed strains were subsequently tested for their ability to grow on oleate. Such experiments might reveal whether the proper stoichiometry of a protein is essential for peroxisome function. As shown in Figure 6A, overexpression of Pex13p under the control of the strong CTA1-promoter in wild-type cells leads to growth inhibition. Similarly, when Pex14p is expressed under the control of the CTA1 promoter, growth on oleate is also inhibited. However, simultaneous overexpression of Pex13p and Pex14p allows normal growth on oleate, whereas cooverexpression of the nonfunctional pex13 mutant E320K and Pex14p inhibits growth on oleate. Overexpression of Pex5p does not affect growth and is also not able to rescue the inhibitory effect of Pex13p or Pex14p overexpression on oleate (Figure 6B). We conclude that stoichiometry of Pex13p and Pex14p is required for correct peroxisomal function, which indicates close cooperation between these two peroxins.
Figure 6.
Growth characteristics on oleate of wild-type cells overexpressing Pex13p, Pex14p, and Pex5p under the control of the CTA1 promoter. Peroxins were expressed in wild-type cells either separately (A) or in combination (B) as indicated. Transformants are indicated with “+ Pex.” Positive and negative controls for growth are untransformed wild-type cells and the pex13Δ strain, respectively.
In Vivo Effects of pex5 Mutations F208L and E212V
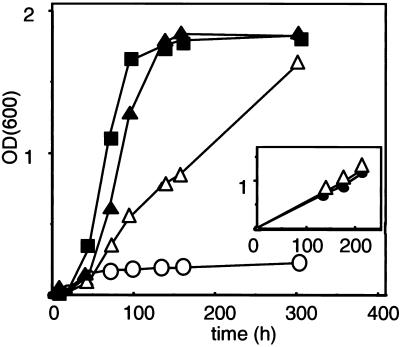
Wild-type and mutant pex5 alleles were cloned downstream of the PEX5 promoter in a yeast expression plasmid. These plasmids were transformed to a pex5Δ strain and transformants were cultured on oleate. The growth rate of cells expressing Pex5p(F208L) was approximately fourfold reduced compared with that of wild-type Pex5p, whereas growth of Pex5p(E212V) cells was less affected (Figure 7). Growth on glucose or glycerol media was unaffected for all transformants (our unpublished results). In addition, we constructed a pex5 mutant with three amino acid substitutions in the region involved in Pex13-SH3 domain binding: F208L, E212V, and E214G. This triple mutant showed growth rates on oleate comparable to the single F208L mutant (Figure 7, inset). These results are in line with the binding studies and suggest an essential role for F208 in the interaction with Pex13-SH3.
Figure 7.
Growth on liquid oleate medium of pex5Δ cells (○) expressing wild-type Pex5p (▪), Pex5p(F208L) (▵), Pex5p(E212V) (▴), Pex5p(F208L; E212V; E214G)(●; see inset). Cells were grown to mid-log phase in 0.3% glucose medium and inoculated at OD600 of 0.001 in liquid oleate medium. Growth was followed with time by measuring the optical density at 600 nm (OD600).
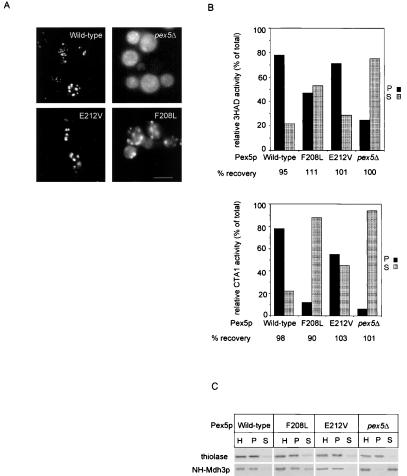
We expressed the GFP fused to PTS1 (GFP-SKL) to measure PTS1 protein import in these mutants. GFP-SKL expression was visualized using fluorescence microscopy (Figure 8A). In pex5Δ cells expressing Pex5p(F208L) a punctated pattern of labeling could be detected on top of a diffuse, cytosolic fluorescence, suggesting a partial mislocalization of GFP-SKL. Pex5p wild-type and Pex5p(E212V) transformants showed an exclusively punctated pattern (Figure 8A).
Figure 8.
Localization of peroxisomal matrix proteins in pex5Δ cells expressing wild-type Pex5p, Pex5p(F208L), or Pex5p(E212V). Subcellular distribution of GFP-SKL visualized by fluorescence microscopy (A). Bar, 10 μm. Subcellular distribution of CTA1 and 3HAD (B) and 3-ketoacyl-CoA thiolase and Mdh3p (C). After subcellular fractionation equivalent volumes of the 600 × g postnuclear supernatant (homogenate [H]), 20,000 × g pellet (P) and 20,000 × g supernatant (S) were analyzed by measuring enzyme activities (B) or by Western blotting (C). Antibodies were directed against the proteins as indicated. Recoveries varied between 90 and 110%.
The apparent mislocalization of PTS1 proteins in pex5Δ cells expressing Pex5p(F208L) was substantiated by subcellular fractionation experiments. pex5Δ transformants were homogenized and a postnuclear supernatant was centrifuged at 20,000 × g. Equivalent volumes of the pellet and the supernatant fractions were analyzed for the presence of peroxisomal proteins by using enzyme assays (Figure 8B: CTA1 and 3HAD) or Western blotting (Figure 8C: Mdh3p, 3-ketoacyl-CoA thiolase). In cells expressing wild-type Pex5p, 3HAD, CTA1, and Mdh3p were recovered almost exclusively from the pellet fraction. In cells expressing Pex5p(F208L) 3HAD, and Mdh3p were partially mislocalized to the supernatant, whereas CTA1 was completely mislocalized to the supernatant fraction. The protein import defect of CTA1 could not be rescued by replacing its PTS1 SKF by the canonical PTS1 SKL (our unpublished results), suggesting that the failure of Pex5(F208L) cells to import CTA1 is not reflected by its PTS1 composition. In Pex5p(E212V) cells, CTA1 was partially mislocalized to the supernatant, whereas other PTS1 proteins showed a wild-type distribution. The distribution of the PTS2 protein 3-keto-acyl-CoA thiolase was comparable in wild-type, Pex5p(E212V), and Pex5p(F208L) cells (Figure 8C), implying that the defect in protein import in pex5(F208L) cells is specific for the PTS1 import pathway. Moreover, these results suggest that loss of SH3-Pex5p interaction can be partially compensated for in vivo. This is born out by an in vitro reconstitution experiment. GST-Pex5p(F208L) could be coeluted with MBP-SH3 when 6xHis-Pex14p was first bound to the immobilized MBP-SH3 column (Figure 4, lane 4). These results show that Pex14p contains two different binding sites: one for Pex13-SH3 and another for Pex5p, and that these proteins can bind Pex14p simultaneously in vitro, resulting in a complex formed by Pex5p, Pex14p and Pex13-SH3.
Pex5p(F208L) and Pex5p(E212V) Are Still Associated with Peroxisomes
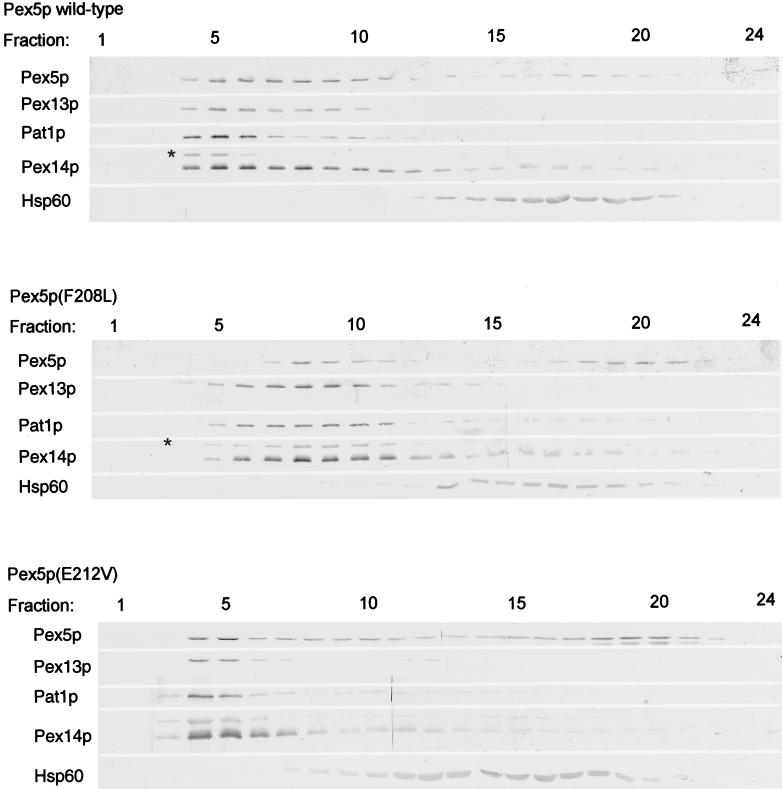
Because Pex5p(F208L) and Pex5p(E212V) are disturbed in binding to the Pex13-SH3 domain, we investigated whether the subcellular distribution of the pex5 mutants is affected. Subcellular fractionation of pex5Δ cells expressing mutant or wild-type Pex5p revealed that Pex5p(F208L) and Pex5p(E212V), like wild-type Pex5p, were partially associated with the 20,000 × g pellet fraction (our unpublished results). To investigate whether Pex5p present in the pellet fractions was associated with peroxisomes these fractions were analyzed by equilibrium density centrifugation. Fractions were collected and analyzed for Pex5p and marker proteins for peroxisomes (Pex13p, Pex14p, and Pat1p) and mitochondria (Hsp60) by using SDS-PAGE and Western blotting. Cells expressing Pex5p(F208L) contained peroxisomes equilibrating at lower density in a Nycodenz gradient than peroxisomes from wild-type cells, which may reflect the partial loss of matrix protein import in Pex5p(F208L) cells. Both Pex5p(E212V) and Pex5p(F208L) were localized in the peroxisomal peak fractions (Figure 9). These results suggest that in vivo, although interaction with the SH3 domain of Pex13p is impaired, Pex5p can still associate with peroxisomes. Based on our in vitro binding experiments Pex14p is a likely candidate to fulfill this function.
Figure 9.
Pex5p mutants are associated with peroxisomes. 20,000 × g pellet fractions of pex5Δ cells expressing wild-type Pex5p, Pex5p(F208L), or Pex5p(E212V) were loaded on top of a continuous Nycodenz gradient and centrifuged at 29,000 × g in a vertical rotor for 2.5 h. Fractions of 0.5 ml were collected and analyzed by SDS-PAGE and Western blotting with antibodies specific for Pex5p; the peroxisomal membrane markers Pex13p, Pex14p, and Pat1p; and the mitochondrial marker Hsp60. Fraction 1 is the bottom of the gradient. The asterisk indicates a cross-reacting band.
DISCUSSION
Proteins containing a PTS need to be targeted after synthesis in the cytoplasm to the peroxisomal membrane for subsequent import into the peroxisomal matrix. Many proteins (peroxins) have been discovered that are involved in this targeting and membrane-translocation process, some of which are active in the soluble phase (targeting), whereas others are integral or peroxisomal membrane-associated proteins acting as components of the protein-translocation machinery. Pex5p is the soluble receptor that recognizes PTS1 proteins and targets these PTS1 proteins to the membrane-located peroxins (Pex13p, Pex14p, and Pex17p). Here we have investigated the region of Pex5p important for association with the SH3 domain of Pex13p.
Pex5p mutants were selected in a two-hybrid setup that had lost the ability to bind to Pex13-SH3 but that retained the ability to interact with other proteins. The screen revealed at least three residues important for Pex13-SH3 interaction, F208, E212, and E214. Mutation of F208 (to leucine) had a strong down effect, whereas mutation of either E212 or E214 (to valine and glycine, respectively) showed diminished binding capacity with Pex13-SH3 (Table 3). The properties of the mutants in the two-hybrid system could be reproduced in an in vitro reconstituted system with bacterially expressed fusion proteins, thus excluding possible contributions of other yeast proteins. The mutations are located close to each other in a region N-terminal of the TPR-containing domain of Pex5p. Here we find the motif W204XXQF208, conserved among Pex5 proteins ranging from yeast to human. Mutation of the strictly conserved tryptophan (W204) in this motif also compromised the interaction with Pex13-SH3 (Table 3), indicating a central role for this motif in Pex13-SH3 binding. A second motif with a similar sequence (WSQEF) is present ∼90 amino acids N-terminal of the WXXQF motif. Mutations in this second motif do no affect the interaction of Pex5p with Pex13-SH3 (our unpublished results). Recently, it was shown that a peptide containing amino acids 100–213 of Pichia pastoris Pex5p is able to interact with the SH3 domain of PpPex13p in vitro (Urquhart et al., 2000). This peptide includes the conserved WXXQF motif, suggesting that the SH3 binding region in Pex5p is conserved between different yeast species. Whereas ScPex5p contains only two WXXXF motifs, human Pex5p contains seven of these motifs. Based on in vitro binding studies with HsPex5p and a fragment of HsPex14p (amino acids 1–78), Schliebs et al. (1999) have suggested a role for these motifs in Pex14p binding. We have not been able to find support for this suggestion in yeast. Mutation of either of these motifs in ScPex5p did not specifically affect Pex14p binding (Table 3; our unpublished results). Because pex5 mutants with severely disturbed binding to the Pex13-SH3 domain are still able to interact with Pex14p in the two-hybrid system (Table 3) and in vitro (Figure 4), we conclude that there are separate binding regions in Pex5p for Pex14p and Pex13-SH3.
A consensus SH3-binding motif (PTLPHR) is present in the primary sequence of Pex14p. Girzalsky et al. (1999) demonstrated by mutating the two prolines in the PXXP motif of Pex14p that these residues are essential for interaction with Pex13-SH3. The other Pex13-SH3 binding partner, Pex5p, does not contain a PXXP binding motif or a degenerated version thereof. Moreover, in our screen for mutants that had lost the interaction with Pex13-SH3 we did not find any mutations in proline residues, which suggests that Pex5p contains a novel, non-PXXP-related, SH3-binding motif. This is underscored by the differential effect of the W349A and E320K mutations in the Pex13-SH3 domain on the interaction with Pex5p and Pex14p. Pex13-SH3 (W349A) is mutated in one of the conserved aromatic residues that form the hydrophobic binding cleft of the SH3 domain and Pex13-SH3(E320K) contains a mutation in the RT loop of the SH3 domain. Both mutations abrogated interaction with Pex14p but interaction with Pex5p was not affected, either in the two-hybrid assay or in in vitro reconstitution experiments. Because both the hydrophobic binding cleft and the RT loop of the SH3 domain are part of the canonical PXXP ligand-binding region (Lim et al., 1994; Lee et al., 1995), the results suggest a novel binding mode for Pex5p with Pex13-SH3. This is supported by two other observations. First, our in vivo overexpression studies showed that overproduction of Pex5p had no noticeable effect on the ability of cells to grow on oleate, suggesting that Pex5p does not compete with Pex14p for Pex13-SH3 domain binding. Second, we found in the two-hybrid system that the presence of Pex5p cooperatively stimulated Pex13-SH3-Pex14p interaction. Both observations are in line with the existence of separate binding sites for Pex14p and Pex5p on the Pex13-SH3 domain.
We tested the effects of the mutations in Pex5p in cells with respect to growth and import of proteins into peroxisomes. Growth of Pex5 (F208L) was clearly retarded on oleate as sole carbon source, but growth of pex5 (E212V) was only mildly affected. A triple mutant of Pex5p containing all three SH3 loss-of-interaction mutations (F208L, E212V, and E214G) showed the same growth defect on oleate as the single F208 mutant, suggesting that F208 identifies the most important position for interaction with Pex13-SH3. Considering the clear deficiencies we observed with these mutants in the yeast two-hybrid and in vitro reconstitution experiments it is very unlikely that the mild phenotypes in vivo are due to residual binding of Pex5p to Pex13-SH3. It rather suggests that in vivo alternative ways exist to dock Pex5p with its PTS1 protein load. Pex5p not only binds to Pex13-SH3 but also to Pex14p. Indeed, Pex14p may substitute for Pex13p as docking site. This notion is based on the in vitro experiments, which show that binding of Pex5 (F208L) mutant protein to immobilized Pex13-SH3 can be rescued when Pex14p is mixed in. It suggests that Pex14p can function as a bridge between Pex13-SH3 and the mutant version of Pex5p. Indeed, our fractionation experiments showed that Pex5p(F208L) was still able to associate with peroxisomes, which indicates that in the absence of Pex13-SH3 interaction, Pex5p is tethered to the peroxisome membrane in an alternative way, most likely through the interaction with Pex14p.
The combined roles of Pex13p and Pex14p in forming a docking platform for Pex5p-mediated PTS1 protein delivery was underlined by experiments in which Pex5p, Pex13p, and Pex14p were overproduced. Overexpression of Pex14p or Pex13p individually impaired growth of cells on oleate-containing medium. A similar phenotype has been reported for Hansenula polymorpha cells overexpressing Pex14p (Komori et al., 1997). Overexpression of both Pex13p and Pex14p together, however, restored normal growth. Disruption of the Pex13p-Pex14p interaction had the same effect in vivo: yeast cells containing the E320K mutation in the RT loop of Pex13-SH3, which abrogated Pex14p association, were unable to grow on oleate-containing medium (Elgersma et al., 1996; Girzalsky et al., 1999). Together, these results show that both the association and the stoichiometry of Pex13p and Pex14p in a cell are important, which implies that they fulfill their role in protein import as a well-defined pair.
Import of PTS1 proteins was differentially affected in vivo in the Pex5p(F208L) mutant context. As expected, import of 3-keto-acyl-CoA thiolase (a PTS2 protein) was normal, but 3HAD and Mdh3p (both PTS1 proteins containing the PTS1 SKL) were only partially mislocalized to the cytosol, whereas CTA1 (containing the PTS1 SKF) was completely mislocalized to the cytosol. The PTS1 consensus sequence is rather degenerate and this may be related to its efficiency to function as targeting signal. We swapped PTS1 motifs between Mdh3p and catalase A to investigate whether the composition of the PTS1 could explain the observed partial versus complete import efficiencies of Mdh3p and catalase A in the Pex5p(F208L) mutant; no support was found for the notion that the PTS1 composition of catalase determines the import efficiency (our unpublished results).
It is noteworthy that mild peroxisome biogenesis phenotypes are also observed in humans. Analysis of the fibroblasts of a patient suffering from the peroxisome biogenesis disorder neonatal adrenoleukodystrophy revealed that most peroxisomal matrix proteins were partially mislocalized to the cytosol, whereas catalase was found exclusively in the cytosol (Liu et al., 1999; Shimozawa et al., 1999), a phenotype similar to that of the yeast Pex5p(F208L) mutant. These observations underscore the notion that mild import deficiencies can affect normal cellular function, thereby leading to a diseased state of the organism. Interestingly, the mild phenotype in this adrenoleukodystrophy patient is caused by a missense mutation, I326T, in the SH3 domain of Pex13p. Introduction of the analogous mutation in Pex13p of the yeast P. pastoris also resulted in a mild peroxisome biogenesis deficiency (Liu et al., 1999). The effects of this mutation on the interaction between Pex13p and its partner proteins have not yet been determined, nor is it clear from the location of the mutation in the SH3 domain which interaction might be affected. Given that I326 of human Pex13p is conserved in S. cerevisiae Pex13p it will be of interest to include this mutation in future studies. Particularly, in vitro interaction studies because we observed that deficiencies show up more clearly in the simple reconstituted state than in vivo.
ACKNOWLEDGMENTS
We thank Aldo Stein and Carlo van Roermund for assistance with two-hybrid and Nycodenz density gradients analyses. We are grateful to Dr. P. van der Sluys for providing the NH-antibodies and Dr. S. Rospert for providing the Hsp60 antibodies. This work was supported by grants from the Netherlands Organization of Scientific Research (NWO) and the European Community (BIO4-97-2180).
Abbreviations used:
- AD
activation domain
- DB
DNA-binding domain
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- MBP
maltose binding protein
- NH
N-terminal hemagglutinin
- PTS
peroxisomal targeting signal
- SH3
Src homology 3
REFERENCES
- Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel JAKW, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- Arold S, O'Brien R, Franken P, Strub M-P, Hoh F, Dumas C, Ladbury JE. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brocard C, Kragler F, Simon MM, Schuster T, Hartig A. The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem Biophys Res Commun. 1994;204:1016–1022. doi: 10.1006/bbrc.1994.2564. [DOI] [PubMed] [Google Scholar]
- Brocard C, Lametschwandtner G, Koudelka R, Hartig A. Pex14p is a member of the protein linkage map of Pex5p. EMBO J. 1997;16:5491–5500. doi: 10.1093/emboj/16.18.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Warren DS, Sacksteder KA, Gould SJ. PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol. 1999;147:761–773. doi: 10.1083/jcb.147.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevray PM, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G, Braverman N, Wong C, Moser A, Moser HW, Watkins P, Valle D, Gould SJ. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995;9:115–125. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Elgersma-Hooisma M, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. A mobile PTS2 receptor for peroxisomal protein import in Pichia pastoris. J Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak HF, Distel B. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1-containing proteins. J Cell Biol, 1996a;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, van den Berg M, Tabak HF, Distel B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisomal assembly mutants of Saccharomyces cerevisiae. Genetics, 1993;135:731–740. doi: 10.1093/genetics/135.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Vos A, van den Berg M, van Roermund CWT, van der Sluijs P, Distel B, Tabak HF. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996b;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Veenhuis M, Kunau W-H. Peroxisomes: organelles at the crossroads. Trends Cell Biol. 1997;7:400–407. doi: 10.1016/S0962-8924(97)01126-4. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fransen M, Terlecky SR, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast Escherichia coli shuttle vectors constructed with in vitro mutagenised yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Girzalsky W, Rehling P, Stein K, Kipper J, Blank L, Kunau W-H, Erdmann R. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144:1151–1162. doi: 10.1083/jcb.144.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Distel B, Tabak HF. Import of proteins into peroxisomes. Biochim Biophys Acta. 1999;1451:17–34. doi: 10.1016/s0167-4889(99)00087-7. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Girzalsky W, van den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae Pex3p and Pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Ruigrok CCM, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I. The cytosolic DnaJ-like protein Djp1 is involved specifically in peroxisomal protein import. J Cell Biol. 1998;142:421–434. doi: 10.1083/jcb.142.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, van Roermund CWT, Distel B, van den Berg M, Vilela C, Rodrigues-Pousada C, Wanders RJA, Tabak HF. The A.B.C. transporter proteins Pat1 and Pat2 are required for import of long-chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996;15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Honsho M, Tamura S, Shimozawa N, Suzuki Y, Kondo N, Fujiki Y. Mutation in PEX16 is causal in the peroxisome-deficient Zellweger syndrome of complementation group D. Am J Hum Genet. 1998;63:1622–1630. doi: 10.1086/302161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau W-H. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Ghaedi K, Shimozawa N, Wanders RJA, Matsuzono Y, Imanaka T, Okumoto K, Suzuki Y, Kondo N, Fujiki Y. Newly identified Chinese hamster ovary cell mutants are defective in biogenesis of peroxisomal membrane vesicles (peroxisomal ghosts), representing a novel complementation group in mammals. J Biol Chem. 1998;273:24122–24130. doi: 10.1074/jbc.273.37.24122. [DOI] [PubMed] [Google Scholar]
- Komori M, Rasmussen SW, Kiel JAKW, Baerends RJS, Cregg JM, van der Klei IJ, Veenhuis M. The Hansenula polymorpha PEX14 gene encodes a novel peroxisomal membrane protein essential for peroxisome biogenesis. EMBO J. 1997;16:44–53. doi: 10.1093/emboj/16.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lee C-H, Leung B, Lemmon MA, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the SH3 domain of Hck determines its high affinity and specificity in binding to HIV-1 Nef protein. EMBO J. 1995;14:500–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- Lim WA, Richards FM, Fox RO. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Björkman J, Urquhart A, Wanders RJA, Crane DI, Gould SJ. PEX13 is mutated in complementation group 13 of the peroxisome biogenesis disorders. A J Hum Genet. 1999;65:621–634. doi: 10.1086/302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucke H. In: In: Methods of Enzymatic Analysis. Bergmeyer HU, editor. New York: Academic Press; 1963. pp. 885–894. [Google Scholar]
- Marzioch M, Erdmann R, Veenhuis M, Kunau W-H. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJA, Suzuki Y, Kondo N, Fujiki Y. Human PEX19: cDNA cloning of functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci USA. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Fiore PPD. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- Pisabarro MT, Serrano L, Wilmanns M. Crystal structure of the Abl-SH3 domain complexed with a designed high-affinity peptide ligand: implications for SH3-ligand interactions. J Mol Biol. 1998;281:513–521. doi: 10.1006/jmbi.1998.1932. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Lazarow PB. Peroxisomal biogenesis: multiple pathways of protein import. J Biol Chem. 1994;269:30065–30068. [PubMed] [Google Scholar]
- Rehling P, Skaletz-Rorowski A, Girzalsky W, Voorn-Brouwer T, Franse MM, Distel B, Veenhuis M, Kunau W-H, Erdmann R. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor Pex5p. J Biol Chem. 2000;275:3593–3602. doi: 10.1074/jbc.275.5.3593. [DOI] [PubMed] [Google Scholar]
- Ren R, Mayer BJ, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- Schliebs W, Saidowsky J, Agianian B, Dodt G, Herberg FW, Kunau W-H. Recombinant human peroxisomal targeting signal receptor PEX5: structural basis for interaction of PEX5 with PEX14. J Biol Chem. 1999;274:5666–5673. doi: 10.1074/jbc.274.9.5666. [DOI] [PubMed] [Google Scholar]
- Shimozawa N, Suzuki Y, Zhang Z, Imamura A, Toyama R, Mukai S, Fujuki Y, Tsukamoto T, Osumi T, Orii T, Wanders RJA, Kondo N. Nonsense and temperature sensitive mutations in PEX13 are the cause of complementation group H of peroxisome biogenesis disorders. Hum Mol Genet. 1999;8:1077–1083. doi: 10.1093/hmg/8.6.1077. [DOI] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Snyder WB, Faber KN, Wenzel TJ, Koller A, Luers GH, Rangell L, Keller GA, Subramani S. Pex19p interacts with Pex3p and Pex10p and is essential for peroxisome biogenesis in Pichia pastoris. Mol Biol Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South ST, Gould SJ. Peroxisome synthesis in the absence of preexisting peroxisomes. J Cell Biol. 1999;144:255–266. doi: 10.1083/jcb.144.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Braakman I, Distel B. Peroxisomes: simple in function but complex in maintenance. Trends Cell Biol. 1999;9:447–453. doi: 10.1016/s0962-8924(99)01650-5. [DOI] [PubMed] [Google Scholar]
- Urquhart AJ, Kennedy D, Gould SJ, Crane DI. Interaction of Pex5p, the type I peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J Biol Chem. 2000;275:4127–4136. doi: 10.1074/jbc.275.6.4127. [DOI] [PubMed] [Google Scholar]
- Van der Klei IJ, Hilbrands RE, Kiel JAKW, Rasmussen SW, Cregg JM, Veenhuis M. The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J. 1998;17:3608–3618. doi: 10.1093/emboj/17.13.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I, Franse MM, Elgersma Y, Distel B, Tabak HF. PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:11782–11786. doi: 10.1073/pnas.90.24.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Leij I, van den Berg M, Boot R, Franse MM, Distel B, Tabak HF. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 1992;119:153–162. doi: 10.1083/jcb.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Ijst L, Gennip AHV, Jacobs C, Jager JPD, Dorland L, Duran M. Long-chain 3-hydroxyacyl-CoA. dehydrogenase deficiency: identification of a new inborn error of mitochondrial fatty acid beta-oxidation. J Inherited Metab Dis. 1990;13:311–314. doi: 10.1007/BF01799383. [DOI] [PubMed] [Google Scholar]
- Wiemer E, Nuttley WM, Bertolaet BL, Li X, Francke U, Wheelock MJ, Anne UK, Johnson KR, Subramani S. The human PTS1 receptor restores peroxisomal protein import deficiency in cells from patients with fatal peroxisomal disorders. J Cell Biol. 1995;130:51–65. doi: 10.1083/jcb.130.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–601. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]