Abstract
Activation of the Wnt-signaling pathway is known to play a crucial role in carcinogenesis of various human organs including the colon, liver, prostate, and endometrium. To investigate the mechanisms underlying hepatocellular carcinogenesis, we attempted to identify genes regulated by β-catenin/Tcf complex in a human hepatoma cell line, HepG2, in which an activated form of β-catenin is expressed. By means of cDNA microarray, we isolated a novel human gene, termed MARKL1 (MAP/microtubule affinity-regulating kinase-like 1), whose expression was downregulated in response to decreased Tcf/LEF1 activity. The transcript expressed in liver consisted of 3529 nucleotides that contained an open reading frame of 2256 nucleotides, encoding 752 amino acids homologous to human MARK3 (MAP/microtubule affinity-regulating kinase 3). Expression levels of MARKL1 were markedly elevated in eight of nine HCCs in which nuclear accumulation of β-catenin was observed, which may suggest that MARKL1 plays some role in hepatocellular carcinogenesis.
Keywords: MARKL1, β-catenin, microarray, expression profile, hepatocellular carcinoma
Introduction
Wnt-signaling pathway is considered to be essential for development and organogenesis. Abrogation of this pathway due to genetic alteration in the adenomatous polyposis coli (APC), β-catenin (CTNNB1) or Axin (AXIN1) gene has been reported in various human cancers in the colon, liver, prostate, and endometrium [1–7].β-catenin, a key component of this pathway, was shown to be phosphorylated by a complex of APC, Axin, and glycogen synthase kinase 3β (GSK-3β), and subsequently underwent degradation in cytoplasm through a ubiquitin-proteasome pathway [8]. Moreover, a recent study has clarified that APC functions as a nuclear export transporter of β-catenin [9]. Therefore, mutation of APC, AXIN1, or CTNNB1 itself leads to accumulation of β-catenin in the cytoplasm and/or nucleus. Accumulated β-catenin interacts with the transcription complex of T-cell factor/lymphoid enhancer-binding factor (Tcf/LEF) family [10] and transactivates downstream genes that include c-myc [11], cyclin D1 [12], matrilysin [13], WISP [14], c-jun, fra-1, uPAR [15], and NBL4 [16]. However, target molecules of Tcf/LEF1 complex and involvement of their altered expression in hepatocarcinogenesis remain to be fully elucidated.
We previously reported that gene transfer of wild-type AXIN1 or the β-catenin binding domain of wild-type APC could reduce the activity of Tcf/LEF complex in hepatoma cells [7]. Using this model system, we compared the expression profiles between HepG2 hepatoma cells infected with adenovirus containing LacZ, a part of APC corresponding to β-catenin binding domain, and AXIN1 by cDNA microarray. In this paper, we report isolation of a novel human gene, MARKL1, whose expression correlates to the activity of Tcf/LEF1 complex in HepG2 cells, and its involvement in human hepatocellular carcinogenesis.
Materials and Methods
Cell Lines
A human embryonic kidney cell line, HEK293, and human hepatoma cell lines, HepG2 and Alexander, were obtained from the American Type Culture Collection (ATCC). SNU423 and SNU475 cells were obtained from the Korea cell-line bank. All cell lines were grown in monolayers in appropriate media: Dulbecco's modified Eagle's medium for HepG2 and Alexander; Eagle's minimum essential medium for HEK293; and RPMI1640 for SNU423 and SNU475, supplemented with 10% fatal bovine serum and 1% antibiotic/antimycotic solution (Sigma, St. Louis, MO) and maintained at 37°C in air containing 5% CO2 and humidity.
Construction and Infection of Recombinant Adenovirus
Expression of the 20-amino-acid repeat domain of APC that binds to β-catenin [17] or that of the entire coding region of AXIN1 was shown to downregulate β-catenin [7,11]. Therefore, we constructed adenoviral vectors containing the 20-amino-acid repeat region of wild-type APC (Ad-APC), the entire coding region of AXIN1 (Ad-Axin) and LacZ (Ad-LacZ) into pAd-Bg/ll vector. The recombinant adenoviruses, constructed as described previously [7], were propagated in HEK293 cells and purified by two rounds of CsCI density centrifugation. HepG2 cells were infected with the viral solutions at 100 MOI and incubated at 37°C for 1 hour, with brief agitation every 15 minutes, followed by addition of culture medium. The infected cells were maintained at 37°C for 72 hours.
Tcf-4 Reporter Assays
The reporter assays were performed as described previously [7]. Briefly, the reporter plasmid containing four copies of Tcf4-specific DNA-binding sequence (TBE2) in the upstream region of the luciferase gene in pGL3-Basic vector (Promega, Madison, Wl) or mock pGL3- Basic vector (Promega) was cotransfected with a plasmid vector, pRL-TK (Promega), using FuGENE™ 6 reagent (Boehringer) according to the supplier's recommendations. The reporter gene activities were measured 72 hours after transfection using dual-luciferase reporter assay system (Promega) according to the manufacturer's protocol.
Preparation of RNA
Total RNAs were isolated from HepG2 cells infected with Ad-APC, Ad-Axin or Ad-LacZ using Trizole reagent (Life Technologies) according to the manufacturer's method.
cDNA Microarray Production
The fabrication of cDNA microarray slides has been described elsewhere [20]. Two sets of cDNA microarray slides containing a duplicate set of 9216 cDNA spots were used for each analysis of expression profiles to reduce the experimental fluctuation of each signal.
For the preparation of probes, after treatment with 10 units of DNase I, 1 µg of polyA RNA was reverse-transcribed and second-strand synthesis was performed. The double-stranded cDNA was then amplified with Ampliscribe T7 transcription kit (Epicentre Technologies, Madison, Wl) and the amplified RNA was labeled during reverse transcription with Cy-dye (Amersham Pharmacia Biotech, Piscataway, NJ). RNA from HepG2 cells with Ad-LacZ was labeled with Cy3 and that from HepG2 cells with Ad-APC or Ad-Axin was labeled with Cy5.
The Cy3-labeled cDNA derived from HepG2 cells with Ad-LacZ and Cy5-labeled cDNA from cells with Ad-APC or Ad-Axin were mixed with microarray hybridization solution (Amersham Pharmacia Biotech) and formamide at a final concentration of 50%. Hybridization was performed using an automated slide processor (Amersham Pharmacia Biotech) for 16 hours at 42°C. After hybridization, the glass slides were washed in 2xSSC, 0.1% SDS for 10 minutes at 55°C, 0.2xSSC, 0.1% SDS for 10 minutes at 55°C, and 0.1xSSC for 1 minute at room temperature. Subsequently the slides were scanned by an Array Scanner (Molecular Dynamics) and fluorescence intensities of Cy3 and Cy5 for each gene were evaluated by Array Vision software (Amersham Pharmacia Biotech). After subtraction of background signal, the quadruplicated values were averaged for each spot and adjusted so that the mean Cy3 and Cy5 intensities of 52 housekeeping genes for each slide were equal. After the normalization, genes were excluded from further investigation if the intensities of both Cy3 and Cy5 were below 100,000 fluorescence units.
Preparation of A6555 cDNA and Isolation of the Entire Coding Sequence of MARKL1
The A6555 cDNA spotted on the microarray was prepared by RT-PCR using a primer set, A6555F (5′-CCTAAAGA-CTGGAGAATCTGG-3′), and A6555R (5′-AAGAAGTGGC-TTAGAGTCCAGTG-3′) designed from the sequence of Hs.8312 in UniGene database (http://www.ncbi.nlm.nih.gov/UniGene/). By a computer search in the human EST database in NCBI with the BLAST program, we identified 58 EST sequences. To obtain the sequence of the 5′ region of A6555, we further searched for genomic sequences in genomic databases (http://www.ncbi.nlm.nih.gov/blast/blast.cgi), and found three sequences. By the use of exon prediction programs, GENSCAN (http://ccr-081.mit.edu/GENSCAN.html) and Gene Recognition and Assembly Internet Link (http://grail.genome.ad.jp/Grail-1.3/), we could identify 10 exon-like sequences. RT-PCR experiments using a primer set, A6555F2 (5′-ATGT-CTTCGCGGACGGTG-3′) and A6555R2 (5′-CCAGATT-CTCCAGTCTTTAGG-3′), which were designed in the exon-like sequence and the sequence of Hs. 8312, respectively, produced a 2733-base product and subsequently its sequence was determined.
5′ Rapid Amplification of cDNA Ends (5′RACE)
5′RACE using Marathon cDNA amplification kit (Clontech, Palo Alto, CA) was performed according to the manufacturer's instructions. For the amplification of the 5′ part of MARKL1 cDNA, a gene-specific reverse primer (5′-GCTGGGATTCAGCTGGGTTTTGTCG-3′) and the AP1 primer supplied in the kit were used. The cDNA template was synthesized from human testis mRNA. The nucleotide sequences of the PCR products were determined directly by the use of the gene-specific reverse primer with an ABI PRISM 377 DNA sequencer (Applied Biosystems) according to the manufacturer's instructions.
Northern Blot Analysis
Human multiple-tissue blots (Clontech) were hybridized with a 32P-labeled A6555 cDNA. Prehybridization, hybridization, and washing were performed according to the supplier's recommendations. The blots were autoradiographed with intensifying screens at -80°C for 24 hours.
Immunocytochemical Analysis
The entire coding region of MARKL1 was amplified by RT-PCR with a primer set, MARKL1EGFP-F (5′-CCCCGTCGACCCGGAGAAGATGTCTTCGCGG-3′) and MARKL1EGFP-R (5′-TCAGAATTCGAGGTCGTTGGA-GATGCGGGTG-3′). The product was digested with SalI and EcoRI, and cloned into the appropriate cloning site of a plasmid vector, pEGFP-N2 (Clontech). Another product amplified with a different primer set, MARKL1FLAG-F (5′-CCCGGAATTCGGAGAAGATGTCTTCGCG-GACGG-3′) and MARKL1FLAG-R (5′-CTCAGTCGAC-GAGGTCGTTGGAGATGCGGGTG-3′) was digested with EcoRI and SalI, and cloned into pFlag-CMV-5a (Kodak). HEK293 cells were transfected with either pEGFP-MARKL1 or pFlag-MARKL1 and fixed with PBS containing 4% paraformaldehyde. Cells tranfected with pFlag-MARKL 1 was stained by anti-Flag monoclonal antibody (Sigma) and by rhodamine-conjugated anti-mouse secondary antibody. Nuclei were stained with 4′,6′-diamidine-2′-phenylindole dihydrochloride (DAPI; Boehringer Mannheim). Subcellular localization of the protein was visualized by fluorescent image using Eclipse E800 (Nikon).
Clinical Materials and RT-PCR
Twenty sets of primary HCCs and their corresponding noncancerous liver tissues were obtained with informed consent from surgical specimens of patients who underwent hepatectomy in Kyoto University Hospital. Among them, nine cases of HCCs revealed nuclear accumulation of β-catenin by immunohistochemical staining. Total RNAs were extracted from these nine HCCs and their corresponding noncancerous tissues. Ten micrograms of total RNA was reversely transcribed for single-stranded cDNAs using d(T)12–18 primer (Amersham Pharmacia Biotech) with Superscript II reverse transcriptase (Life Technologies). Each single-stranded cDNA was diluted for subsequent PCR amplification by monitoring glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a quantitative control. The primers used for amplification were GAPDHF (5′-ACAACAGCCTCAAGATCATCAG-3′), GAPDHR (5′-GGT-CCACCACTGACACGTTG-3′), MARKL1F (5′-CCTAAA-GACTGGAGAATCTGG-3′) and MARKL1R (5′-AAGA-AGTGGCTTAGAGTCCAGTG-3′). Each PCR was carried out in a 20-µl volume of PCR buffer (TAKARA), and amplified for 4 minutes at 94°C for initial denaturing, followed by 25 (for GAPDH) or 30 (for MARKL1) cycles of 94°C for 30 seconds, 57°C for 30 seconds and 72°C for 30 seconds, in the Gene Amp PCR system 9600 (Perkin-Elmer, Foster City, CA).
Colony-Formation Assay
Alexander, SNU423, and SNU475 cells were transfected with sense S-oligonucleotide (5′-TCGGACACGGTGAG-TG-3′) or antisense S-oligonucleotide (5′-CACTCACCGT-GTCCGA-3′) encompassing the first exon-intron boundary designed to suppress the expression of MARKL1 using Lipofectin Reagent (Gibco BRL, Grand Slam, NY). Cells were maintained for 1 week, and then fixed with 100% methanol and stained by Giemsa solution.
Results
Identification of Genes Whose Expression Was Altered in Response to Introduction of APC or AXINI into HepG2 Cells
As shown in Figure 1A, transactivational activity of Tcf/LEF in HepG2 cells was markedly reduced 72 hours after adenovirus-mediated gene transfer of a part of APC corresponding to the β-catenin binding domain (Ad-APC) or AXIN1 (Ad-Axin). Introduction of AXIN1 into HepG2 cells, in which an activated form of β-catenin is expressed and accumulated in the nucleus, sequestered β-catenin in the cytoplasm by unknown mechanisms. Compared with the high activity of Tcf/LEF in LacZ (Ad-LacZ)-infected HepG2 cells, the activity in Ad-APC or Ad-Axin-infected cells was significantly reduced, but the effect was much stronger in Ad-Axin-infected cells than in Ad-APC-infected cells. To identify genes associated with the Tcf/LEF activity, we analyzed expression profiles of HepG2 cells infected with Ad-LacZ, Ad-APC or Ad-Axin by cDNA microarray representing 9216 genes. Because the gene transfer of Ad-APC or Ad-Axin reduced the activity of Tcf/LEF complex, expression of target genes were expected to be downregulated in response to the infection of Ad-APC or Ad-Axin. Compared with the expression profile of HepG2 cells infected with Ad-LacZ, we identified 356 genes in cells with Ad-APC, and 78 genes in cells with Ad-Axin, whose expression levels were reduced to less than 50%. Among the genes selected, expression levels of 21 genes including 9 ESTs were more significantly reduced in Ad-Axin infected cells than Ad-APC infected cells. Therefore we further analyzed those 21 genes as candidates to be downregulated in response to the reduced Tcf/LEF activity.
Figure 1.
(A) Effect of gene transfer of the β-catenin binding region of APC or the entire coding region of AXIN1 on Tcf/LEF transcriptional activity in HepG2 cells. Cells infected with Ad-LacZ, Ad-APC, or Ad-Axin at 100 MOI were subsequently transfected with a reporter plasmid containing the Tcf4-binding element (TBE2). As a control, cells were transfected with pGL3-Basic plasmid (mock). Each bar represents the mean relative luciferase activities of TBE2 to vector control (mock) with unbiased standard deviation by a triplicate experiment. (B) Expression of MARKL1 in nine primary HCCs. RT-PCR experiments were performed with a MARKL1-specific primer set using RNA extracted from HCCs with nuclear accumulation of β-catenin and their matched noncancerous liver tissue. The expression of GAPDH served as an internal control. (C) The expression levels of MARKL1 in HepG2 cells infected with Ad-LacZ, Ad-APC, or Ad-Axin were determined by RT-PCR. The lower panel shows the expression of GAPDH.
Isolation of a Novel Human Gene, MARKL1
Among the 21 genes, we first focused on a gene, A6555, corresponding to an EST (Hs. 8312), because its expression was significantly upregulated in eight of nine clinical HCCs that had a high level of nuclear accumulation of β-catenin compared with the corresponding noncancerous liver tissues (Figure 1B). RT-PCR experiment reconfirmed the positive correlation of its expression levels to the Tcf/LEF1 activity in HepG2 cells (Figure 1C). Multitissue Northern blot analysis using the cDNA as a probe showed a 3.6-kb transcript expressed ubiquitously in all tissues examined, with a highly abundant expression in testis (Figure 2).
Figure 2.
Northern blot analysis of MARKL1 in various human tissues. The molecular size of the transcript is approximately 3.6 kb. The expression of β-actin served as an internal control.
To further investigate the role of this gene in hepatocellular carcinogenesis, we attempted to isolate a cDNA covering the entire coding region of this gene. Because the predicted size of the transcript was approximately 3.6 kb and no EST clones containing the 5′ part of this gene was identified in EST databases, we searched for genomic sequences corresponding to the cDNA in the genomic databases and found that three cosmid sequences (AC005781, AC005581.1, and AC006126.1) assigned to chromosomal band 19q13.2 included this gene. Using GENSCAN and Gene Recognition and Assembly Internet Link programs, we predicted candidate-exon sequences and performed exon-connection. In addition, we carried out 5′ RACE to determine the sequence of the 5′ region of the transcript. As a result, we could obtain an assembled sequence of 3609 nucleotides that contained an open reading frame of 2064 nucleotides encoding 688 amino acids (DNA sequences are available from GenBank, accession number AB049127). Comparison of the cDNA sequence with the cosmid sequences disclosed that this gene consists of 18 exons. Because the coding sequence of this gene shared 56% identity with MARK3 (MAP/microtubule affinity-regulating kinase 3), we termed this gene as MARKL1 (MAP/microtubule affinity-regulating kinase-like 1). During the process, we also detected an alternative-spliced transcript consisting of 3529 nucleotides. The smaller transcript is generated by alternative splicing and lacks exon 16. In normal testis as well as in HCC cell lines, the 3529-bp transcript was exclusively expressed, and the 3609-bp transcript was observed specifically in the brain (data not shown).
Characterization of MARKL1
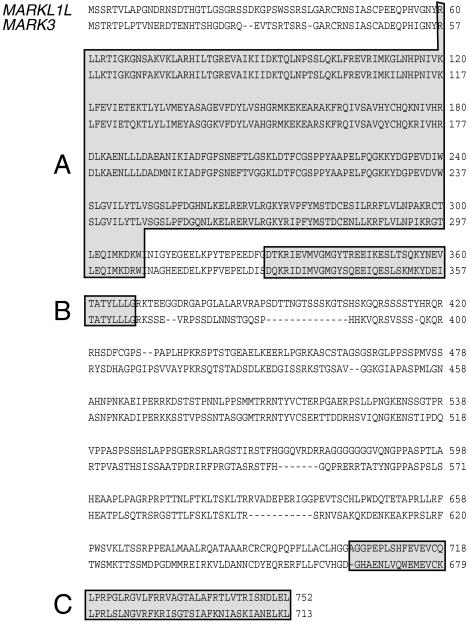
The cDNA sequences of the two alternatively spliced transcripts and their deduced amino acid sequences are shown in Figure 3. The predicted 688 (MARKL1S) and 752 (MARKL1L) amino acids of MARKL1 revealed 60% and 62% homology to MARK3 protein, respectively. Both MARKL1 peptides contained a serine/threonine kinase domain and a ubiquitin-associated domain in their N-terminal portion whereas only MARKL1L protein contained a KM kinase-associated domain in the C-terminal portion. The serine/threonine protein kinase domain, ubiquitin-associated domain and KM kinase-associated domain shared 89%, 69%, and 43% identity with those of MARK3 (Figure 3).
Figure 3.
Homology between the deduced amino acid sequence of MARKL 1L and MARK3 protein. The serine/threonine kinase domain (A), ubiquitin-associated domain (B) and KA1 kinase-associated domain (C) showed 89%, 69%, and 43% identity, respectively.
The entire coding region corresponding to MARKL1L was cloned into a pEGFP-N2 vector and a pFlag-CMV-5a vector, and these constructs were transfected into HEK293 cells. Both MARKL1-EGFP fusion protein and Flag-tagged MARKL1 protein were detected homogeneously in cytoplasm by fluorescent immunocytochemistry (Figure 4). Similar results were obtained when they were transfected into COS7 cells (data not shown).
Figure 4.
Subcellular localization of MARKL1 detected by immunocytochemistry. (A) HEK293 cells were transfected with pEGFP-MARKL1. Green fluorescent signals indicating EGFP-MARKL1 fusion protein were observed in the cytoplasm. (B) DAPI was used to indicate the nuclei of the cells shown in (A). (C, D) HEK293 cells tranfected with pFlag-MARKL1 were stained with anti-Flag monoclonal antibody and subsequently visualized by rhodamine-conjugated anti-mouse antibody. The Flag-tagged MARKL 1 protein was also detected in the cytoplasm.
Colony-Formation Assay
To assess a role of MARKL1 in hepatocarcinogenesis, we synthesized various sense and antisense S-oligonucleotide of MARKL1 and transfected each of them into Alexander, SNU423, and SNU475 cells. A comparison of the number of colonies in each dish revealed that the antisense S-oligonucleotide of MARKL1 encompassing the first exon-intron boundary significantly suppressed colony formation of SNU475 cells, compared with its complementary sense S-oligonucleotide (Figure 5, A and B). This result was reconfirmed by three independent experiments. Significant difference in the cell number between sense-transfected and antisense-transfected cells was not observed in Alexander or SNU423 cells. This antisense oligonucleotide specifically suppressed the expression of MARKL1 in SNU475 cells (Figure 5C).
Figure 5.
(A) Colony-formation assay using S-oligonucleotides was performed in SNU475 cells. The antisense S-oligonucleotide designed to suppress the expression of MARKL 1 reduced the growth of SNU475 cells. (B) The number of colonies larger than 1 mm in cells with the antisense oligonucleotide and the control sense oligonucleotide. Each bar represents the mean±SD of colonies with unbiased standard deviation from three independent experiments (sense: 541.3±153.7, antisense: 260.0±56.4). (C) Expression of MARKL1 in the cells treated with either sense or antisense oligonucleotide for 12 hours was examined by RT-PCR. Expression of GAPDH served as an internal control.
Discussion
In this study, we identified a novel gene, MARKL1, homologous to human MARK3 using cDNA microarray technique, which facilitates comparison of expression profiles between various materials [18–20]. The evidences that MARKL1 was reduced by the infection of Ad-APC or Ad-Axin into cancer cells, and that its expression was elevated in the majority of primary HCCs having accumulated β-catenin in the nuclei, indicated that MARKL1 is a good candidate molecule in the β-catenin/Tcf/LEF signaling pathway. Because the predicted amino acid sequence contained the serine/threonine kinase domain which is observed commonly in proteins involved in signal transduction, MARKL1 is considered to play a role as a messenger of the Wnt-signaling pathway.
C-TAK1, a product of MARK3, has been reported to bind to and phosphorylate serine-216 of cdc25C, which inhibits mitotic entry of cells by promoting an association with 14-3-3 proteins in the cytoplasm [21,22]. The high similarity of serine/threonine kinase domain, ubiquitin-associated domain and kinase-associated domain of MARKL1 to those of C-TAK1 prompted us to examine a possible association of MARKL1 with cdc25C in vitro, but we did not obtain any evidence to support their direct interaction (data not shown). Nevertheless, it may be possible that MARKL1 can associate with and phosphorylate cdc25C as a complex and regulate cell cycle at G2-M. In accordance with the subcellular localization of C-TAK1 [22], immunohistochemical staining of MARKL1 protein was also observed homogeneously in the cytoplasm. However, because we did not examine a function of the protein encoded by the other transcript, MARKL1S, which is exclusively expressed in the brain, the physiological role of this protein that lacks the KA1-associated domain remains unresolved.
We suspected that enhanced expression of MARKL1L provides a growth advantage to cells. This notion was in line with the result of colony-formation assay in SNU475 cells. Suppression of MARKL1 using antisense S-oligonucleotide resulted in a significant deduction in the number of colonies for SNU475 cells, but not for the other two cell lines, implying that redundancy of other similar molecules may compensate for the reduction of MARKL1. Although the role(s) of MARKL1 in hepatocellular carcinogenesis needs to be investigated further, the identification of frequently activated molecules in HCC will shed light on discovering novel therapeutic targets, making more sensitive and adequate diagnoses, and finding a preventive strategy against HCC.
Acknowledgements
We appreciate the help of Jun-ichi Okutsu, Hitoshi Zenbutsu, Renpei Yanagawa, Kenji Hirotani, Hiroko Bando, Noriko Nemoto, and Noriko Sudo for the fabrication of cDNA microarray and that of Meiko Takahashi for preparation of the manuscript.
Footnotes
This work was supported in part by Research for the Future Program Grant #96L00102 from the Japan Society for the Promotion of Science.
References
- 1.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 2.Chan EF, Gat U, McNiff JM, Fuchs E. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet. 1999;21:10–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 3.Miyoshi Y, Iwao K, Nagasawa Y, Aihara Y, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. Activation of the β-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58:2524–2527. [PubMed] [Google Scholar]
- 4.Voeller HJ, Truica CI, Gelmann EP. β-Catenin mutations in human prostate cancer. Cancer Res. 1998;58:2520–2523. [PubMed] [Google Scholar]
- 5.Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res. 1998;58:896–899. [PubMed] [Google Scholar]
- 6.Iwao K, Nakamori S, Kameyama M, Imaoka S, Kinoshita M, Fukui T, Ishiguro S, Nakamura Y, Miyoshi Y. Activation of the β-catenin gene by interstitial deletions involving exon 3 in primary colorectal carcinomas without adenomatous polyposis coli mutations. Cancer Res. 1998;58:1021–1026. [PubMed] [Google Scholar]
- 7.Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, Sasaki Y, Imaoka S, Murata M, Shimano T, Yamaoka Y, Nakamura Y. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 8.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–3804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosin-Arbesfeld R, Townsley F, Bienz M. The APC tumor suppressor has a nuclear export function. Nature. 2000;406:1009–1012. doi: 10.1038/35023016. [DOI] [PubMed] [Google Scholar]
- 10.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 11.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 12.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 13.Crawford HC, Fingleton BM, Rudolph-Owen LA, Goss KJ, Rubinfeld B, Polakis P, Matrisian LM. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18:2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 14.Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, Taneyhill LA, Deuel B, Lew M, Watanabe C, Cohen RL, Melhem MF, Finley GG, Quirke P, Goddard AD, Hillan KJ, Gurney AL, Botstein D, Levine AJ. WISP genes are members of the connective tissue growth factor family that are up-regulated in wnt-1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA. 1998;95:14717–14722. doi: 10.1073/pnas.95.25.14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Llyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, Hanski C. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci USA. 1999;16:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiguro H, Furukawa Y, Daigo Y, Miyoshi Y, Nagasawa Y, Nishiwaki T, Kawasoe T, Fujita M, Satoh S, Miwa N, Fujii Y, Nakamura Y. Isolation and characterization of human NBL4, a gene involved in the β-catenin/Tcf signaling pathway. Jpn J Cancer. 2000;91:597–603. doi: 10.1111/j.1349-7006.2000.tb00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochim Biophys Acta. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 18.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 19.Iyel VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JCF, Trent JM, Staudt LM, Hudson J, Jr, Boguski MS, Lashkari D, Shalon D, Botstein D, Brown PO. The transcriptional program in the response of human fibroblasts to serum. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 20.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 21.Peng CY, Graves PR, Ogg S, Thoma RS, Byrnes MJ, III, Wu Z, Stephenson MT, Piwnica-Worms H. C-TAK1 protein kinase phosphorylates human Cdc25C on serine 216 and promotes 14-3-3 protein binding. Cell Growth Differ. 1998;9:197–208. [PubMed] [Google Scholar]
- 22.Dalai SN, Schweizer CM, Gan J, Decaprio JA. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol Cell Biol. 1999;19:4465–4479. doi: 10.1128/mcb.19.6.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]