Abstract
The way in which cytogenetic aberrations develop in prostate cancer (CaP) is poorly understood. Spectral karyotype (SKY) analysis of CaP cell lines has shown that they have unstable karyotypes and also have features associated with chromosomal instability (CIN). To accurately determine the incidence of de novo structural and numerical aberrations in vitro in CaP, we performed SKY analysis of three independent clones derived from one representative cell line, DU145. The frequent generation of new chromosomal rearrangements and a wide variation in the number of structural aberrations within two to five passages suggested that this cell line exhibited some of the features associated with a CIN phenotype. To study numerical cell-to-cell variation, chromosome 8 aneusomy was assessed in the LNCaP, DU145, and PC-3 cell lines and a patient cohort of 15 CaP primary tumors by interphase fluorescence in situ hybridization (FISH). This analysis showed that a high frequency of numerical alteration affecting chromosome 8 was present in both in vitro and in CaP tissues. In comparison to normal controls, the patient cohort had a statistically significant (P<.05), greater frequency of cells with one and three centromere 8 copies. These data suggest that a CIN-like process may be contributing towards the generation of de novo numerical and structural chromosome abnormalities in CaP.
Keywords: karyotype evolution, translocation, aneuploidy, ploidy, molecular cytogenetics
Introduction
Prostate cancer (CaP) has the highest cancer incidence in men and the second most common cause of male cancer mortality [1]. The tumorigenic process has slow onset pathology occurring over a few decades during the lifetime of the individual [2]. Our understanding of the disease process that underlies the progression of CaP has been complicated by both genotypic and phenotypic heterogeneity [3]. A model of CaP progression based on the well-established model of colon cancer progression [4,5] involves the accumulation of multiple genetic alterations. This model is essentially descriptive and does not address the mechanism leading to the early steps of CaP tumorigenesis.
Recent spectral karyotyping (SKY) analyses of the three CaP cell lines (LNCaP, DU145, PC-3) have demonstrated aneuploid karyotypes with many chromosomal aberrations including complex chromosomal rearrangements and a high degree of karyotype instability [6,7]. In contrast, most early-stage CaP tumors appear to be karyotypically normal diploid with the most common chromosomal changes affecting chromosome 8 [8]. In a small but significant number of cases, the disease progresses to advanced stages, giving the transition from a diploid to aneuploid chromosomal constitution a greater degree of karyotype complexity [9,10].
Karyotype instability can be defined as a progressive alteration of the karyotype affecting a cell population, either in vivo or in vitro [11]. It implies the selective transmission of chromosomal alterations through cell generations, resulting in clonal cell populations derived from a single cell but not necessarily completely homogeneous. Karyotype instability is distinct from chromosomal instability (CIN) in which an excess of chromosome alterations occurs at each cell generation and, without selective force, these alterations need not necessarily be transmitted through each cell generation [12,13]. CIN is thought to arise as a result of aberrations in the mitotic machinery and/or structural integrity of the chromosome constitution, leading to excessive numerical and structural chromosomal changes [12–15]. The current model for CaP progression does not provide experimental approaches for understanding why in vitro CaP cell lines have such complex karyotypes in comparison to the relatively simple karyotypes observed by direct analysis of CaP [8] nor has the role of CIN in this tumor type been rigorously assessed.
In this study, we have employed a comprehensive molecular cytogenetic analysis of CaP including SKY [16] analysis of three DU145-derived single cell clones (SCCs) and fluorescence in situ hybridization (FISH) to assess numerical variation of chromosome 8 in 15 early-stage low-grade patient tumors. SKY is a “24-color” FISH approach that uniquely identifies each chromosome based on its specific spectral color composition [16], and allows for the unambiguous identification of the diversity of each chromosomal region present in aberrations and marker chromosomes on a cell-by-cell basis. Therefore, SKY is ideal for assessing both the qualitative and quantitative chromosomal changes associated with tumors expressing the CIN phenotype. Our findings suggest that a CIN-like process may be contributing significantly towards the generation of de novo numerical and structural chromosome abnormalities in CaP.
Materials and Methods
CaP Cell Lines and Clone Selection
LNCaP (CRL-1740), DU145 (HTB-81), and PC-3 (CRL-1435) were obtained from the American Type Culture Collection (ATCC, Rockville, MD). LNCaP, an androgen-dependent cell line originating from a lymph node metastasis [17], was grown in RPMI 1640 with 2 mM l-glutamine, 1.5 g/l sodium bicarbonate, 4.5 g/l glucose, 10 mM HEPES, 1.0 mM sodium pyruvate, and 10% fetal bovine serum. PC-3, an androgen-independent cell line originating from a brain metastasis [18], was grown in Ham's F12K with 2 mM l-glutamine, 1.5 g/l sodium bicarbonate, and 10% fetal bovine serum.
DU145, also an androgen-independent cell line and obtained from a metastasis to the bone [19], was grown in F15K Minimum Essential Medium with 1.5 g/l sodium bicarbonate and 10% fetal bovine serum. Three individual subclones were derived from single cells selected from the parental DU145 flask (passage 83). Briefly, the parental culture was incubated in trypsin (Gibco BRL, Gaithersburg, MD), washed in medium, and dissociated by gentle titration. Following microscopic examination to measure complete dissociation of cells, the cell suspension was serially diluted to the approximate concentration of 100 cells/ml. Ten microliters of the suspension was seeded into multiwell flasks and examined by phase contrast microscopy by two independent observers. Three wells containing a single cell were maintained as SCC1, SCC2, and SCC3. After a period of 2 weeks, the SCC cells were dissociated and reseeded into flasks, and maintained for three, two, and five passages (SCC1, SCC2, SCC3, respectively) prior to SKY and FISH analyses.
Tissue Accrual, Tissue Culture, and Cytogenetic Preparations
CaP patients who underwent radical prostatectomy at the University Health Network (Toronto, ON, Canada) and had no previous radiotherapy or chemotherapy were evaluated for study eligibility based on tumor stage (pT1-T2), low tumor grade, prostate-specific antigen (PSA) levels, and past biopsy history. The surgeon dissected a small wedge (approximately 1–2 cm3) of tumor tissue from the excised prostate, and the resected prostatic capsule was analyzed for extracapsular tumor extension by the pathologist. The tissue wedge was quick-sectioned and hematoxylin- and eosin-stained, and the histopathology was assessed. Samples from 15 patients which showed high tumor content (>80%) were included in this study, and their apposing regions obtained for analysis.
The tissue was digested in 250 U/ml collagenase IV (Gibco BRL) in tissue culture media (RPMI 1640, 10% fetal bovine serum, antibiotics) for 2 to 3 hours. The resulting cell suspension was centrifuged gently and washed with phosphate buffer saline. The cells were then either directly harvested or harvested following a short-term culture (<1 week) for FISH analysis. Tissue sections from the tumor samples were not used since it is well established [20] that a large proportion of nuclei is bisected during preparation, leading to loss of FISH signals.
SKY
The SKY KIT probe cocktail from Applied Spectral Imaging (ASI, Carlsbad, CA) was hybridized to metaphase slides for the DU145 and DU145-derived subclones according to standard protocols [16] and the manufacturer's instructions (ASI).
Results in the figures were reported using an abbreviated format of the International System for Human Cytogenetic Nomenclature (ISCN) for chromosomal aberrations, omitting breakpoint information [21]. In keeping with ISCN conventions, irrespective of ploidy, an individual chromosomal aberration (structural or numerical change) is considered a clonal gain when it is observed at least twice, and clonal loss when missing in at least three cells [21]. The frequency of nonclonal changes were taken to reflect the propensity of aberration within the cell line. Clonal changes, however, indicate the perpetuation of aberrations through successive cell divisions.
FISH
Cytogenetic slides were prepared as previously described [22] from a total of 15 patients as well as the CaP cell lines using 1.5-hour colcemid treatment and 75 mM KCI hypotonic treatment. While metaphase and interphase nuclei were readily produced from the CaP cell lines, only interphase nuclei were obtained from the patient samples. Normal cytogenetic control slides bearing metaphases and interphase nuclei were made from phytohemagglutinin-stimulated normal male lymphoblasts. The centromere 8 D8Z1 alphoid centromeric sequence genomic clone was obtained from the ATCC (#59904, Rockville, MD) and labeled with biotin-14-dATP (Gibco BRL). A minimum of 100 cells was evaluated in each cytogenetic sample for assessing the centromere 8 frequencies.
Statistical Analysis
Statistical analysis using the chi-square test evaluated each of the patient samples versus the normal control, with 3 df (α=0.05). A similar comparison was made between each of the DU145 subclones and the DU145 parental cell line (4 df, α=0.001).
Results
Ploidy
As previously reported for this early passage number [6,7,19,23], our analysis by SKY demonstrated the DU145 parental cell line to have a range of 55 to 63 chromosomes per cell (Figure 1A). Because this range is less than three times the haploid chromosome count and most chromosomes were present in three copies, this cell line is probably derived from a hypotriploid stem line. The SCC2 and SCC3 cells were also observed to be hypotriploid, with ranges of 59 to 63 (Figure 1C) and 60 to 65 (Figure 1D), respectively. The SCC1 cells, however, had two distinct populations with different ploidy levels. In addition to the hypotriploid (56–61) cells, the SCC1 subclone had cells with double this ploidy (108–122) or hypohexaploid (Figure 4B). These results were corroborated by interphase and metaphase FISH on the DU145 cell lines (Table 1).
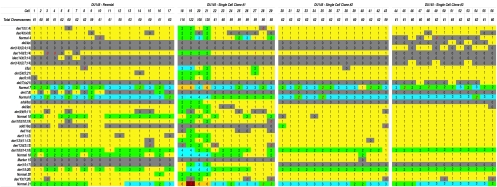
Figure 1.
Overview of the frequency of numerical change of chromosomal aberrations in the three DU145 subclones. In this figure, a color grid has been used to depict numerical change of the various aberrations that are characteristic of DU145 and its three subclones. The leftmost panel is the parental DU145 culture obtained from the ATCC. The three panels to its right depict copy number change in each of the three independently derived DU145 subclones. Each cell studied is represented by a column and each one of the 33 chromosomal aberrations of DU145 is identified as separate row. Colors have been used to depict copy number (1=yellow; 2=green; 3=light blue; etc.) so that it is easy to see that SCC2 and SCC3 exhibit a more homogeneous pattern of numerical change than SCC1 based on the more uniform appearance of the colored grid in the two rightmost panels. All cells in this analysis were hypotriploid except for cells 18 to 21 in SCC1, which had double the chromosome number. Copy numbers written in red text indicate that a nonclonal structural rearrangement of the chromosomal aberration was present in that particular cell.
Table 1.
Interphase and Metaphase FISH Analyses of the Centromere 8 Copy Number in the CaP Cell Lines and the Three DU145-Derived SCC Cells.
| Cell Line | Centromere 8 Copy Number | |||||
| 1 (%) | 2 (%) | 3 (%) | 4 (%) | 5 (%) | ≥6 (%) | |
| LNCaP | 3.0 | 67.0 | 6.0 | 20.5 | 1.5 | 2.0 |
| PC-3 | - | 79.4 | 5.9 | 10.3 | 2.2 | 2.2 |
| DU145 Parental | - | 0.7 | 70.0 | 21.3 | 1.3 | 6.7 |
| DU145 SCC1 | - | 5.1 | 79.2 | 2.0 | 2.5 | 11.2 |
| DU145 SCC2 | - | 5.5 | 89.0 | 2.8 | 1.4 | 1.4 |
| DU145 SCC3 | - | 1.9 | 88.3 | 0.6 | 3.1 | 6.2 |
In each cell line, the copy number with the highest observed frequency is in boldface. There is variability of the centromere 8 copy number around the modal values.
The LNCaP cell line was observed by interphase and metaphase FISH analysis to have two populations with diploid and tetraploid centromere 8 signals, reflecting previous data that demonstrated a mixed ploidy in this cell line [6,24]. The PC-3 cells were shown to have predominantly two copies of centromere 8 signals, which is in accordance with our previous data that showed it to be hypotriploid but with only two copies of chromosome 8 [6]. FISH results for LNCaP and PC-3 are also reported in Table 1.
Since the patient samples used in this study were derived from early-stage, low-grade primary tumors, it is probable that they have a diploid karyotype [25,26]. In keeping with this observation, our interphase FISH observations showed predominantly two centromere 8 copies in all the tumor samples (Table 2).
Table 2.
Interphase FISH Analyses of the Centromere 8 Copy Number in the CaP Patients.
| Patient | Centromere 8 copy number | |||
| 1 (%) | 2 (%) | 3 (%) | ≥4 (%) | |
| Normal blood | 1.0 | 96.0 | - | 3.0 |
| 1 | 5.4 | 79.6 | 8.2 | 6.8 |
| 2 | 4.9 | 84.2 | 4.9 | 6.1 |
| 3 | 2.7 | 90.3 | 6.0 | 1.1 |
| 4 | 0.8 | 86.3 | 7.3 | 5.7 |
| 5 | 1.5 | 85.1 | 10.5 | 3.0 |
| 6 | 0.7 | 89.8 | 7.5 | 2.0 |
| 7 | 2.5 | 91.5 | 5.9 | - |
| 8 | 3.6 | 48.7 | 44.1 | 3.6 |
| 9 | 10.2 | 83.2 | 4.4 | 2.2 |
| 10 | 7.2 | 75.5 | 5.0 | 12.2 |
| 11 | 4.9 | 76.1 | 18.6 | 0.4 |
| 12 | 3.0 | 92.3 | 2.7 | 1 |
| 13 | 9.7 | 80.2 | 6.9 | 3.2 |
| 14 | 7.6 | 69.6 | 14.3 | 8.5 |
| 15 | 5.0 | 86.1 | 7.5 | 1.5 |
In each patient, the copy number with the highest observed frequency is in boldface. There is variability of the centromere 8 copy number around the diploid modal value.
Aneusomy
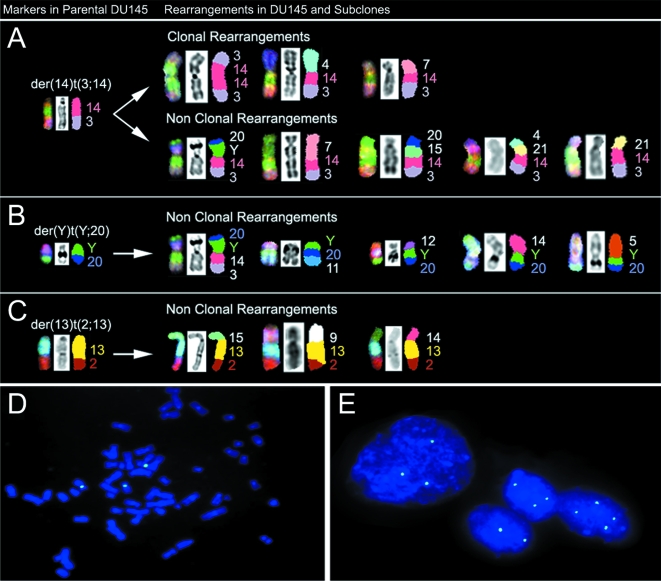
Our SKY analysis of aneusomies drew attention to a number of consistent features associated with karyotype evolution in each of the subclones (Figure 1). With the exception of SCC1, all the cell lines had two copies of chromosome 4. In the parental DU145 and SCC2 cells, the two copies were comprised of the normal 4 and derivative 4 chromosomes. As outlined in Figure 1, in parental cells where these chromosomes were not present, for instance in cells 9 and 10, the der(4)t(4;6) was replaced with a der(4)t(4;15) and the normal 4 was replaced with a der(4)t(4;9), respectively, to maintain the total of two copies of chromosomes 4. Similarly, in SCC3, two copies of chromosome 4 or derivative 4 chromosomes were maintained by having any two combinations of der(4)t(4;6), normal 4, del(4p), or der(14)t(3;4;14). The exception to this was cell 49, where a der(4)t(4;7) was observed in place of the normal chromosome 4 (Figure 1D).
Several clonal rearrangements were observed by SKY analyses that were specific to the parental or subclone cell lines. These results are summarized in Figure 1. For example, the isochromosome derivative 14 chromosome, ider(14)t(3;14), was observed in 2/17 parental cells, but not in any of the subclones. The der(7;8), which was a centromeric fusion of the short arm of chromosome 7(7p) and long arm of chromosome 8(8q), was observed in the parental cells (7/17) but was absent in the subclones. The clonal addition of chromosome 10 material to 10q, add(10q), was present in 3/17 parental cells, absent in the SCC1 cells, and ubiquitous in the SCC2 and SCC3 cells. Several novel chromosomal rearrangements that were not previously observed in the parental cells were identified in the subclones. The der(15;17), and the clonal addition of chromosome 8 material to 8q, add(8q), were specific to the SCC1 cells. A small derivative chromosome 15 rearranged with another minute indeterminable partner chromosome, designated Marker 15, was observed in only SCC2 and SCC3 cells. The del(7) (p21), and the previously described der(14)t(3;4;14), der(14)t(3;7;14), and del(4p) chromosomes were observed clonally and only in the SCC3 cells.
In addition to the observed structural changes, chromosomal aneusomy was also evident as numerical changes specific to the parental or subclone cell lines. As outlined in Figure 1, SKY analysis revealed that chromosome 10 was observed in one or two copies in the parental cells (4/17 and 13/17 cells, respectively), but SCC2 and SCC3 cells consistently had one copy. Similarly, while the parental cells had one, two, or three copies of chromosome 21 (4/17, 9/17, and 4/17, respectively), trisomy was a more dominant feature than disomy in the SCC2 (12/14 and 2/14, respectively) and SCC3 cells (9/13 and 4/13, respectively). Furthermore, the variability of chromosome 18 copy number in the parental line (13/17 disomy, 4/17 monosomy) was eliminated in the SCC2 and SCC3 cells which consistently exhibited a single copy.
Structural Aberrations
SKY analysis of the DU145 cell lines was able to identify all the complex chromosomal rearrangements within the DU145 cell lines. Of interest was the der(14)t(3;14), which was present in 13/17 cells of the DU145 parental cells (Figure 1). The evolution of the der(14)t(3;14) is particularly interesting (Figure 2A). Where this rearrangement was absent, rearrangements involving the derivative 14 chromosome were observed: e.g., der(Y)t(Y;3;14;20) (cell 3),der(14)t(3;14;21)(cell 4), and isoderivative chromosomes ider(14)t(3;14) (cells 5 and 7). Interestingly, the der(14)t(3;14) was also present in SCC1 (12/12) and SCC2 (14/14), but absent in SSC3 (0/13). In SCC3 cells, the der(14)t(3;14) usually presented as der(14)t(3;4;14) (8/13) or der(14)t(3;7;14) (3/13) rearrangements; exceptions to this observation were cells 51 and 56, in which these chromosomes were lost. Neither the novel der(14)t(3;4;14)nor der(14)t(3;7;14) chromosomes were observed in the SCC1, SCC2, or the parental cell lines. The occurrence of this der(14)t(3;14) in several different rearrangements in DU145 and its subclones is remarkable and suggests that it may confer a selective advantage.
Figure 2.
Evidence of a CIN-like process in DU145 cells as shown by structural and numerical aberrations. Panels A, B, C: Representative marker chromosomes observed in parental DU145 cell line [der( 14) t(3; 14); der(Y) t(Y;20); der(13)t(2;13)], and putative structural derivatives thereof (clonal and nonclonal) in the parental and subclone cell lines. Note the maintenance of the original marker chromosome in each of the structural derivatives and progression from a simple to a more complex rearrangement, suggestive of marker evolution. Panels D, E: Representative metaphase (Panel D) and interphase nuclei (Panel E) FISH image using centromere 8 probe (green) from the DU145 parental cell line. Analysis on a cell-by-cell basis revealed frequent chromosome 8 copy number change in a subpopulation of DU145 cells.
In Figure 2B, the der(Y)t(Y;20) was also observed to participate in five different nonclonal structural translocations, suggesting that the Y chromosome may be preferentially involved in rearrangement. Similarly, the distal part of the der(13)t(2;13) (Figure 2C) was involved in three different aberrations. It is noteworthy that the breakpoints involved in the marker evolution of der(14)t(3;14), der(13)t(2;13), and der(Y)t(Y;20) are all at regions of repetitive DNA.
The SCC1 cells demonstrated a greater number of nonclonal structural rearrangements and gains and losses of chromosomes than the other two subclones (Figure 1B). For instance, in cell 18, there were 11 novel, unique chromosomal rearrangements (not shown). Furthermore, while in the SCC2, SCC3, and DU145 parental cells there were consistent trends such as the total chromosome 4 and der (14) t (3;14) content reported above, this consistency was not as apparent in the SCC1 cells. Interestingly, more marked variation was observed even when comparing between the hypohexaploid cells (cells 18–21) of the SCC1 cell line. For instance, cell 18 had many more nonclonal structural and numerical aberrations than cell 21 (not shown).
By SKY analysis, it was apparent that dicentric chromosomes were present within the DU145 cell lines. As demonstrated in Figure 2C, the der(13)t(2;13) translocation, which is present in almost every cell of the DU145 cell lines, has two centromeres. Also depicted are other nonclonal dicentric chromosomes. There was no evidence of double minutes, ring chromosomes, or other chromosomal features associated with gross CIN in the DU145 cell lines.
Interphase FISH
Results for the FISH analysis of the CaP cell lines are given in Table 1, and representative metaphase and interphase FISH images of the DU145 parental cells are depicted in Figure 2D and E, respectively. The DU145 parental cells were observed to have a major population of cells with three copies of chromosome 8 (70%) and a smaller population with four copies (21%). SKY analysis of the DU145 parental cells demonstrated a similar frequency of cells having three copies of chromosome 8, and cells having four copies of chromosome 8 centromere when counted together with the additional der(7;8) chromosome. The SCC2 and SCC3 cells demonstrated by FISH had less centromere 8 copy number variation than the parental DU145 cells, with the majority of cells having three copies (89% and 88%, respectively). While the SCC1 cells had similar frequencies of three and four centromere copy numbers (79% and 2%, respectively) as the other two subclones, it also had a relatively high frequency of cells with at least six copies (11%), suggesting more genomic variability than the other two subclones. The reduction in centromere 8 copy number variation in the DU145 subclones versus the DU145 parental cells was statistically significant (P<.001). It should be noted that in the DU145 parental, SCC1, and SCC3 cells, there were no greater than 8, 14, and 8 centromere 8 copies, respectively. Notably, although by FISH it appeared that the majority of cells in the DU145 cell lines had three copies of chromosome 8, SKY analysis distinguished that in SCC2 and SCC3 cells, these were all normal copies of chromosome 8, but that in DU145 parental cells one of the chromosome 8 copies may be involved in a der(7;8) translocation, and in the SCC1 cells there was additional chromosome 8 material at the qter in one of the copies.
FISH analysis of the LNCaP cells showed most cells to have two copies of centromere 8 signals (67%), and a smaller percentage of cells to have four copies (20%). The PC-3 cells had predominantly two copies of centromere 8 (79%), and a smaller percentage of cells with four copies (10%), most likely representing cells in G2 phase of the cell cycle. Interestingly, both cell lines were observed to have variation of centromere 8 copy number (Table 1).
Examination of the centromere 8 copy number by interphase FISH in the CaP patient cohort (Table 2) demonstrated that the majority of cells had predominantly two centromere 8 copies. Furthermore, as compared to the normal control, the patient cohort had a statistically significant (P<.05), greater frequency of cells with one and three centromere 8 copies. While many cells also had some frequency of four copies, it is indeterminate without further analysis if these cells are in the G2 phase of the cell cycle, similar to the normal control, or are truly aberrant. Of interest, four patients had a trisomy of centromere 8 of greater than 10%, with patient 10 having the greatest frequency of 44%. Also, one patient had a frequency of monosomy of the centromere 8 signal at 10%.
Discussion
Features expected of tumors expressing CIN include aneuploidy and random chromosomal alterations, including aneusomy and structural rearrangements [27,28]. CIN may be associated with aberrations in mitotic spindle checkpoints [29] and genes such as hBUB1 and MAD2 [14,15], aberrant sister chromatid exchange [30], DNA repair pathways [31], and abnormal centrosome copy numbers or amplification [32–35]. In a more recent study, breakage-fusion-bridge cycles have been implicated in generating CIN [36]. Interestingly, some tumors exhibit both micro-satellite instability as well as CIN [15,37,38] as is the case for DU145 [39]. However, to date, there has been no direct evidence implicating CIN in CaP.
Giemsa banding and SKY analysis of DU145, reported previously [6,7], demonstrated that this cell line had a complex karyotype with some evidence of numerical variation suggestive that a CIN-like process may be operational. SKY analysis was performed on the DU145 parental cells, and as outlined in Figure 1, the pattern of chromosomal aberrations exhibits a high degree of cytogenetic heterogeneity. To investigate whether this karyotypic heterogeneity was due to the presence of multiple clones within the DU145 cell population, or had arisen de novo because of CIN, three DU145-derived subclones were studied by SKY analysis.
Since the three subclones were initially seeded as single cells derived from the parental cell population, it would be expected that they should maintain a high degree of karyotypic clonality and concordance with each other unless inherent instability was present. While it is possible that the high proliferation rate of in vitro growth may exaggerate inherent instability, it is clear that significant deviations between the subclones implicate a CIN-like process. As summarized in Figure 1, while these clones demonstrated an increased homogeneity of structural and numerical aberrations as compared to the DU145 parental cells, there were indications that de novo translocations occurred and that numerical aberrations were generated or lost within two to five passages following subclone generation. These include: del(4p), del(7)(q21), der(7;8) add(8q), der(10q), ider(14)t(3;14), der(14)t(3;14), der(14)t(3;4;14), der (14)t(3;7;14), Marker 15, der(15;17), and copy number changes for chromosomes 4, 10, 18, and 21. Furthermore, from Figure 1, it is apparent that SCC1 demonstrated a more heterogeneous karyotype than the SCC2 and SCC3 cells, with a bimodal population of hypotriploid and hypohexaploid cells and a greater overall number of chromosomal aberrations (Table 3).
Table 3.
Quantification of Clonal and Nonclonal Changes Per DU145 Cell Line.
| Cell Line | Clonal Changes | Nonclonal Changes | |
| DU145 Parental | Total | 11 | 40 |
| Normalised per cell | 0.65 | 2.35 | |
| DU145 SCC1 | Total | 8 | 23 |
| Normalised per cell | 0.67 | 1.92 | |
| DU145 SCC2 | Total | 1 | 13 |
| Normalised per cell | 0.07 | 0.93 | |
| DU145 SCC3 | Total | 8 | 14 |
| Normalised per cell | 0.62 | 1.08 | |
For each cell line, the total numbers of clonal and nonclonal changes were determined and normalised by dividing by the total number of cells analyzed (17, 12, 14, and 13, respectively). These results objectively support that there is more homogenous pattern of numerical changes in SCC2 than in SCC3 and SCC1. Note that the rate of nonclonal changes approaches that of the parental DU145 cell line, indicating the presence of a CIN-like process.
Detailed examination of the relative distribution of marker chromosomes in the three subclones also identified some preferred patterns of chromosomal aberration as the respective karyotypes evolved. For instance, while there was copy number variation of chromosome 4 in all the DU145 cell lines, in general there were at least two copies of chromosome 4 present per cell either as normal copies or involved in a derivative chromosome 4 rearrangement. Similarly, although the der(14)t(3;14) was present in the SCC1, SCC2, and DU145 parental cells, it was involved in novel clonal rearrangements in SCC3 cells (Figure 2A), suggesting that selective forces favor acquisition of specific markers or combinations of certain chromosomal regions. In this regard, it is noteworthy that rearrangement often took place at sites rich in repetitive DNA such as distal Yp, 13p, and 14p. Overall, there appeared to be some consistent features associated with the chromosomal constitution within each subclone, but closer analysis of the aberrations by SKY indicated that chromosomal gain may be achieved by both simple numerical gain and/or unbalanced structural translocation. These results also draw attention to the potential limitations of comparative genomic hybridization (CGH) in the analysis of tumors exhibiting a high degree of complexity of rearrangements, since it is usually only possible to determine the average level of chromosomal imbalance with this method [40].
Our results suggested that intrinsic aneusomy would also be measurable by FISH analysis. Cell-by-cell analysis of numerical change as determined by FISH using the centromere 8 probe with LNCaP, DU145, PC-3, and DU 145-derived subclones correlated well with the findings concerning chromosomal loss and gain identified by SKY (Table 1) [6]. Furthermore, it was apparent that there was variation in both the mean chromosome number and the range or spread of centromere 8 signals observed in all cell lines and subclones, suggesting that chromosomal segregation errors may be a general feature of CaP cell lines. Examination of the levels of aneusomy determined by centromere 8 FISH analysis of cells derived from primary tumor tissue revealed a modal distribution of frequencies with the majority of cells being close to diploid. Even if some bias towards diploidy — due to the unavoidable admixture of normal stromal and epithelial components with the tumor epithelial cells in these preparations — is considered, then the level of monosomy and trisomy is excessive in several tumor samples. Taken together, the levels of aneusomy for chromosome 8 present in cell line and primary tumor tissue are suggestive that CIN may be an early feature of the disease process in CaP.
In summary, the results suggest that 1) SKY is a valuable screening tool for the delineation of genomic instability in tumor cells; 2) a CIN-like process is an intrinsic feature of the DU145 cell line leading to excessive numerical and structural alterations to chromosomes; 3) there appears to be preferred sites of rearrangement at chromosomal regions rich in repetitive DNA; 4) the combination of such a CIN-like process and cell selection will lead to rapid acquisition of novel combinations of chromosomal aberrations within a given tumor cell population; 5) there may be different inherent rates of this CIN-like process, as demonstrated by the variation within the three subclones; and 6) excessive aneusomy of chromosome 8 in early-stage CaP tumors suggests that a CIN-like process may initiate the numerical variation upon which selective forces subsequently operate. In this regard, the adaptive capacity of a tumor may be defined by its higher intrinsic rate of instability. Given that no specific tumor-suppressor genes or dominant oncogenes have been associated with CaP to date, the role of CIN in CaP tumorigenesis and perhaps other tumor systems in general may warrant further investigation as an alternate model of oncogenesis.
Acknowledgements
The authors gratefully acknowledge the technical expertise of Salomon Minkin and Jane Bayani.
Abbreviations
- add
additional material to chromosome
- CIN
chromosomal instability
- del
deleted chromosome
- der
derivative chromosome
- ider
isochromosome derivative
- SCC
single cell clone
Footnotes
Financial support for this project is funded by a research grant from the Concern Foundation. P.C.P. was supported by a grant from the AFUD/AUA Research Scholar Program and Imclone Systems.
References
- 1.Ekman P, Adolfsson J, Gronberg H. Textbook of Prostate Cancer Pathology, Diagnosis and Treatment. London: Blackwell Sciences; 1999. The natural history of prostate cancer. [Google Scholar]
- 2.Bookstein R. Tumor-suppressor genes in prostatic oncogenesis. J Cell Biochem Suppl. 1994;19:217–223. [PubMed] [Google Scholar]
- 3.Macintosh CA, Stower M, Reid N, Maitland NJ. Precise microdissection of human prostate cancers reveals genotypic heterogeneity. Cancer Res. 1998;58:23–28. [PubMed] [Google Scholar]
- 4.Fearon ER, Hamilton SR, Vogelstein B. Clonal analysis of human colorectal tumors. Science. 1987;238:193–197. doi: 10.1126/science.2889267. [DOI] [PubMed] [Google Scholar]
- 5.Dong JT, Isaacs WB, Isaacs JT. Molecular advances in prostate cancer. Curr Opin Oncol. 1997;9:101–107. doi: 10.1097/00001622-199701000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Beheshti B, Karaskova J, Park PC, Squire JA, Beatty BG. Identification of a high frequency of chromosomal rearrangements in the centromeric regions of prostate cancer cell lines by sequential Giemsa banding and spectral karyotyping. Mol Diagn. 2000;5:23–32. doi: 10.1007/BF03262019. [DOI] [PubMed] [Google Scholar]
- 7.Pan Y, Kytola S, Farnebo F, Wang N, Lui WO, Nupponen N, Isola J, Visakorpi T, Bergerheim US, Larsson C. Characterization of chromosomal abnormalities in prostate cancer cell lines by spectral karyotyping. Cytogenet Cell Genet. 1999;87:225–232. doi: 10.1159/000015432. [DOI] [PubMed] [Google Scholar]
- 8.Ozen M, Pathak S. Genetic alterations in human prostate cancer: a review of current literature. Anticancer Res. 2000;20:1905–1912. [PubMed] [Google Scholar]
- 9.Oesterling J, Fuks Z, Lee CT, Scher HI. Cancer: Principles and Practice of Oncology. Philadelphia: Lippincott-Raven Publishers; 1997. Cancer of the prostate; pp. 1322–1385. [Google Scholar]
- 10.Verma RS, Manikal M, Conte RA, Godec CJ. Chromosomal basis of adenocarcinoma of the prostate. Cancer Invest. 1999;17:441–447. doi: 10.3109/07357909909021436. [DOI] [PubMed] [Google Scholar]
- 11.Dutrillaux B. DNA Alterations in Cancer. MA, USA: Eaton, Natwick; 2000. Chromosome and karyotype instability in human cancers and cancer-predisposing syndromes; pp. 369–382. [Google Scholar]
- 12.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 13.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and Darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–M60. [PubMed] [Google Scholar]
- 14.Pihan GA, Doxsey SJ. The mitotic machinery as a source of genetic instability in cancer. Semin Cancer Biol. 1999;9:289–302. doi: 10.1006/scbi.1999.0131. [DOI] [PubMed] [Google Scholar]
- 15.Cahill DP, da Costa LT, Carson-Walter EB, Kinzler KW, Vogelstein B, Lengauer C. Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics. 1999;58:181–187. doi: 10.1006/geno.1999.5831. [DOI] [PubMed] [Google Scholar]
- 16.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes. Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 17.Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L, Kim U, Chai LS, Kakati S, Arya SK, Sandberg AA. The LNCaP cell line — a new model for studies on human prostatic carcinoma. Prog Clin Biol Res. 1980;37:115–132. [PubMed] [Google Scholar]
- 18.Kaighn ME, Narayan KS, Ohnuki Y, Lechner JF, Jones LW. Establishment and characterization of a human prostatic carcinoma cell line (PC-3) Invest Urol. 1979;17:16–23. [PubMed] [Google Scholar]
- 19.Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145) Int J Cancer. 1978;21:274–281. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- 20.Aubele M, Zitzelsberger H, Szucs S, Werner M, Braselmann H, Hutzler P, Rodenacker K, Lehmann L, Minkus G, Hofler H. Comparative FISH analysis of numerical chromosome 7 abnormalities in 5-micron and 15-micron paraffin-embedded tissue sections from prostatic carcinoma. Histochem Cell Biol. 1997;107:121–126. doi: 10.1007/s004180050096. [DOI] [PubMed] [Google Scholar]
- 21.Mitelman F. ISCN (1995): International System for Human Cytogenetic Nomenclature. New York: S. Karger; 1995. [Google Scholar]
- 22.Dracopoli NC. Current Protocols in Human Genetics. New York, USA: John Wiley and Sons; 2000. [Google Scholar]
- 23.Bernardino J, Bourgeois CA, Muleris M, Dutrillaux AM, Malfoy B, Dutrillaux B. Characterization of chromosome changes in two human prostatic carcinoma cell lines (PC-3 and DU145) using chromosome painting and comparative genomic hybridization. Cancer Genet Cytogenet. 1997;96:123–128. doi: 10.1016/s0165-4608(96)00258-0. [DOI] [PubMed] [Google Scholar]
- 24.Gibas Z, Becher R, Kawinski E, Horoszewicz J, Sandberg AA. A high-resolution study of chromosome changes in a human prostatic carcinoma cell line (LNCaP) Cancer Genet Cytogenet. 1984;11:399–404. doi: 10.1016/0165-4608(84)90020-7. [DOI] [PubMed] [Google Scholar]
- 25.Micale MA, Mohamed A, Sakr W, Powell IJ, Wolman SR. Cytogenetics of primary prostatic adenocarcinoma. Clonality and chromosome instability. Cancer Genet Cytogenet. 1992;61:165–173. doi: 10.1016/0165-4608(92)90082-j. [DOI] [PubMed] [Google Scholar]
- 26.Lieber MM. DNA ploidy: early malignant lesions. J Cell Biochem Suppl. 1992;16H:44–46. doi: 10.1002/jcb.240501210. [DOI] [PubMed] [Google Scholar]
- 27.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 29.Skibbens RV, Hieter P. Kinetochores and the checkpoint mechanism that monitors for defects in the chromosome segregation machinery. Annu Rev Genet. 1998;32:307–337. doi: 10.1146/annurev.genet.32.1.307. [DOI] [PubMed] [Google Scholar]
- 30.Dhillon VS, Dhillon IK. Chromosome aberrations and sister chromatid exchange studies in patients with prostate cancer: possible evidence of chromosome instability. Cancer Genet Cytogenet. 1998;100:143–147. doi: 10.1016/s0165-4608(97)00022-8. [DOI] [PubMed] [Google Scholar]
- 31.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salisbury JL, Whitehead CM, Lingle WL, Barrett SL. Centrosomes and cancer. Biol Cell. 1999;91:451–460. [PubMed] [Google Scholar]
- 33.Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes, Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehead CM, Salisbury JL. Regulation and regulatory activities of centrosomes. J Cell Biochem Suppl. 1999;32–33:192–199. doi: 10.1002/(sici)1097-4644(1999)75:32+<192::aid-jcb23>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR, Sen S. Tumour-amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20:189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 36.Saunders WS, Shuster M, Huang X, Gharaibeh B, Enyenihi AH, Petersen I, Gollin SM. Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl Acad Sci USA. 2000;97:303–308. doi: 10.1073/pnas.97.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung SY, Yuen ST, Chan TL, Chan AS, Ho JW, Kwan K, Fan YW, Hung KN, Chung LP, Wyllie AH. Chromosomal instability and p53 inactivation are required for genesis of glioblastoma but not for colorectal cancer in patients with germline mismatch repair gene mutation. Oncogene. 2000;19:4079–4083. doi: 10.1038/sj.onc.1203740. [DOI] [PubMed] [Google Scholar]
- 38.Ohshima K, Haraoka S, Yoshioka S, Hamasaki M, Fujiki T, Suzumiya J, Kawasaki C, Kanda M, Kikuchi M. Mutation analysis of mitotic checkpoint genes (hBUB1 and hBUBR1) and microsatellite instability in adult T-cell leukemia/lymphoma. Cancer Lett. 2000;158:141–150. doi: 10.1016/s0304-3835(00)00512-7. [DOI] [PubMed] [Google Scholar]
- 39.Boyer JC, Umar A, Risinger JI, Lipford JR, Kane M, Yin S, Barrett JC, Kolodner RD, Kunkel TA. Microsatellite instability, mismatch repair deficiency, and genetic defects in human cancer cell lines. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 40.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes, Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]