Abstract
After exploring evolutionary aspects of branched-chain amino acid biosynthesis, the review focuses on the extended leucine biosynthetic pathway as it operates in Saccharomyces cerevisiae. First, the genes and enzymes specific for the leucine pathway are considered: LEU4 and LEU9 (encoding the α-isopropylmalate synthase isoenzymes), LEU1 (isopropylmalate isomerase), and LEU2 (β-isopropylmalate dehydrogenase). Emphasis is given to the unusual distribution of the branched-chain amino acid pathway enzymes between mitochondrial matrix and cytosol, on the newly defined role of Leu5p, and on regulatory mechanisms governing gene expression and enzyme activity, including new evidence for the metabolic importance of the regulation of α-isopropylmalate synthase by coenzyme A. Next, structure-function relationships of the transcriptional regulator Leu3p are addressed, defining its dual role as activator and repressor and discussing evidence in support of the self-masking model. Recent data pointing at a more extended Leu3p regulon are discussed. An overview of the layered controls of the extended leucine pathway is provided that includes a description of the newly recognized roles of Ilv5p and Bat1p in maintaining mitochondrial integrity. Finally, branched-chain amino acid biosynthesis and its regulation in other fungi are summarized, the question of leucine as metabolic signal is addressed, and possible directions of future research in this area are outlined.
INTRODUCTION
There has in recent years been a renewed interest in the intricacies of branched-chain amino acid biosynthesis, particularly leucine biosynthesis, sparked by a number of new and sometimes surprising observations in the yeast Saccharomyces cerevisiae, the organism of choice for many investigators in this field. These observations include the following. (i) First is the unusual partitioning of the pathway enzymes between the mitochondrial matrix and the cytoplasm and its consequences (Fig. 1). While the general aspects of this distribution pattern have been known for a long time, there have been recent refinements, especially with respect to the α-isopropylmalate synthase isoenzymes. In addition, hitherto unknown transport systems have come to light that are essential for efficient branched-chain amino acid biosynthesis, as well as for other processes. They include a transporter required for the accumulation of coenzyme A (CoA) inside the mitochondria; a system for the export of Fe-S clusters from the mitochondrial matrix to the cytoplasm, involving proteins whose primary function is to serve as branched-chain amino acid aminotransferases; and a hypothetical transporter that facilitates the exit of α-isopropylmalate from the mitochondria. (ii) Second is completion of the Saccharomyces genome sequencing project and the resulting definitive information on the number and structural relatedness of isoenzymes, questions of particular importance for the first specific step in leucine biosynthesis. It is now evident that there are two very closely related genes that encode α-isopropylmalate synthase. Together, they are capable of producing three isoforms of the enzyme, two located in the mitochondrial matrix and one that remains cytoplasmic. (iii) Third are new findings regarding metabolic regulation, which emphasize the signal character of the leucine pathway intermediate α-isopropylmalate and also assign a role to the transcriptional regulator Leu3p that reaches beyond branched-chain amino acid biosynthesis. Great strides have been made toward understanding the mechanism of action of Leu3p; it appears to be a “self-masking” regulator that represses transcription in the absence of α-isopropylmalate and becomes a strong transcriptional activator in its presence. Remarkably, the Leu3p-α-isopropylmalate system works as well when expressed in mammalian cells as it does in yeast. Since mammals do not have the capacity to produce branched-chain amino acids, the yeast Leu3p-α-isopropylmalate system may be useful as a novel hormone- and antibiotic-independent regulator of therapeutic genes.
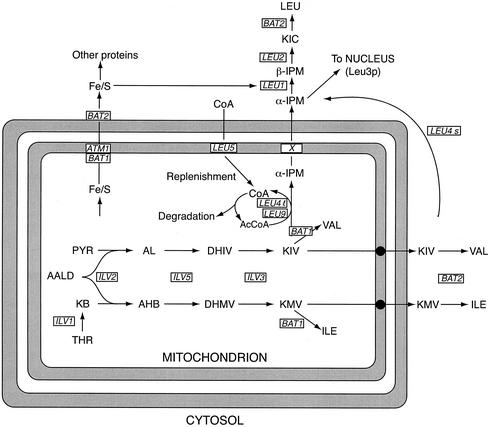
FIG. 1.
Compartmentation of the branched-chain amino acid biosynthetic pathways of S. cerevisiae. The boxed characters refer to the genes involved directly or indirectly in the pathways. Their protein products are as follows: ILV1, threonine deaminase; ILV2, acetohydroxy acid synthase catalytic subunit; ILV5, acetohydroxy acid reductoisomerase; ILV3, dihydroxy acid dehydratase; BAT1, mitochondrial branched-chain amino acid aminotransferase; BAT2, cytosolic branched-chain amino acid aminotransferase; LEU4l, α-isopropylmalate synthase I, long form; LEU4s, α-isopropylmalate synthase I, short form; LEU9, α-isopropylmalate synthase II; X, hypothetical α-isopropylmalate transporter; LEU1, isopropylmalate isomerase; LEU2, β-isopropylmalate dehydrogenase; LEU5, protein necessary for the accumulation of CoA within the mitochondria; presumably an importer of CoA or precursor thereof; ATM1, ABC transporter involved in exporting Fe-S clusters to the cytosol; the implied interaction between Bat1p and Atmp is hypothetical (58, 66). Abbreviations: KB, α-ketobutyrate; AALD, active acetaldehyde; PYR, pyruvate; AL, acetolactate; AHB, α-aceto-α-hydroxybutyrate; DHIV, α,β-dihydroxyisovalerate; DHMV, α,β-dihydroxy-β-methylvalerate; KIV, α-ketoisovalerate; KMV, α-keto-β-methylvalerate; α-IPM, α-isopropylmalate; β-IPM, β-isopropylmalate; KIC, α-ketoisocaproate; Fe/S, iron-sulfur cluster. Not shown for reasons of clarity: transport of leucine and/or KIC back into the mitochondria.
In this review, an attempt is made to critically evaluate earlier results and use them together with recent developments to give the reader a comprehensive picture of the extended leucine pathway and its metabolic ramifications in S. cerevisiae. In addition, to gain a wider perspective, information available on branched-chain amino acid biosynthesis in other fungi is summarized.
EVOLUTIONARY ASPECTS
As depicted in Fig. 1 (using the nomenclature for S. cerevisiae), the biosynthesis of the branched-chain amino acids consists of a common pathway that leads from pyruvate and α-ketobutyrate to valine and isoleucine and a branch that leads from the immediate precursor of valine, α-ketoisovalerate, to leucine. These pathways operate in eubacteria (106), archaebacteria (112), fungi (41, 59), and green plants (43, 62, 79). They are not found in mammals, a fact that was important in the development of herbicides that specifically inhibit branched-chain amino acid biosynthesis (24). Whenever nucleotide or amino acid sequences of genes or enzymes involved in the branched-chain amino acid pathways were compared in different species, significant similarities were found (an example is shown in Fig. 2). There are also strong functional similarities among enzymes from different species that relate to such properties as pH optima, kinetics, and cation requirements. It has been concluded, based on these observations, that the branched-chain amino acid pathways are very ancient and may have been derived from a common ancestor of the three major lineages (112). However, while the basic catalytic functions of the enzymes that make up these pathways have remained largely unchanged throughout evolution, differences with respect to other properties such as gene organization, gene and enzyme regulation, and pathway compartmentation clearly exist. For example, the genes involved in branched-chain amino acid biosynthesis in Escherichia coli are organized in operons whereas those of S. cerevisiae or Neurospora crassa are found throughout the genome, often on different chromosomes. The ilvGMEDA, ilvBN, and leuABCD operons of E. coli are regulated by attenuation of transcription in response to the availability of the cognate aminoacyl-tRNAs (106, 113); the ilvIH operon is under the control of Lrp, the leucine-responsive regulatory protein (19). By contrast, the transcription of the ILV and LEU genes of S. cerevisiae and N. crassa is individually controlled by a regulatory protein (Leu3p) in conjunction with the leucine precursor α-isopropylmalate and is also subject to other, more general controls (see below). Comparing compartmentation of the branched-chain amino acid biosynthetic enzymes in different eukaryotic species again reveals significant differences that demonstrate how difficult it is to foretell the way(s) in which problems of adaptation to new environments have been solved. For example, all of the enzymes of the common pathway plus most of the α-isopropylmalate synthase activity of S. cerevisiae are found in the mitochondrial matrix while the two remaining enzymes specific for leucine synthesis are cytoplasmic (Fig. 1; also see below). By contrast, the enzymes for leucine biosynthesis in spinach are located entirely to the chloroplasts (43); subfractionation showed that spinach α-isopropylmalate synthase is tightly associated with the thylakoid membranes, site of the light reactions.
FIG. 2.
Alignment of the R-regions (boxed) of known α-isopropylmalate synthase sequences. Amino acid sequence identities are indicated by bold letters, similarities are indicated by asterisks. Abbreviations and references: ScI, S. cerevisiae α-isopropylmalate synthase I (10; the amino acid positions shown at the top refer to this sequence); Sc II, S. cerevisiae α-isopropylmalate synthase II (107); Cg, Corynebacterium glutamicum (82); Ba, Buchnera aphidicola (16); Hi, Haemophilus influenzae (34); Mj, Methanococcus jannaschii (18); St, Salmonella enterica serovar Typhimurium (90); Ll, Lactococcus lactis (38); Ta, Thermus aquaticus (GenBank accession number U52907). Reprinted from reference 23 with permission of the copyright owner.
As an aside, the leucine pathway may have played a more important role in evolution than is evident from its present “peripheral” function. Miller and coworkers (54) have proposed that it may have served as the forerunner of the Krebs cycle (which, in its ancient form, is thought to have arisen as a means of supplying the cell with glutamate, aspartate, and related amino acids). This scenario assumes that the leucine pathway-specific genes were duplicated and mutated so that their protein products would accommodate the respective substrates of the Krebs cycle. Assuming that an α-isopropylmalate synthase homologue would act as citrate synthase, that isopropylmalate isomerase homologues would function as aconitase and fumarase and that β-isopropylmalate dehydrogenase homologues would act as isocitrate dehydrogenase and malate dehydrogenase, and also that a fatty acid reductive carboxylase was available to catalyze the interconversion of succinate and α-ketoglutarate, the only additional enzyme required to complete the cycle would then have been succinate dehydrogenase. It was also assumed that the Krebs cycle-related amino acids, because of their apparent great abundance under prebiotic conditions, would have been depleted later than the less abundant branched-chain amino acids, implying that branched-chain amino acid biosynthesis preceded that of glutamate and aspartate.
α-ISOPROPYLMALATE SYNTHASE OF S. CEREVISIAE: ISOFORMS, COMPARTMENTATION, AND REGULATION
The Leu4p and Leu9p Isoforms and Their Intracellular Localization
It had long been surmised on genetic grounds that yeast cells elaborate two independent α-isopropylmalate synthases (7). With the completion of the sequencing of the S. cerevisiae genome, it became clear that there are indeed two, and only two, genes, designated LEU4 and LEU9, that encode α-isopropylmalate synthases (21, 107). LEU4 is located on chromosome XIV (27), and LEU9 is located on chromosome XV (107). The primary structures of the two isoenzymes Leu4p (α-isopropylmalate synthase I) and Leu9p (α-isopropylmalate synthase II) are 83% identical. In wild-type cells, Leu4p accounts for about 80% of the total synthase activity. It is also the better understood of the two isoenzymes.
An interesting aspect of the LEU4 gene is that it itself can generate two forms of α-isopropylmalate synthase. This conclusion was first arrived at when it was observed that two of four major transcription start sites utilized in vivo are located downstream from the ATG at the beginning of the open reading frame (ORF) of LEU4 (10), which suggested that LEU4 mRNAs might be translated into full-length and shortened versions of the isoenzyme. This was subsequently shown to be true. As it turns out, the full-length form of α-isopropylmalate synthase I contains at its N terminus a signal sequence that directs the enzyme to the mitochondrial matrix; the short form, starting at what would be the second in-frame AUG of the long mRNA, lacks the 30 N-terminal residues of the long form and hence the mitochondrial import sequence. It remains in the cytoplasm (11). These observations provided an early example of how differential transcription can be used to generate mitochondrial and nonmitochondrial forms of the same enzyme.
Why should α-isopropylmalate synthase be localized in two cellular compartments? Its presence in the mitochondrial matrix can be rationalized by arguing that both of the enzyme's substrates, acetyl-CoA and α-ketoisovalerate, are prevalent in that compartment. The reason that α-ketoisovalerate accumulates in mitochondria stems from the fact that all of the enzymes required to convert pyruvate and active acetaldehyde to α-ketoisovalerate are also located in this organelle (93) (Fig. 1). Given this logic, it is not immediately obvious why α-isopropylmalate synthase should also be available in the cytoplasm. It has been speculated that the reason for this may have to do with the ability of S. cerevisiae to grow both aerobically and anaerobically, i.e., both with and essentially without functional mitochondria. Under anaerobic conditions, the mitochondrial form of α-isopropylmalate synthase might be unstable or nonfunctional for other reasons and the cytosolic form might take over. A similar situation might arise under glucose repression, a condition in which cells also elaborate only very few mitochondria with poorly developed cristae (68). These questions of compartmentation seem to be limited to Leu4p, since Leu9p is apparently found only in the mitochondria (86).
While the synthesis of α-isopropylmalate occurs largely within the mitochondria, its conversion to leucine takes place in the cytoplasm (94), as discussed in more detail below and shown in Fig. 1.
The Unexpected Role of Leu5p
An early tetrad analysis (7) had indicated that α-isopropylmalate synthase activity in yeast depended on two separate genes, which were designated LEU4 and LEU5. (A leu4 leu5 strain is a leucine auxotroph.) While LEU4 was identified as a structural gene, LEU5 was thought to either directly encode the second synthase or provide some function needed for the expression of a second structural gene. Subsequent experiments showed that strains deficient in LEU5 behaved as petite strains, i.e., did not grow well on most nonfermentable carbon sources, and that LEU5 was probably not the second structural gene (32). The conundrum of LEU5 was solved by the recent surprising demonstration that the gene encodes a carrier protein located in the mitochondrial inner membrane that is required for the buildup of CoA inside the mitochondria (86). It presumably serves to transport CoA, or a precursor thereof, from the cytosol, its likely site of synthesis, to the mitochondrial matrix to replenish the CoA pool and allow efficient synthesis of acetyl-CoA Fig. (1). The fact that a Δleu5 strain exhibits a 15-fold drop in the mitochondrial CoA level, accompanied by a slight increase in the cytoplasmic CoA level, is consistent with the carrier role of Leu5p and might also explain the petite-like nature of leu5 cells. Additional support for a carrier role of Leu5p comes from the observation that intact mitochondria from leu5 cells are essentially incapable of synthesizing α-isopropylmalate, even though they contain α-isopropylmalate-synthesizing activity that can easily be detected after lysis of the mitochondria (86).
LEU5 was later joined by several additional genes whose absence in a leu4 background substantially decreases the capacity to synthesize α-isopropylmalate (33). The additional genes (complementation groups) were designated LEU6, LEU7, and LEU8. They have not been analyzed, and we do not know the functions of their protein products. It is entirely possible, though, that either LEU7 or LEU8 (neither of which shows petite-like deficiencies when mutated) is identical to LEU9.
Regulation of α-Isopropylmalate Synthases
Regulation of LEU4 expression.
The regulation of LEU4 and its major gene product is more complex than might be expected if the synthesis of α-isopropylmalate were to serve the leucine pathway only. Gene expression control, studied with the aid of a LEU4-lacZ fusion, was found to depend on three distal elements and one proximal element of the LEU4 promoter (52). Of these, the most distal element corresponded to a binding site for the transcriptional regulator Leu3p (UASLEU). Mutation of UASLEU resulted in the loss of control by Leu3p. The other two elements in this region were identified as likely Gen4p binding sites; their inactivation by mutation eliminated general amino acid control of LEU4. Transcriptional activation from all three distal elements was additive, implying that the regulatory proteins bound to these elements do not compete with one another or act synergistically. The fourth, proximal regulatory element showed similarity to the Bas2p response element of the HIS4 promoter (102). Bas2p (Phos2p) is a “global,” homeobox-containing regulatory protein that is required, together with Bas1p, for incremental HIS4 transcriptional activation and also activates transcription of the secreted acid phosphatases. LEU4 gene expression decreased by as much as 45% when the inorganic phosphate concentration in the growth medium was increased from a depleted level to 5 mM. This response was absent when the presumed BRE of the LEU4 promoter was made nonfunctional by mutation (52). Elimination of the BRE also caused a drop in LEU4 expression under phosphate sufficiency.
Regulation of synthase activity.
The enzymatic activity of α-isopropylmalate synthase is subject to two major controls by small molecules. The first is feedback (end product) inhibition by leucine. The apparent inhibitor constants for l-leucine were determined to be about 0.1 mM for the long form of α-isopropylmalate synthase I (26), about 0.4 mM for the short form of synthase I (11), and about 1 mM for α-isopropylmalate synthase II (27), all at pH 7.2. The second type of activity regulation of α-isopropylmalate synthase is Zn2+-dependent reversible inactivation by CoA (44, 103, 104, 105). While CoA is a product of the reaction catalyzed by α-isopropylmalate synthase, the inactivation by CoA clearly is not simple product inhibition. Rather, it requires a second binding site for CoA that, in contrast to the substrate/product site, opens up only in the presence of zinc ions (104). In the absence of zinc and other divalent metal ions, CoA (bound at the product site) is a competitive inhibitor with respect to acetyl-CoA, with an apparent inhibitor constant of 65 to 70 μM; binding to the second CoA site in the presence of 50 μM Zn2+ occurs with an apparent dissociation constant of about 35 μM and causes a rapid, albeit noninstantaneous inactivation of the enzyme. Acetyl-CoA neither prevents nor reverses this inactivation. However, experiments performed in situ, i.e., in permeabilized cells, demonstrated that reactivation is possible with physiological concentrations of ATP or ADP. ATP is also capable of protecting α-isopropylmalate synthase against CoA inactivation. As might be expected from these results, protection was found to increase with increasing adenylate energy charge values (44).
Further support for the physiological importance of the CoA inactivation and its occurrence in vivo comes from a recent demonstration that strains in which α-isopropylmalate synthase I is no longer regulated by CoA are resistant to 5′,5′,5′-trifluoroleucine, owing to overproduction and secretion of leucine (23). Two such strains were identified within a group of seven spontaneous, trifluoroleucine-resistant mutants; the other five were shown to contain an α-isopropylmalate synthase I that had largely or completely lost its sensitivity to leucine. Remarkably, all seven mutations (six point mutations and one codon deletion) map to a short, 39-residue region near the C terminus of the enzyme, designated the R region by the authors. The R region is also present in α-isopropylmalate synthase II, with only six conservative changes, and is found largely conserved (69 to 56% similarity) in the synthases from seven bacterial species (Fig. 2). It has not been established whether the R region contains part or all of the leucine and CoA-Zn2+ binding sites or whether it is involved in the signal propagation caused by these molecules. The fact that leucine can partially protect yeast α-isopropylmalate synthase against CoA inactivation (105) is consistent with both interpretations. Whatever the mechanism, the observations by Cavalieri et al. (23) clearly show that the R region of α-isopropylmalate synthase contains subregions that respond to either leucine or CoA-Zn2+ and that both types of control must function to prevent overproduction of leucine.
Yet another hint at the metabolic importance of the CoA-Zn2+ inactivation of yeast α-isopropylmalate synthase comes from the work of Pronk et al. (87). These authors observed that a null mutant lacking PDA1, the gene encoding the E1α subunit of the pyruvate dehydrogenase complex, exhibited a partial leucine requirement. Since this requirement could not be satisfied by valine, the basis for this phenotype was thought to lie within the leucine pathway itself. Knowing that deletion of the PDA1 gene eliminates pyruvate dehydrogenase activity and sharply elevates the CoA/acetyl-CoA ratio in the mitochondria, the authors suggested that the Leu− phenotype of the pda1 null mutant might stem from increased CoA inactivation of α-isopropylmalate synthase. It has not been ruled out, however, that the diminished availability of acetyl-CoA itself might contribute to the partial leucine auxotrophy.
In a wider metabolic context, it is noteworthy that CoA inactivation is not limited to α-isopropylmalate synthase but is also observed with homocitrate synthase, the enzyme catalyzing the first committed step in lysine biosynthesis, and with β-hydroxy-β-methylglutaryl-CoA reductase, a key enzyme in the biosynthesis of sterols (101, 103). (In the latter case, the actual inactivating species is probably CoA disulfide [37].) By contrast, citrate synthase is entirely resistant to inactivation by CoA or derivatives thereof. This scenario was interpreted to mean that under conditions where the intramitochondrial CoA/acetyl-CoA ratio is high due to a low acetyl-CoA level, the remaining acetyl-CoA would be channeled into the citric acid cycle by limiting its consumption in biosynthetic pathways (44).
Why is the production of α-isopropylmalate subject to such elaborate controls? This question can be answered at least in part by pointing to the likely involvement of α-isopropylmalate, when complexed with Leu3p, in metabolic processes that go distinctly beyond the biosynthesis of the branched-chain amino acids (see below). The CoA-Zn2+ inactivation of α-isopropylmalate synthase may then be looked on not just as an acetyl-CoA saving device but also as an important link between those metabolic processes and central energy metabolism.
ISOPROPYLMALATE ISOMERASE (LEU1P) AND β-ISOPROPYLMALATE DEHYDROGENASE (LEU2P) OF S. CEREVISIAE
Intracellular Localization and the Question of an Isomerase-Dehydrogenase Complex
Of several reasons to lump together the two yeast enzymes needed to convert α-isopropylmalate to α-ketoisocaproate, their intracellular localization is perhaps the most compelling. In contrast to the enzymes responsible for the conversion of α-ketobutyrate and pyruvate to the α-ketoacid precursors of isoleucine and valine and to α-isopropylmalate, which are largely mitochondrial, isopropylmalate isomerase and β-isopropylmalate are entirely cytoplasmic (94). A recent independent confirmation of the cytoplasmic localization of the isomerase came from work done by Lill and coworkers on the maturation of Fe-S proteins (57, 58, 67, 75, 85). These authors observed that lesions in the ATM1 gene lead to leucine auxotrophy. They explained this behavior by pointing out that Atm1p, a mitochondrial ATP binding cassette (ABC) transporter, is required for the efficient generation of extramitochondrial (but not intramitochondrial) Fe-S clusters and that, in the absence of functional Atm1p, isopropylmalate isomerase, an Fe-S protein, would not be able to function if it were cytoplasmic.
The spatial separation of (most of) the α-isopropylmalate synthases from the isomerase and the dehydrogenase means that α-isopropylmalate would have to be transported from the mitochondrial matrix to the cytoplasm. The molecule also would have to enter the nucleus in order to form an active complex with Leu3p. The possibility that α-isopropylmalate binds to Leu3p in the cytoplasm cannot be dismissed but is considered unlikely since Leu3p localizes to the nucleus and interacts with its target promoters irrespective of whether α-isopropylmalate is present (17, 56). So far, no α-isopropylmalate transporter has been identified.
It has long been known that both isopropylmalate isomerase and β-isopropylmalate deydrogenase are unusually labile when removed from their natural environment (13, 14, 48). For example, isomerase precipitated from crude extract by ammonium sulfate fractionation has a half-life of only 2 to 3 h. The enzyme can be stabilized by a combination of cryoprotectants and, to some extent, by the presence of β-isopropylmalate. The dehydrogenase is exceptionally sensitive to low temperatures (and to dilution), but this occurs only when it is outside the cells; intact yeast cells can be stored frozen for months without loss of dehydrogenase activity. Strikingly, when LEU2 is expressed in E. coli, the yeast dehydrogenase is cold sensitive even in intact cells (60). This behavior of the two enzymes has led to the speculation that they might exist as a mutually stabilizing complex in vivo (59). It should be pointed out in this context that when a polyfunctional complex of isomerase and dehydrogenase was sought in N. crassa because of the coordinate expression of the corresponding genes, none was found (41).
Regulation of LEU1 and LEU2 Expression
Regulation of the synthesis of isopropylmalate isomerase and β-isopropylmalate dehydrogenase is achieved by controlling the rate of transcription of LEU1 and LEU2, mainly through Leu3p-α-isopropylmalate. The levels of the encoded enzymes and the cognate mRNAs respond similarly to genetic or physiological changes that affect LEU3 or alter the intracellular concentration of α-isopropylmalate (3, 7, 50). In addition to being controlled by Leu3p-α-isopropylmalate, LEU1 appears to be mildly up-regulated by Gcn4p, although the effect may be indirect (76). The response of LEU2 to Gen4p is atypical (17). There is no up-regulation by Gcn4p, but Gcn4p is needed to maintain a low but significant level of expression (basal level I) when Leu3p is absent. By an unknown mechanism, UASLEU-bound, activation-incompetent Leu3p abolishes this Gcn4p effect and reduces LEU2 expression to basal level II, a very low level of expression (see also the next section).
LEU3P: A DUAL-FUNCTION TRANSCRIPTION FACTOR
Leu3p as Transcriptional Activator and Repressor
The LEU3 gene of S. cerevisiae was originally identified as a positive regulator of the expression of LEU1 and LEU2, the genes encoding isopropylmalate isomerase and β-isopropylmalate dehydrogenase, respectively (7). The activation of these genes was found to depend not only on the presence of an intact LEU3 gene product but also on the rate at which α-isopropylmalate is produced. In the absence of α-isopropylmalate synthase activity, the activities of the LEU1 and LEU2 gene products were less than 10% of those found in wild-type cells. On the other hand, when α-isopropylmalate was synthesized in largely uncontrolled fashion (in a LEU4fbr strain that produces feedback-resistant α-isopropylmalate synthase), the LEU1 and LEU2 gene product activities rose three- to fivefold over those seen in wild-type cells. When, in addition, α-isopropylmalate was made to accumulate to even higher concentrations by preventing it from being metabolized (using LEU4fbr leu1 and LEU4fbr leu2 strains), the activities of the LEU2 or the LEU1 gene product increased about 15- and 10-fold, respectively. It was concluded that, by analogy to the pattern seen previously with N. crassa (41), the expression of LEU1 and LEU2 is subject to induction by a Leu3p-α-isopropylmalate complex. Subsequent refinement of the analysis of LEU2 expression using a LEU2-lacZ fusion confirmed this conclusion and demonstrated, furthermore, that Leu3p is a dual regulator (17): in the presence of α-isopropylmalate, it is a transcriptional activator; in its absence, it acts as a repressor. To understand this behavior, one has to realize that in the total absence of Leu3p, LEU2 gene expression is not zero but still proceeds at about 8% of that of the wild-type control, rendering Δleu3 strains leaky (basal level I expression). Leu3p molecules that are activation-incompetent but retain an intact DNA binding function decrease basal level I expression of LEU2 by another fourfold, to basal level II, a very low level of expression also seen in cells that are wild type with respect to LEU3 but are essentially unable to produce α-isopropylmalate (e.g., in a leu4 leu5 strain). The switch from basal level I to basal level II defines the repressive effect of Leu3p. It follows from these observations that Leu3p should bind to its cognate DNA irrespective of the presence or absence of α-isopropylmalate. That was indeed shown to be the case (17, 99). It was shown independently that Leu3p localizes to the nucleus (and binds to target sites) under a variety of growth conditions which included the presence and the absence of α-isopropylmalate (56).
Structure-Function Relationships: the Self-Masking Model
Structural aspects of Leu3p.
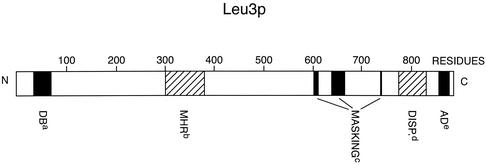
Important regions along the primary structure of Leu3p and their significance are shown schematically in Fig. 3.
FIG. 3.
Regions of interest along the primary structure of Leu3p of S. cerevisiae.aDNA binding region, residues 37 to 67, containing the cysteine motif for Zn(II)2Cys6 cluster formation;bMHR, residues 300 to 380;cthree clusters of residues (604 to 611, 643 to 664, and 738 to 741) potentially involved in activation domain masking;dregion dispensable for all known functions, residues 774 to 854;etranscriptional activation domain, residues 857 to 875; N, N terminus; C, C terminus. Note that the 11 C-terminal residues (residues 876 to 886), while not needed for the activation function, are essential for the masking process. See the text for further details.
Leu3p is a homodimeric DNA binding protein with 886 residues per monomer and a deduced monomer molecular weight of close to 100,000 (89, 118). It binds to cognate promoters at upstream activating sequences (UASLEU) with the structure 5′-CCGN4CGG-3′ (45). This structure is not entirely invariable. For example, the element found in the promoter of the LEU4 gene fits the above palindrome only when one mismatch is allowed (10, 45). Also, the sequence of the spacing nucleotides is not random but provides additional information for binding specificity (78). The DNA binding region of Leu3p is of the Zn(II)2Cys6 binuclear cluster type and is located near the N terminus. The DNA binding motif is Cys-Xxx2-Cys-Xxx6-Cys-Xxxn-Cys-Xxx2-Cys-Xxx6-Cys, where Xxx is any amino acid. This motif is found in at least 79 fungal proteins, including at least a dozen transactivators (95). Apparent dissociation constants of Leu3p-DNA complexes using purified Leu3p and a LEU2 promoter-derived 30-mer containing the UASLEU were determined to be in the low nanomolar range (97), values very close to those seen with other yeast transcriptional factors and with mammalian hormone receptors. Circular permutation and cyclization experiments showed that binding of Leu3p to UASLEU causes DNA bending (H. Guo and G. B. Kohlhaw, unpublished results). The apparent flexure angle, calculated from circular permutation assays, was found to be 47° with full-length Leu3p and 25° with a Leu3p fragment containing residues 17 to 147, i.e., little more than the DNA binding region. This result suggests that interaction between Leu3p and its cognate DNA not only depends on the DNA binding motif but also is influenced by other regions of the protein.
The transcriptional activation domain of Leu3p is located near the C terminus. This region is promiscuous: it is functional across species and when attached to other DNA binding domains (42, 98, 110). There is very little sequence homology among the activation domains of this class of transactivators. Rather, what seems to be important is the presence (and probably the spatial arrangement) of both acidic and hydrophobic residues. The activation domain of Leu3p is surprisingly small. The 30-C-terminal-amino-acid residue region originally identified as encompassing the activation function was subsequently whittled down to only 19 residues; even a Leu3p molecule lacking 17 of the 30 C-terminal residues retains about 25% of the activation potential of the wild-type protein (109). Reassuringly, its activation domain still contains three acidic and four hydrophobic residues.
In agreement with the notion that the regions of Leu3p that are responsible for DNA binding, dimerization, and transcription activation operate largely autonomously, a large internal segment of Leu3p that encompasses more than two-thirds of the entire protein can be deleted without infringing on the above functions. However, the shortened protein now behaves as a constitutive activator that maximally stimulates the expression of genes under its control irrespective of the presence or absence of α-isopropylmalate (35, 117). These results suggest that some aspects of the internal region (consisting of residues 174 to 773) are essential for the modulation of Leu3p by α-isopropylmalate. Additional deletion analysis showed that an 81-residue region on the N-terminal side of the activation domain is dispensable for all known functions of Leu3p (119). Since deletions often led to instability, other strategies were used to elucidate the mechanism by which the activation domain of Leu3p is masked in the absence of α-isopropylmalate and unmasked in its presence. One important question was whether auxiliary factors were needed for the masking-unmasking process, as is the case with some other modulated transactivators, e.g., Gal4p (63, 64) and possibly Hap1p (115, 116).
Are auxiliary factors needed to render the activation domain of Leu3p ineffective?
In a first approach to answer this question, the LEU3 gene was expressed in mammalian cells (42). To this end, mouse preadipocytes were transfected with two plasmids, one containing the LEU3 gene behind the human cytomegalovirus major intermediate-early promoter and the other carrying a luciferase reporter gene with four consecutive UASLEU sequences in its promoter. Cultured cells strongly expressed Leu3p, irrespective of whether α-isopropylmalate was added to the cell suspension. A basal level of reporter gene expression was seen in the absence of LEU3. This level decreased by about fourfold when LEU3 was present. When both LEU3 and α-isopropylmalate were present, reporter gene expression increased about 17-fold compared to the “repressed” level. Similar results were obtained in in vitro transcription assays using nuclear extract from mouse preadipocytes and highly purified Leu3p. These assays showed in addition that induction by α-isopropylmalate was specific since it was not observed when β-isopropylmalate was used instead. Several conclusions may be drawn from these experiments. First, Leu3p assumes its masked (and repressive) configuration when expressed in the absence of the coactivator. This is in striking contrast to what was observed when Lac9p, a Gal4p-related activator from Kluyveromyces lactis, was expressed in mammalian cells (96). In that case, it was necessary to coexpress the inhibitory yeast Gal80 protein in order to prevent transcription activation of a reporter gene. Second, both repression and activation by Leu3p occur in mammalian cells very much like they do in yeast. Third, since LEU3 was the only yeast gene present in the mammalian cell experiment and since mammalian cells do not elaborate branched-chain amino acid pathways, masking and unmasking of the activation domain of Leu3p very probably occur without the participation of additional Leu-specific factors.
More direct support for the idea that masking and unmasking of the activation domain are intra-Leu3p events comes from extensive mutant analyses and physical interaction studies (109, 110). Permutation of the activation domain showed that the modulation function was much more sensitive to amino acid changes and deletions than was the activation function. The effects seen on modulation could be classified as either permanent unmasking of the activation domain (i.e., generation of constitutive activators), diminished masking, or intensified masking. Of particular interest was a doubly mutated Leu3p (Leu3p[D872N/D874N], designated Leu3-dd) that is essentially unresponsive to α-isopropylmalate, even though it is able to interact with the coactivator and possesses strong intrinsic activation capability. The latter was demonstrated by creating a Leu3-dd molecule from which the large internal segment recognized earlier as important for modulation had been deleted; it was fully active. To further investigate whether the “permanent mask” for the activation domain of Leu3-dd was provided by the Leu3 protein itself, a modified two-hybrid experiment was designed in which a Leu3p wild-type molecule minus its activation domain served as “bait” and separate activation domains served as potential “prey.” The results were consistent with a self-masking model. It was found that (i) both wild-type and Leu3-dd activation domains interacted with the remainder of the Leu3p molecule; (ii) the interaction between the Leu3-dd activation domain and the bulk of Leu3p was much stronger than the interaction between the wild-type activation domain and the remainder of Leu3p, which, nevertheless, was clearly visible; (iii) most importantly, the interaction in both cases depended on the α-isopropylmalate concentration, i.e., took place only when little or no coactivator was present, demonstrating that the interactions were specific and physiologically meaningful. It should be noted that interaction between the activation domain and an internal region of Leu3p might well involve both subunits of the protein; i.e., activation domain masking could be an intradimer rather than an intramonomer event.
As expected, the Leu3-dd protein caused repression, and cells containing Leu3-dd grew only very slowly on minimal medium, opening the door to select for faster-growing suppressors. Methods were developed to select for intragenic suppressors of the Leu3-dd phenotype that were limited to an internal region of LEU3 covering residues 172 to 772 of Leu3p (109, 110). Interestingly, all nine missense mutations thus identified mapped to three very short regions located within the C-terminal one-third of Leu3p and extending from residues 604 to 611, 643 to 664, and 738 to 741, respectively (Fig. 3). Every one of the nine mutations restored substantial activation potential and various degrees of modulation, i.e., response to α-isopropylmalate, to the Leu3-dd molecule. When transferred to wild-type Leu3p, each of the nine mutations gave rise to constitutive, highly active activators, apparently abolishing masking altogether. It has been argued that at least some of the nine residues, in their wild-type form, might be directly involved in trapping the activation domain. However, more structural knowledge is needed to answer that question.
Cha4p is a serine/threonine-responsive transcriptional activator in yeast with an arrangement of functional and structural regions along the primary structure that is not unlike that of Leu3p (47). Domain swapping between Leu3p and Cha4p demonstrated that each activation domain requires its cognate internal segment in order to be eclipsed (110). This requirement does not extend to the DNA binding domains since normal regulatory behavior was seen in both cases when only these regions were swapped.
It is not known exactly where and how α-isopropylmalate interacts with Leu3p. It has, however, been possible to determine apparent binding constants for the coactivator, using in vitro transcription assays. The values obtained were 0.2 mM using an S. cerevisiae system (99) and 0.25 mM using a nuclear extract from mammalian cells (42). No interaction was detected with β-isopropylmalate.
Masking versus repression.
The masked form of Leu3p is also the form that represses gene expression below the basal level. However, masking and repression are two entirely different processes. While it is now clear that masking of the activation domain requires (part of) the central region of the protein and does not require any part of the extended DNA binding domain (defined as residues 1 to 173), it has been shown that repression is as efficient with a peptide encompassing residues 17 to 147 of Leu3p as it is with the full-length protein (98). The region of Leu3p from residues 17 to 147 contains the cysteine residues that give rise to the Zn(II)2Cys6 cluster and a heptad repeat that is similar to the one observed in Gal4p (72) and is therefore probably involved in dimerization. The fragment from residues 17 to 147 is identical to the one that has been shown to bind to and bend UASLEU-containing DNA (Guo and Kohlhaw, unpublished), and repression could be a (direct or indirect) consequence of this change in DNA structure. It is also possible, however, that the region of Leu3p from residues 17 to 147 recruits another protein(s) that in turn exerts a negative effect on transcription. It has been shown that repression by Leu3p requires the presence of Mot1p (108). Mot1p is an evolutionarily conserved repressor of transcription by RNA polymerase II that acts by displacing the TATA box binding protein from its sites, perhaps through ATP-dependent conformational changes (1, 6). However, since it is unclear how Leu3p and Mot1p communicate, the exact pathway by which Leu3p exerts its repressive effect remains unresolved.
The ‘middle homology region' of Leu3p.
Many of the Zn(II)2Cys6 cluster-forming proteins, Leu3p included, contain an internal region known as the middle homology region (MHR). This homology was first identified by Chasman and Kornberg (28) and was thereafter reported to be present in as many as 50 such proteins but not in other protein classes (95). The MHR of Leu3p is located between residues 300 and 380. No conclusive function has been established for the MHR, but it has been speculated that it might be involved in decreasing the affinity of Zn(II)2Cys6 cluster proteins for “wrong” genomic binding sites, based mainly on the observation that binding site specificity of full-length Gal4p in vivo is stricter than that of the Gal4p binding domain in vitro (72, 73). As far as Leu3p is concerned, it is possible that the much stronger bending of cognate DNA by the full-length molecule than by the Leu3p [17-147] fragment is due to the presence of the MHR, but direct evidence to support this idea is not available.
HOW LARGE IS THE LEU3P REGULON?
Genetic and biochemical analyses have demonstrated that the expression of seven S. cerevisiae genes is, at least in part, under the control of Leu3p (Table 1, first category). Five of these genes belong to the branched-chain amino acid pathways. The product of the sixth gene (BAP2) belongs to a family of permeases concerned primarily with the uptake of branched-chain amino acids (reference 77 and references therein). The seventh gene (GDH1) is responsible in a major way for the assimilation of ammonia in S. cerevisiae. To date, no systematic DNA microarray study relating to Leu3p has been carried out. There are, however, two recent items that bear on the potential range of Leu3p-mediated regulation. The first of these is a genome-wide analysis of regions located upstream of ORF's with regard to Leu3p binding and occupancy (69). These authors determined the equilibrium dissociation constants of 43 variants of the consensus Leu3p binding site which made it possible to predict an equilibrium dissociation constant for all potential binding sites in the genome and to calculate the fractional occupancy of these sites at an assumed intranuclear concentration of Leu3p. A total of 6,276 ORFs were analyzed in this way and ranked according to the number and quality of Leu3p binding sites present in their potential 5′-“regulatory” regions (defined as the 600 bp 5′ to an ORF). Over a range of free Leu3p concentrations between 1 and 100 nM, all seven genes known to be regulated by Leu3p were found among the top 5% of all yeast genes (with four of them being among the top 0.2%), demonstrating the predictive power of the method. The second item concerns results from a limited DNA microarray experiment (L. Breeden, personal communication). In this experiment, which was a by-product of an unrelated project, the following genotypes were compared: leu2 HAA1 with LEU2 haa1 and LEU2 HAA1 with LEU2 haa1. Cells with an intact LEU2 gene accumulate much less α-isopropylmalate than do cells with a deficient LEU2 gene and would therefore be expected to show lesser induction of genes controlled by the Leu3p-α-isopropylmalate complex. Genes whose expression was specifically depressed when LEU2 was intact included not only LEU1, ILV3, ILV5, and GDH1 but also about two dozen other genes and hypothetical ORFs. Four of the additional genes (BAT1, GAP1, OAC1, and MAE1) were also found to rank within the top 5% of all genes in the above occupancy calculation (three of them ranked within the top 0.5%). Since the likelihood that these four genes are regulated by Leu3p is based on the results of two very different approaches, they are included as a second category (“potential Leu3p targets”) in Table 1. Also added to this category was LEU9, a close homologue of LEU4 that likewise ranked very high in the occupancy analysis. The 12 genes listed in Table 1 can be assigned to three classes. The first and largest class encompasses nine genes involved in branched-chain amino acid biosynthesis and amino acid uptake into the cell (LEU1, LEU2, LEU4, LEU9, ILV2, ILV5, BAT1, BAP2, and GAP1). The second class includes two genes that function in central nitrogen and carbohydrate metabolism (GDH1 and MAE1). Finally, the one gene of the third class (OAC1) produces an inner mitochondrial membrane-based carrier protein for oxaloacetate and sulfate. It is relatively easy to see connections between these classes. For example, the well-established activation of GDH1 by Leu3p-α-isopropylmalate may be looked on as a signal transduction from the periphery of nitrogen metabolism (represented by the leucine pathway) to an essential early step in nitrogen metabolism (the assimilation of ammonia). The product of the MAE1 gene, mitochondrial malic enzyme, functions in the intramitochondrial synthesis of pyruvate and may therefore be considered a supplier of an important precursor in the biosynthesis of valine, leucine, and isolecine within the very compartment where most of these biosynthetic reactions occur (15). Also, given the sensitivity of α-isopropylmalate synthase I (and probably II as well) to changes in the mitochondrial CoA/acetyl-CoA ratio and the dependence of α-isopropylmalate synthase II on functioning CoA import into the mitochondria, it is perhaps not surprising to find that the Leu3p-α-isopropylmalate complex might regulate a gene like OAC1. In terms of the number of genes controlled, the presumptive extended Leu3p regulon is comparable to several other yeast regulons. For example, the Zap1p zinc-responsive regulon encompasses some 40 genes (70), Mac1p activates seven genes of a copper regulon (39), and a multidrug resistance regulon consists of 26 targets controlled by Pdr1p-Pdr3p (31).
TABLE 1.
Targets for regulation by Leu3p-α-isopropylmalate in S. cerevisiae
| Gene | Function | Reference(s) |
|---|---|---|
| First category: established targets | ||
| LEU1/YGL009c | Isopropylmalate isomerase | 7, 36 |
| LEU2/YCL018w | β-Isopropylmalate dehydrogenase | 7, 17, 36 |
| LEU4/YNL104c | α-Isopropylmalate synthase I | 36, 52, 83 |
| ILV2/YMR108w | Acetohydroxyacid synthase | 36 |
| ILV5/YLR355c | Acetohydroxyacid reductoisomerase | 36 |
| BAP2/YBR068c | Branched-chain amino acid permease | 77 |
| GDH1/YOR375c | Glutamate dehydrogenase (NADP+) | 51 |
| Second category: potential targetsa | ||
| BAT1/YHR208w | Branched-chain amino acid aminotransferase | |
| GAP1/YKR039w | General amino acid permease | |
| OAC1/YKL120w | Mitochondrial oxaloacetate, sulfate carrier | |
| MAE1/YKL029c | Mitochondrial malic enzyme | |
| LEU9/YOR108w | α-Isopropylmalate synthase II |
The first four of these genes meet two criteria: first, they rank very high in a genome-wide Leu3p binding-occupancy analysis (see text); second, they show diminished expression, as determined by a DNA microarray experiment, in cells whose genetic background allows only very little accumulation of α-isopropylmalate compared to cells whose genetic background allows strong accumulation of the coactivator (see text). The fifth gene (LEU9) meets only the first criterion.
MULTIPLE LAYERS OF CONTROL
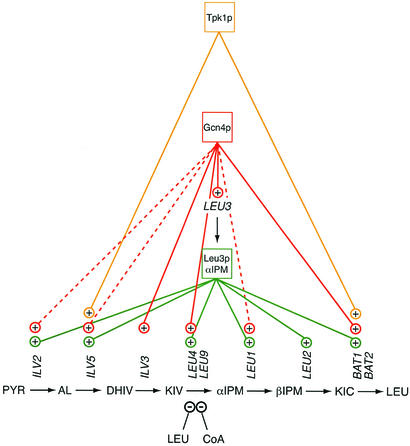
As illustrated in Fig. 4, the most direct control of the genes that define the extended leucine pathway is exerted by the Leu3p-α-isopropylmalate complex, which functions at six of the seven steps shown (ILV2, ILV5, LEU4, LEU1, LEU2, and BAT1), with the caveat that Leu3p control of BAT1 has not yet been directly demonstrated. Superimposed on this leucine-specific regulation is general amino acid control, mediated by Gcn4p. The effect of Gcn4p is twofold. First, it increases the Leu3p level, as inferred from experiments showing an increase in the rate of production of a Leu3p-β-galactosidase fusion protein in a manner typical for the general amino acid control system (118). This probably has physiological consequences since increased production of Leu3p can lead to increased target gene expression (110). Second, it acts directly on at least four genes (ILV3, LEU4, and BAT1-BAT2) of the extended leucine pathway; the effect on three more genes (ILV2, ILV5, and LEU1) may be indirect, through Leu3p (49, 52, 76). The Leu3p regulon thus becomes part of a remarkable regulatory network that encompasses at least 539 bona fide targets, among them a total of 26 genes that encode DNA binding transcription factors (76). By stimulating LEU3 expression, Gcn4p would amplify its impact in cascade-type fashion. Its simultaneous stimulation of LEU3 and LEU4 ensures that both components of the Leu3p-α-isopropylmalate complex will be made at increased rates. It can be imagined that, in a typical sequence of events, starvation for amino acids (or purines or glucose) would elicit an overriding response in a multitude of pathways, through the Gcn4p general amino acid control system. The Leu3p regulon would obey these starvation signals and augment them. In addition, it would respond independently to other signals, e.g. changes in the CoA/acetyl-CoA ratio, the cell's adenylate energy charge, or the leucine pool, that have an effect on the intracellular α-isopropylmalate level. This would give the yeast cell even more flexibility in its adjustment to environmental changes and demands.
FIG. 4.
Major regulatory mechanisms impacting the extended leucine pathway of S. cerevisiae. For abbreviations of intermediates and the protein products of the genes shown, see the legend to Fig. 1. See the text for further explanation.
Yet another layer of control that acts on genes of the extended leucine pathway is represented by Tpk1p (Fig. 4). This protein is one of three catalytic subunits of yeast protein kinase A. Like Tpk2p and Tpk3p, Tpk1p is released from an inactive complex under conditions that raise the intracellular cyclic AMP level, for example when cells leave stationary phase after gaining access to glucose or other fermentable carbon sources. The functions of the three subunits were recently studied by genome-wide transcriptional profiling, and it was established by comparing a TPK1 mutant strain with the wild type that there was a significant reduction in the expression of BAT1 and ILV5 in the TPK1 mutant in YPD medium (91). Why should BAT1 and ILV5 be regulated by Tpk1p? It turns out that the enzymes encoded by these two genes play other important roles besides their catalytic functions in the branched-chain amino acid pathways, and it is likely that the stimulatory effect of Tpk1p is aimed at these other functions, which have to do with mitochondrial integrity.
The pathway function of Bat1p is to serve as mitochondrial branched-chain amino acid aminotransferase; Bat2p is an isoenzyme located in the cytosol (58, 85). Cells deficient in both proteins do not grow on minimal media unless supplied with all three branched-chain amino acids. However, even in the presence of these amino acids, growth remains sluggish. A possible explanation for this behavior comes from the observation that the Bat proteins, in particular Bat1p, perform an essential function in iron homeostasis by being involved in the efficient transfer of Fe-S clusters from the mitochondria, where the clusters are synthesized, to the cytosol, a process that also involves the mitochondrial ABC transporter Atm1p. In fact, the BAT1 gene was isolated as a suppressor of a temperature-sensitive ATM1 mutant, and elevated levels of Bat1p were able to stabilize mutant Atm1p at the nonpermissive temperature. This suggested direct interaction between Bat1p and Atm1p, but other possibilities have not been ruled out (57, 58, 66) (Fig. 1). An important point is that a disturbance in the process of maintaining iron homeostasis can lead to a severe accumulation of iron in the mitochondria with subsequent damage and loss of mitochondrial DNA (mtDNA).
Ilv5p, which normally functions as acetohydroxy acid reductoisomerase, is also involved in enhancing the stability of mtDNA (114). This feature of Ilv5p was first discovered when ILV5 was identified as a suppressor of the mtDNA instability phenotype of Δabf2 cells grown on rich glucose medium (Abf2p is required for maintenance of mtDNA under these conditions). Suppression was achieved with a relatively small increase in ILV5 copy number or increased expression of the ILV5 gene in a strain with a constitutively active GCN4 allele. The fact that an ILV5 null mutant showed a strong tendency to produce ρ− petite strains further supported the notion that Ilv5p stabilizes mtDNA. More recently, it was shown that Gcn4p-mediated activation of ILV5 expression results in increased formation of nucleoids (mtDNA-protein complexes considered to be the basic unit of mtDNA segregation) and that such an increase in nucleoid number also increases the transmission of mtDNA to daughter cells (71). It was proposed that the simultaneous activation of amino acid biosynthesis and nucleoid formation by Gcn4p might enhance the chance of progeny survival.
By increasing the expression of BAT1 and ILV5, Gcn4p and Tpk1p are likely to achieve similar results with respect to preservation of mtDNA, parsing of mtDNA into nucleoids, and respiratory competence. Apparently, the action of Tpk1p is reinforced by Gcn4p since the TPK1 gene is controlled, at least in part, by Gcn4p (76). Also, given the possibility that Gcn4p regulation of ILV5 actually works through the Leu3p-α-isopropylmalate complex, the latter might likewise contribute to mtDNA maintenance.
BRANCHED-CHAIN AMINO ACID BIOSYNTHESIS IN OTHER FUNGI
Neurospora crassa
Our knowledge of leucine as well as isoleucine-valine biosynthesis and its regulation in N. crassa stems to a large extent from the pioneering biochemical and genetic work of S. Gross and coworkers. The three leucine pathway-specific enzymes of N. crassa are encoded by three genes: leu-4 (α-isopropylmalate synthase), leu-2 (isopropylmalate isomerase), and leu-1 (β-isopropylmalate dehydrogenase). These genes are not only unlinked but actually located on different linkage groups. It was observed early on (40, 41) (i) that the synthesis of the leu-2 and leu-1 gene products was proportional to the amount of leu-4 product generated; (ii) that leu-4 mutants unable to make synthase would also produce very little isomerase and dehydrogenase; (iii) that leu-2 mutants, which accumulate α-isopropylmalate, produce relatively large amounts of the leu-1 and leu-4 gene products (and that leu-1 mutants overproduce leu-2 and leu-4 gene products); and (iv) that the expression of leu-2 and leu-1 was reduced to very low levels in leu-3 mutants. It was suggested on the basis of these and other results that α-isopropylmalate, in conjunction with the leu-3 gene product, induces the expression of leu-2 and leu-1. Variations in the intracellular concentration of α-isopropylmalate were initially achieved by indirect means. Later, isolation of an α-isopropylmalate-permeable strain allowed direct demonstration of the induction of isopropylmalate isomerase by the leucine precursor (88). Additional work showed that leu-4 as well as four genes involved in isoleucine-valine biosynthesis and encoding threonine deaminase, acetohydroxy acid synthase, acetohydroxy acid isomeroreductase, and dihydroxy acid dehydratase require the leu-3 gene and α-isopropylmalate for full expression (80, 84).
Of considerable interest in terms of defining a “regulon” for leu-3 of N. crassa was the discovery of a connection between the leucine and histidine pathways (55). A connection was suspected after it was observed that the degree of resistance to 3-amino-1,2,4-triazole exactly paralleled the ability of the cell to generate leu-3 product and α-isopropylmalate. It was then established (again by using the α-isopropylmalate-permeable mutations mentioned above) that the target for regulation was his-1, which encodes imidazoleglycerol-phosphate dehydratase, a key enzyme in histidine biosynthesis. There thus exists a second layer of control for histidine biosynthesis in addition to what is kown as “cross-pathway control,” a cpc-1-governed mechanism that affects the histidine pathway and several other amino acid biosynthetic pathways by responding to amino acid starvation and that is analogous to the GCN4 system of S. cerevisiae (20, 81). The physiological importance of the N. crassa leu-3 product-α-isopropylmalate complex was underscored even further by the observation that the complex is able to cause transient repression of stable RNA and overall protein synthesis, an effect reminiscent of the “stringent response” of bacteria (5). While the mechanism of the negative effect of the complex on macromolecular synthesis is not known, it was established that the onset of repression is very rapid compared to the Neurospora doubling time and that one of the consequences is a significant efflux of Ca2+ from the cells, which is also transient.
Viewing the information gathered for both S. cerevisiae and N. crassa with regard to the metabolic effects of the Leu3p-α-isopropylmalate complex, one would seem justified in concluding that the fungal regulator known as Leu3p plays a rather more comprehensive role than its name suggests.
As far as the intracellular localization of branched-chain amino acid biosynthetic enzymes in N. crassa is concerned, it appears that only the enzymes of the isoleucine-valine pathway have been studied in this respect. Of these, threonine deaminase is apparently cytosolic while most, if not all, of a cell's acetohydroxy acid synthase, acetohydroxy acid isomeroreductase, and dihydroxy acid dehydratase is localized to the mitochondrial matrix (22, 80).
Other Species
There are very few studies with fungi other than S. cerevisiae and N. crassa that deal with the full set of genes and gene products concerned with the biosynthesis of all three branched-chain amino acids or even just the leucine branch. One exception is a report on leucine auxotrophs of the basidiomycete Phanerochaete chryosporium, an organism that is potentially useful in schemes for the bioprocessing of lignocellulose (74). The authors of this report found three complementation groups corresponding to three genes encoding α-isopropylmalate synthase, isopropylmalate isomerase, and β-isopropylmalate dehydrogenase. Mutations in any one of these genes caused the loss of the encoded enzyme only. In auxotrophs that lack synthase, the activities of the isomerase and the dehydrogenase were found to be 3- to 4-fold higher than in wild-type cells; auxotrophs that lack isomerase had 2-fold-elevated synthase and unchanged dehydrogenase levels; and auxotrophs deficient in dehydrogenase had 20% more synthase than did wild-type cells and up to 10-fold elevated activities of isomerase. The authors concluded that these results are indicative of leucine pathway regulation but did not elaborate further. However, the regulation seems to differ in at least some respects from that seen in S. cerevisiae and N. crassa, where very little formation of isomerase and dehydrogenase occurs in synthase-minus strains (see above). Another study of leucine biosynthesis was carried out with Candida maltosa, an imperfect yeast that is able to assimilate liquid hydrocarbons for biomass production (9). In this case, 13 leucine auxotrophs were found to constitute five complementation groups (determined by protoplast fusion since C. maltosa lacks a sexual cycle), one affecting α-isopropylmalate synthase, three affecting isopropylmalate isomerase, and one affecting β-isopropylmalate dehydrogenase. Again, a unique pattern of apparent regulation was observed, in that in a synthase-minus strain, isomerase activity was decreased only marginally and dehydrogenase still proceeded at about 25% of wild-type activity. When isomerase was missing, causing significant excretion of α-isopropylmalate, synthase activity increased four- to sixfold while dehydrogenase activity remained essentially unchanged. When dehydrogenase was missing, synthase activity again increased 4- to 6-fold while isomerase activity rose 12- to 20-fold. These data are not easily interpreted in terms of known regulatory mechanisms and appear to be at odds with an earlier report that a cloned gene from C. maltosa that showed 76% homology (at the protein sequence level) to LEU2 from S. cerevisiae (4), and was therefore named C-LEU2, was regulated just like LEU2 from S. cerevisiae when transferred into that organism (100). The interpretation of genetic data from C. maltosa is complicated by evidence of a highly aneuploid genome and the spontaneous occurrence of auxotrophy-prototrophy-auxotrophy changes due to the presence of silent gene copies (8).
Ever since the first successful transformation of yeast, which used cloned LEU2 from S. cerevisiae to complement a leu2 double mutant (46), the LEU2 gene or its homologues have been the selectable marker of choice in developing new host-vector systems. This gene is therefore one of the best-studied genes of the branched-chain amino acid pathway in fungi. A partial list of organisms from which a LEU2 homologue has been cloned, sequenced, or otherwise characterized includes (in addition to the C. maltosa example mentioned above) the yeast Yarrowia lipolytica (29), the methylotrophic yeasts Hansenula polymorpha (2) and Pichia anomala (30), and the industrially used Kluyveromyces marxianus (12) and Aspergillus niger (111). A. niger elaborates two isoenzymes of β-isopropylmalate dehydrogenase (Leu2A and Leu2B) that are only 52% identical at the amino acid level. In fact, Leu2A is more closely related to its S. cerevisiae and N. crassa homologues than to Leu2B. Since A. niger has been shown previously to incorporate DNA from other organisms in its environment, the authors speculate that the gene encoding Leu2B was introduced from another species. In all of the above cases, with the exception of A. niger, where this question was not studied, sequences were found in the 5′-noncoding regions that strongly resemble the consensus binding site for Leu3p, suggesting that Leu3p-α-isopropylmalate regulation of the kind seen in S. cerevisiae and N. crassa may be important in these other yeasts as well.
LEUCINE AS METABOLIC SIGNAL
The use of leucine or a precursor of leucine to coordinate the status of peripheral nitrogen metabolism with other cellular processes is not limited to the fungi. Another prominent example of this type of signal transduction is the leucine-responsive regulatory protein (Lrp) of E. coli. This DNA binding protein controls its own regulon of more than 20 operons that are involved principally in amino acid biosynthesis, the assimilation of ammonia, amino acid degradation, nutrient transport, and the formation of pili (19). (Interestingly enough, Lrp does not directly affect the leuABCD operon which, as pointed out above [see “Evolutionary Aspects”] is controlled by transcription attenuation [61].) Lrp functions as a positive regulator for some operons and as a negative regulator for others. Leucine either potentiates or diminishes the action of Lrp or has no effect in some cases. It has been proposed that the Lrp regulon may serve to facilitate metabolic changes in cells subjected to shifts from feast to famine and back (19). Evidence has been presented suggesting that leucine affects the equilibrium among different oligomeric states of Lrp, thus influencing Lrp occupancy at its target sites (92).
There appears to be no obvious structural similarity between the 165-residue Lrp and the 886-residue Leu3 protein and no similarity in the molecular mechanism of action of the two regulators (for example, the effect of leucine is only to modulate the efficiency of Lrp whereas α-isopropylmalate alters the regulatory properties of Leu3p by changing it from a repressor to an activator). However, there clearly are physiological similarities between the two regulons, in terms of what types of genes (operons) are being regulated. One is led to conclude that leucine was selected at least twice in evolution—once in (mostly enteric) bacteria and once in fungi—to function, directly or indirectly and by very different mechanisms, as a signal molecule in a wider metabolic context.
OUTLOOK
One of the more interesting lessons learned from studying the branched-chain amino acid pathways in fungi has been the realization that it is somewhat artificial to look on them in isolation since components of these pathways fulfill functions that go much beyond the task of supplying the cell with valine, leucine, and isoleucine. A prominent example of the involvement of branched-chain amino acid biosynthetic intermediates or proteins in other cellular processes is the Leu3p-α-isopropylmalate complex. In S. cerevisiae, this complex is involved in the regulation of ammonia assimilation by Gdh1p and is likely to up-regulate carrier functions (both at the plasma membrane and at the inner mitochondrial membrane) as well as mitochondrial malic enzyme. There is a good chance that the current list of genes affected by yeast Leu3p is incomplete. While it is possible that some of even the top-ranking candidates for Leu3p regulation identified by the promoter occupancy calculation of Liu and Clarke (69) will turn out to be statistical noise, as was recently suggested for the occurrence of Gal4p binding sites in the S. cerevisiae genome (65), it is nevertheless reasonable to assume that others will be recognized as true targets for Leu3p-α-isopropylmalate. It is intriguing that among the genes that rank within the upper 1% on the basis of the occupancy calculation (beyond those listed in Table 1), there are three that encode proteins with a mitochondrial function and eight that specify proteins with transport functions. In any event, a systematic search for genes targeted by Leu3p is clearly called for. It would be of interest to do a corresponding study with respect to the Leu3p regulon of N. crassa, where a solid base of information already exists, and to learn whether, and if so, how the use of Leu3p as a “higher-order” metabolic regulator has changed in the course of fungal evolution.
Next, one might ask how widespread among fungi is the surprisingly close relationship between branched-chain amino acid biosynthesis and the mitochondria, with regard to both the subcellular distribution of enzymes and the role of IIv5p and Bat1p or their homologues in maintaining mitochondrial (DNA) integrity.
There are several other loose ends that need to be tied up. One of these concerns the repressor function of Leu3p of S. cerevisiae. How does Mot1p come into play? Are other factors involved? The fact that Leu3p-caused repression of target gene transcription below basal level is seen not only in yeast but also in mammalian cells suggests that a more general mechanism may be at work. It is therefore important to find what factors of the transcription machinery are in direct contact with Leu3p. How do these interactions compare with those seen with other transactivators of the zinc cluster class, such as Gal4p? A recent study using an in vivo protein interaction assay has identified seven proteins as candidates for recognizing the activation domain of Gal4p (25). In addition, the activation domain of Gal4p was shown to contact Gal11p and possibly another component of the mediator complex (53). One also needs to remember that Leu3p exists in phosphorylated and nonphosphorylated states (97). The question whether the degree of phosphorylation of Leu3p has any functional significance has not yet been answered.
Finally, there is the question of how and why α-isopropylmalate synthases and other enzymes are regulated by CoA or a derivative thereof. The identification by Cavalieri et al. (23) of an R region that is involved in, if not responsible for, the CoA inactivation of α-isopropylmalate synthase of S. cerevisiae and is found in the synthases of a variety of bacteria puts this question on a new footing and should be an impetus to further explore the connections between the activity of the leucine pathway and metabolic energy supply as reflected by the CoA/acetyl-CoA ratio.
Acknowledgments
Thanks are due to Michael Hampsey, UMD New Jersey, for many rounds of instructive discussions; to Ron Somerville, Purdue University, for critically reading the manuscript; and to Roland Lill, Philipps Universität Marburg, Germany, whose friendly nudging got this project under way and who also made critical comments on the manuscript. I am also grateful to Linda Breeden, Fred Hutchinson Cancer Research Center, Seattle, Wash., for the communication of unpublished data, and to Neil Clarke, Johns Hopkins School of Medicine, for providing a preprint of a seminal paper.
Work in my laboratory has enjoyed long-term support by the National Institute of General Medical Sciences.
REFERENCES
- 1.Adamkewicz, J. I., C. G. F. Mueller, K. E. Hansen, W. A. Prud'homme, and J. Thorner. 2000. Purification and enzymic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275:21158-21168. [DOI] [PubMed] [Google Scholar]
- 2.Agaphonov, M. O., A. I. Poznyakovski, A. I. Bogdanova, and M. D. Ter-Avanesyan. 1994. Isolation and characterization of the LEU2 gene of Hansenula polymorpha. Yeast 10:509-513. [DOI] [PubMed] [Google Scholar]
- 3.Andreadis, A., Y.-P. Hsu, M. Hermodson, G. Kohlhaw, and P. Schimmel. 1984. Yeast LEU2: repression of mRNA levels by leucine and primary structure of the gene product. J. Biol. Chem. 259:8059-8062. [PubMed] [Google Scholar]
- 4.Andreadis, A., Y.-P. Hsu, G. B. Kohlhaw, and P. Schimmel. 1982. Nucleotide sequence of yeast LEU2 shows 5′-noncoding region has sequences cognate to leucine. Cell 31:319-325. [DOI] [PubMed] [Google Scholar]
- 5.Armaleo, D., M. Fischer, and S. R. Gross. 1985. Effect of α-isopropylmalate on the synthesis of RNA and protein in Neurospora. Mol. Gen. Genet. 200:346-349. [DOI] [PubMed] [Google Scholar]
- 6.Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. [DOI] [PubMed] [Google Scholar]
- 7.Baichwal, V. R., T. S. Cunningham, P. R. Gatzek, and G. B. Kohlhaw. 1983. Leucine biosynthesis in yeast: Identification of two genes (LEU4, LEU5) that affect α-isopropylmalate synthase activity and evidence that LEU1 and LEU2 gene expression is controlled by α-isopropylmalate and the product of a regulatory gene. Curr. Genet. 7:369-377. [DOI] [PubMed] [Google Scholar]
- 8.Becher, D., S. Schulze, A. Kasüske, H. Schulze, I. A. Samsonova, and S. G. Oliver. 1994. Molecular analysis of a leu2 mutant of Candida maltosa demonstrates the presence of multiple alleles. Curr. Genet. 26:208-216. [DOI] [PubMed] [Google Scholar]
- 9.Becher, D., H. Wedler, H. Schulze, R. Bode, A. Kasüske, and I. Samsonova. 1991. Correlation of biochemical blocks and genetic lesions in leucine auxotrophic strains of the imperfect yeast Candida maltosa. Mol. Gen. Genet. 227:361-368. [DOI] [PubMed] [Google Scholar]
- 10.Beltzer, J. P., L. L. Chang, A. E. Hinkkanen, and G. B. Kohlhaw. 1986. Structure of yeast LEU4: the 5′-flanking region contains features that predict two modes of control and two productive translation starts. J. Biol. Chem. 261:5160-5167. [PubMed] [Google Scholar]
- 11.Beltzer, J. P., S. R. Morris, and G. B. Kohlhaw. 1988. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of α-isopropylmalate synthase. J. Biol. Chem. 263:368-374. [PubMed] [Google Scholar]
- 12.Bergkamp, R. M., R. H. Geerse, J. M. A. Verbakel, W. Musters, and R. J. Planta. 1991. Cloning and disruption of the LEU2 gene of Kluyveromyces marxianus CBS 6556. Yeast 7:963-970. [DOI] [PubMed] [Google Scholar]
- 13.Bigelis, R., and H. E. Umbarger. 1975. Purification of yeast α-isopropylmalate isomerase: high ionic strength hydrophobic chromatography. J. Biol. Chem. 250:4315-4321. [PubMed] [Google Scholar]
- 14.Bigelis, R., and H. E. Umbarger. 1976. Yeast α-isopropylmalate isomerase: Factors affecting stability and enzyme activity. J. Biol. Chem. 251:3545-3552. [PubMed] [Google Scholar]
- 15.Boles, E., P. de Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bracho, A. M., D. Martinez-Torres, A. Moya, and A. Latorre. 1995. Discovery and molecular characterization of a plasmid localized in Buchnera sp., a bacterial endosymbiont of the aphid Rhopalosiphum padi. J. Mol. E vol. 41:67-73. [DOI] [PubMed] [Google Scholar]
- 17.Brisco, P. R. G. and G. B. Kohlhaw. 1990. Regulation of yeast LEU2: total deletion of regulatory gene LEU3 unmasks GCN4-dependent basal level expression of LEU2. J. Biol. Chem. 265:11667-11675. [PubMed] [Google Scholar]
- 18.Bult, C. J., O. White, G. J. Olsen, et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed]
- 19.Calvo, J. M., and R. G. Matthews.1994. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol. Rev. 58:466-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carsiotis, M., R. F. Jones, and A. C. Wesseling. 1974. Cross-pathway regulation: histidine-mediated control of histidine, tryptophan and arginine biosynthetic enzymes in Neurospora crassa. J. Bacteriol. 119:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Casalone, E., C. Barberio, D. Cavalieri, and M. Polsinelli. 2000. Identification by functional analysis of the gene encoding α-isopropylmalate synthase II (LEU9) in Saccharomyces cerevisiae. Yeast 16:539-545. [DOI] [PubMed] [Google Scholar]
- 22.Cassady, W. E., E. H. Leiter, A. Bergquist, and R. P. Wagner. 1972. Separation of mitochondrial membranes of Neurospora crassa. II. Submitochondrial localization of the isoleucine-valine biosynthetic pathway. J. Cell Biol. 53:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavalieri, D., E. Casalone, B. Bendoni, G. Fia, M. Polsinelli, and C. Barberio. 1999. Trifluoroleucine resistance and regulation of α-isopropylmalate synthase in Saccharomyces cerevisiae. Mol. Gen. Genet. 261:152-160. [DOI] [PubMed] [Google Scholar]
- 24.Chaleff, R. S., and C. J. Mauvais. 1984. Acetolactate synthase is the site of action of two sulfonylurea herbicides in higher plants. Science 224:1443-1445. [DOI] [PubMed] [Google Scholar]
- 25.Chang, C., F. Gonzalez, B. Rothermel, L. Sun, S. A. Johnston, and T. Kodadek. 2001. The Gal4 activation domain binds Sug2 protein, a proteasome component, in vivo and in vitro. J. Biol. Chem. 276:30956-30963. [DOI] [PubMed] [Google Scholar]
- 26.Chang, L. L., T. S. Cunningham, P. R. Gatzek, W. Chen, and G. B. Kohlhaw. 1984. Cloning and characterization of yeast LEU4, one of two genes responsible for α-isopropylmalate synthesis. Genetics 108:91-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang, L. L., P. R. Gatzek, and G. B. Kohlhaw. 1985. Total deletion of yeast LEU4: further evidence for a second α-isopropylmalate synthase and evidence for tight LEU4-MET4 linkage. Gene 33:333-339. [DOI] [PubMed] [Google Scholar]
- 28.Chasman, D. I., and R. D. Kornberg. 1990. Gal4 protein: purification, association with Gal80 protein, and conserved domain stucture. Mol. Cell. Biol. 10:2916-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidow, L. S., F. S. Kaczmarek, J. R. DeZeeuw, S. W. Conlon, M. R. Lauth, D. A. Pereira, and A. E. Franke. 1987. The Yarrowia lipolytica LEU2 gene. Curr. Genet. 11:377-383. [DOI] [PubMed] [Google Scholar]
- 30.De la Rosa, J. M., J. A. Perez, F. Gutierrez, J. M. Gonzalez, T. Ruiz, and L. Rodriguez. 2001. Cloning and sequence analysis of the LEU2 homologue gene from Pichia anomala. Yeast 18:1441-1448. [DOI] [PubMed] [Google Scholar]
- 31.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 32.Drain, P., and P. Schimmel. 1986. Yeast LEU5 is a PET-like gene that is not essential for leucine biosynthesis. Mol. Gen. Genet. 204:397-403. [DOI] [PubMed] [Google Scholar]
- 33.Drain, P., and P. Schimmel. 1988. Multiple new genes that determine activity for the first step of leucine biosynthesis in Saccharomyces cerevisiae. Genetics 119:13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fleischmann, R. D., M. D. Adams, O. White, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:96-512. [DOI] [PubMed] [Google Scholar]
- 35.Friden, P., C. Reynolds, and P. Schimmel. 1989. A large internal deletion converts yeast LEU3 to a constitutive transcriptional activator. Mol. Cell. Biol. 9:4056-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friden, P., and P. Schimmel. 1988. LEU3 of Saccharomyces cerevisiae activates multiple genes for branched-chain amino acid biosynthesis by binding to a common decanucleotide core sequence. Mol. Cell. Biol. 8:2690-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert, H. F., and M. D. Stewart. 1981. Inactivation of hydroxymethyl-glutaryl-CoA reductase from yeast by coenzyme A disulfide. J. Biol. Chem. 256:1782-1785. [PubMed] [Google Scholar]
- 38.Godon, J., M. Chopin, and S. D. Ehrlich. 1992. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J. Bacteriol. 174:6580-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross, C., M. Kelleher, V. R. Iyer, P. O. Brown, and D. R. Winge. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275:32310-32316. [DOI] [PubMed] [Google Scholar]
- 40.Gross, S. R. 1965. The regulation of synthesis of leucine biosynthetic enzymes in Neurospora. Proc. Natl. Acad. Sci. USA 54:1538-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross, S. R. 1969. Genetic regulatory mechanisms in the fungi. Annu. Rev. Genet. 3:395-424. [Google Scholar]
- 42.Guo, H., and G. B. Kohlhaw. 1996. Regulation of transcription in mammalian cells by yeast Leu3p and externally supplied inducer. FEBS Lett. 390:191-195. [DOI] [PubMed] [Google Scholar]
- 43.Hagelstein, P., and G. Schultz. 1993. Leucine synthesis in spinach chloroplasts: partial characterization of 2-isopropylmalate synthase. Biol. Chem. Hoppe-Seyler 374:1105-1108. [DOI] [PubMed] [Google Scholar]
- 44.Hampsey, D. M., and G. B. Kohlhaw. 1981. Inactivation of yeast α-isopropylmalate synthase by CoA: Antagonism between CoA and adenylates and the mechanism of CoA inactivation. J. Biol. Chem. 256:3791-3796. [PubMed] [Google Scholar]
- 45.Hellauer, K., M. H. Rochon, and B. Turcotte. 1996. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcription activators recognize everted repeats. Mol. Cell. Biol. 16:6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hinnen, A., J. B. Hicks, and G. R. Fink. 1978. Transformation of yeast. Proc. Natl. Acad. Sci. USA 75:1929-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmberg, S., and P. Schjerling. 1996. Cha4p of Saccharomyces cerevisiae activates transcription via serine/threonine response elements. Genetics 144:467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu, Y.-P., and G. B. Kohlhaw. 1980. Leucine biosynthesis in Saccharomyces cerevisiae: purification and characterization of β-isopropylmalate dehydrogenase. J. Biol. Chem. 255:7255-7260. [PubMed] [Google Scholar]
- 49.Hsu, Y.-Y., G. B. Kohlhaw, and P. Niederberger. 1982. Evidence that α-isopropylmalate synthase of Saccharomyces cerevisiae is under the general control of amino acid biosynthesis. J. Bacteriol. 150:969-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu, Y.-P., and P. Schimmel. 1984. Yeast LEU1: repression of mRNA levels by leucine and relationship of 5′-noncoding region to that of LEU2. J. Biol. Chem. 259:3714-3719. [PubMed] [Google Scholar]
- 51.Hu, Y., T. G. Cooper, and G. B. Kohlhaw. 1995. The Saccharomyces cerevisiae Leu3 protein activates expression of GDH1, a key gene in nitrogen assimilation. Mol. Cell. Biol. 15:52-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu, Y., and G. B. Kohlhaw. 1995. Additive activation of yeast LEU4 transcription by multiple cis elements. J. Biol. Chem. 270:5270-5275. [DOI] [PubMed] [Google Scholar]
- 53.Jeong, C.-J., S.-H. Yang, Y. Xie, L. Zhang, S. A. Johnston, and T. Kodadek. 2001. Evidence that Gal11 protein is a target of the Gal4 activation domain in the mediator. Biochemistry 40:9421-9427. [DOI] [PubMed] [Google Scholar]
- 54.Keefe, A. D., A. Lazcano, and S. L. Miller. 1995. Evolution of the biosynthesis of the branched-chain amino acids. Origins Life E vol. Biosphere 25:99-110. [DOI] [PubMed] [Google Scholar]
- 55.Kidd, G. L., and S. R. Gross. 1984. Specific regulatory interconnection between the leucine and histidine pathways of Neurospora crassa. J. Bacteriol. 158:121-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirkpatrick, C. R. and P. Schimmel. 1995. Detection of leucine-independent DNA site occupancy of the yeast Leu3p transcriptional activator in vivo. Mol. Cell. Biol. 15:4021-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kispal, G., P. Csere, C. Prohl, and R. Lill. 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kispal, G., H. Steiner, D. A. Court, B. Rolinski, and R. Lill. 1996. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J. Biol. Chem. 271:24458-24464. [DOI] [PubMed] [Google Scholar]
- 59.Kohlhaw, G. B. 1990. The leucine biosynthetic pathway in yeast: Compartmentation, enzyme regulation, gene expression, p. 33-42. In Z. Barak, D. M. Chipman, and J. V. Schloss (ed.), Biosynthesis of branched-chain amino acids. VCH Verlagsgesellschaft, Weinheim, Germany.
- 60.Kohlhaw, G. B., Y.-P. Hsu, R. D. Lemmon, and T. D. Petes. 1980. Transposed LEU2 gene of Saccharomyces cerevisiae is regulated normally. J. Bacteriol. 144:852-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Landgraf, J. R., J. A. Boxer, and J. M. Calvo. 1999. Escherichia coli Lrp does not directly regulate expression of the leu operon promoter. J. Bacteriol. 181:6547-6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee, Y., and R. G. Duggleby. 2001. Identification of the regulatory subunit of Arabidopsis thaliana acetohydroxyacid synthase and reconstitution with its catalytic subunit. Biochemistry 40:6836-6844. [DOI] [PubMed] [Google Scholar]
- 63.Leuther, K. K., and S. A. Johnston. 1992. Non-dissociation of GAL4 and GAL80 in vivo after galactose induction. Science 256:1333-1335. [DOI] [PubMed] [Google Scholar]
- 64.Leuther, K. K., J. M. Salmeron, and S. A. Johnston. 1993. Genetic evidence that an AD of GAL4 does not require acidity and may form a beta sheet. Cell 72:575-585. [DOI] [PubMed] [Google Scholar]
- 65.Li, Q., and S. A. Johnston. 2001. Are all DNA binding and transcription regulation by an activator physiologically relevant? Mol. Cell. Biol. 21:2467-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lill, R., K. Diekert, A. Kaut, H. Lange, W. Pelzer, C. Prohl, and G. Kispal. 1999. The essential role of mitochondria in the biogenesis of cellular iron-sulfur proteins. Biol. Chem. 380:1157-1166. [DOI] [PubMed] [Google Scholar]
- 67.Lill, R. and G. Kispal. 2000. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem. Sci. 25:352-356. [DOI] [PubMed] [Google Scholar]
- 68.Linnane, A. W., and J. M. Haslam. 1970. The biogenesis of yeast mitochondria. Curr. Top. Cell. Regula. 2:101-172. [Google Scholar]
- 69.Liu, X., and N. D., Clarke. 2002. Rationalization of gene regulation by a eukaryotic transcription factor: calculation of regulatory region occupancy from predicted binding affinities. J. Mol. Biol. 323:1-8. [DOI] [PubMed] [Google Scholar]
- 70.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-response regulon in yeast. Proc. Natl. Acad. Sci. USA 97:7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.MacAlpine, D. M., P. S. Perlman, and R. A. Butow. 2000. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 19:767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marmorstein, R., M. Carey, M. Ptashne, and S. C. Harrison. 1992. DNA recognition by GAL4: structure of a protein-DNA complex. Nature 356:408-414. [DOI] [PubMed] [Google Scholar]
- 73.Marmorstein, R., and S. C. Harrison. 1994. Crystal structure of a PPR1-DNA complex: DNA recognition by proteins containing a Zn2Cys6 binuclear cluster. Genes Dev. 8:2504-2512. [DOI] [PubMed] [Google Scholar]
- 74.Molskness, T. A., M. Alic, and M. H Gold. 1986. Characterization of leucine auxotrophs of the white rot basidiomycete Phanerochaete chryosporium. Appl. Environ. Microbiol. 51:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mühlenhoff, U., and R. Lill. 2000. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta 1459:370-382. [DOI] [PubMed] [Google Scholar]
- 76.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nielsen, P. S., B. van den Hazel, T. Didion, M. de Boer, M. Jorgensen, R. J. Planta, M. C. Kielland-Brandt, and H. A. Andersen. 2001. Transcriptional regulation of the Saccharomyces cerevisiae amino acid permease gene BAP2. Mol. Gen. Genet. 264:613-622. [DOI] [PubMed] [Google Scholar]
- 78.Noël, J., and B. Turcotte. 1998. Zinc cluster proteins Leu3p and Uga3p recognize highly related but distinct DNA targets. J. Biol. Chem. 273:17463-17468. [DOI] [PubMed] [Google Scholar]
- 79.Oaks, A. 1965. The biosynthesis of leucine in maize embryos. Biochim. Biophys. Acta 111:79-89. [DOI] [PubMed] [Google Scholar]
- 80.Olshan, A. R., and S. R. Gross. 1974. Role of the leu-3 cistron in the regulation of the synthesis of isoleucine and valine biosynthetic enzymes of Neurospora. J. Bacteriol. 118:374-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paluh, J., M. J. Orbach, T. L. Legerton, and C. Yanofsky. 1988. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. USA 85:3728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patek, M., K. Krumbach, L. Eggeling, and H. Sahm. 1994. Leucine synthesis in Corynebacterium glutamicum: enzyme activities, structure of LeuA, and effect of LeuA inactivation on lysine synthesis. Appl. Environ. Microbiol. 60:133-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peters, M. H., J. P. Beltzer, and G. B. Kohlhaw. 1990. Expression of the yeast LEU4 gene is subject to four different modes of control. Arch. Biochem. Biophys. 276:294-298. [DOI] [PubMed] [Google Scholar]
- 84.Polacco, J. C., and S. R. Gross. 1973. The product of the leu-3 cistron as a regulatory element for the production of the leucine biosynthetic enzymes of Neurospora. Genetics 74:443-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Prohl, C., G. Kispal, and R. Lill. 2000. Branched-chain amino acid transaminases of yeast Saccharomyces cerevisiae. Methods Enzymol. 324:365-375. [DOI] [PubMed] [Google Scholar]
- 86.Prohl, C., W. Pelzer, K. Diekert, H. Kmita, T. Bedekovics, G. Kispal, and R. Lill. 2001. The yeast mitochondrial carrier Leu5p and its human homolog Graves' disease protein are required for accumulation of CoA in the matrix. Mol. Cell. Biol. 21:1089-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pronk, J. T., H. Y. Steensma, and J. P. Van Dijken. 1996. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607-1633. [DOI] [PubMed] [Google Scholar]
- 88.Reichenbecher, V. E., M. Fischer, and S. R. Gross. 1978. Regulation of isopropylmalate isomerase synthesis in Neurospora crassa. J. Bacteriol. 133:794-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Remboutsika, E., and G. B. Kohlhaw. 1994. Molecular architecture of a Leu3p-DNA complex in solution: a biochemical approach. Mol. Cell. Biol. 14:5547-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ricca, E., and J. M. Calvo. 1990. The nucleotide sequence of leuA from Salmonella typhimurium. Nucleic Acids Res. 18:1290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robertson, L. S., H. C. Causton, R. A. Young, and G. R. Fink. 2000. The yeast A kinases differentially regulate iron uptake and respiratory function. Proc. Natl. Acad. Sci. USA 97:5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosner, C. S., and J. M. Calvo. 2001. Leucine-regulated self-association of leucine-responsive regulatory protein (Lrp) from Escherichia coli. J. Mol. Biol. 312:625-635. [DOI] [PubMed] [Google Scholar]
- 93.Ryan, E., and G. B. Kohlhaw. 1974. Subcellular localization of isoleucine-valine biosynthetic enzymes in yeast. J. Bacteriol. 120:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ryan, E., J. W. Tracy, and G. B. Kohlhaw. 1973. Subcellular localization of the leucine biosynthetic enzymes in yeast. J. Bacteriol. 116:222-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schjerling, P., and S. Holmberg. 1996. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 24:4599-4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schulz, W. A., B. Ebling, A. Hasse, F. Zenke, and K. Breunig. 1993. Highly efficient transactivation by the yeast Kluyveromyces lactis transcription factor LAC9 and its inhibition by the negative regulator GAL80 in mammalian cells. Biol. Chem. Hoppe-Seyler 374:313-318. [DOI] [PubMed] [Google Scholar]
- 97.Sze, J.-Y., and G. B. Kohlhaw. 1993. Purification and structural characterization of transcriptional regulator Leu3 of yeast. J. Biol. Chem. 268:2505-2512. [PubMed] [Google Scholar]
- 98.Sze, J.-Y., E. Remboutsika, and G. B. Kohlhaw. 1993. Transcriptional regulator Leu3 of Saccharomyces cerevisiae: separation of activator and repressor functions. Mol. Cell. Biol. 13:5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sze, J.-Y., M. Woontner, J. A. Jaehning, and G. B. Kohlhaw. 1992. In vitro transcriptional activation by a metabolic intermediate: activation by Leu3 depends on α-isopropylmalate. Science 258:1143-1145. [DOI] [PubMed] [Google Scholar]
- 100.Takagi, M., N. Kobayashi, M. Sugimoto, T. Fujii, J. Watari, and K. Yano. 1987. Nucleotide sequence analysis of a LEU gene of Candida maltosa which complements leuB mutation of E. coli and leu2 mutation of S. cerevisiae. Curr. Genet. 11:451-457. [DOI] [PubMed] [Google Scholar]
- 101.Tan-Wilson, A., and G. B. Kohlhaw. 1978. Specific, reversible inactivation of yeast β-hydroxy-β-methylglutaryl-CoA reductase by CoA. Biochem. Biophys. Res. Commun. 85:70-76. [DOI] [PubMed] [Google Scholar]
- 102.Tice-Baldwin, K., G. R. Fink, and K. Arndt. 1989. BAS1 has a myb motif and activates HIS4 transcription only in combination with BAS2. Science 246:931-935. [DOI] [PubMed] [Google Scholar]
- 103.Tracy, J. W., and G. B. Kohlhaw. 1975. Reversible, coenzyme A-mediated inactivation of biosynthetic condensing enzymes in yeast: a possible regulatory mechanism. Proc. Natl. Acad. Sci. USA 72:1802-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tracy, J. W., and G. B. Kohlhaw. 1977. Evidence for two distinct CoA binding sites on yeast α-isopropylmalate synthase. J. Biol. Chem. 252:4085-4091. [PubMed] [Google Scholar]
- 105.Ulm, E. H., R. Böhme, and G. B. Kohlhaw. 1972. α-Isopropylmalate synthase from yeast: purification, kinetic studies, and effect of ligands on stability. J. Bacteriol. 110:1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Umbarger, H. E.1990. The study of branched-chain amino acid biosynthesis: Its roots and its fruits, p. 1-24. In Z. Barak, D. M. Chipman, and J. V. Schloss (ed.), Biosynthesis of branched-chain amino acids. VCH Verlagsgesellschaft, Weinheim, Germany.
- 107.Voss, H., V. Benes, M. A. Andrade, A. Valencia, S. Rechmann, C. Teodoru, C. Schwager, V. Paces, C. Sander, and W. Ansorge. 1997. DNA sequencing and analysis of 130 kb from yeast chromosome XV. Yeast 13:655-672. [DOI] [PubMed] [Google Scholar]
- 108.Wade, P. A., and J. A. Jaehning. 1996. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol. Cell. Biol. 16:1641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang, D., Y. Hu, F. Zheng, K. Zhou, and G. B. Kohlhaw. 1997. Evidence that intramolecular interactions are involved in masking the activation domain of transcriptional activator Leu3p. J. Biol. Chem. 272:19383-19392. [DOI] [PubMed] [Google Scholar]
- 110.Wang, D., F. Zheng, S. Holmberg, and G. B. Kohlhaw. 1999. Yeast transcriptional regulator Leu3p: self-masking, specificity of masking, and evidence for regulation by the intracellular level of Leu3p. J. Biol. Chem. 274:19017-19024. [DOI] [PubMed] [Google Scholar]
- 111.Williams, B. A., S. Sillaots, A. Tsang, and R. Storms. 1996. Isolation by genetic complementation of two differentially expressed genes for β-isopropylmalate dehydrogenase from Aspergillus niger. Curr. Genet. 30:305-311. [DOI] [PubMed] [Google Scholar]
- 112.Xing, R., and W. B. Whitman. 1991. Characterization of enzymes of the branched-chain amino acid biosynthetic pathway in Methanococcus spp. J. Bacteriol. 173:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yanofsky, C. 1981. Attenuation in the control of expression of bacterial operons. Nature 289:751-758. [DOI] [PubMed] [Google Scholar]
- 114.Zelenaya-Troitskaya, O., P. S. Perlman, and R. A. Butow. 1995. An enzyme in yeast mitochondria that catalyzes a step in branched-chain amino acid biosynthesis also functions in mitochondrial DNA stability. EMBO J. 14:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang, L., and L. Guarente. 1994. The yeast activator HAP1, a GAL4 family member, binds DNA in a directly repeated orientation. J. Biol. Chem. 269:14643-14647. [DOI] [PubMed] [Google Scholar]
- 116.Zhang, L., and L. Guarente. 1995. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 14:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhou, K., Y. Bai, and G. B. Kohlhaw. 1990. Yeast regulatory protein LEU3: a structure-function analysis. Nucleic Acids Res. 18:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou, K., P. R. G. Brisco, A. E. Hinkkanen, and G. B. Kohlhaw. 1987. Structure of yeast regulatory gene LEU3 and evidence that LEU3 itself is under general amino acid control. Nucleic Acids Res. 15:5261-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou, K., and G. B. Kohlhaw. 1990. Transcriptional activator LEU3 of yeast: mapping of the transcriptional activation function and significance of activation domain tryptophans. J. Biol. Chem. 265:17409-17412. [PubMed] [Google Scholar]