Abstract
Through a screen to identify genes that induce multi-drug resistance when overexpressed, we have identified a fission yeast homolog of Int-6, a component of the human translation initiation factor eIF3. Disruption of the murine Int-6 gene by mouse mammary tumor virus (MMTV) has been implicated previously in tumorigenesis, although the underlying mechanism is not yet understood. Fission yeast Int6 was shown to interact with other presumptive components of eIF3 in vivo, and was present in size fractions consistent with its incorporation into a 43S translation preinitiation complex. Drug resistance induced by Int6 overexpression was dependent on the AP-1 transcription factor Pap1, and was associated with increased abundance of Pap1-responsive mRNAs, but not with Pap1 relocalization. Fission yeast cells lacking the int6 gene grew slowly. This growth retardation could be corrected by the expression of full length Int6 of fission yeast or human origin, or by a C-terminal fragment of the fission yeast protein that also conferred drug resistance, but not by truncated human Int-6 proteins corresponding to the predicted products of MMTV-disrupted murine alleles. Studies in fission yeast may therefore help to explain the ways in which Int-6 function can be perturbed during MMTV-induced mammary tumorigenesis.
INTRODUCTION
Initiation of translation in eukaryotes is a highly regulated process requiring more than twenty initiation factor (eIF1) proteins in addition to the ribosome and aminoacylated initiator tRNA (Jackson and Wickens, 1997). Most cellular mRNAs are translated by a mechanism that involves recognition of the 5′ cap by eIF4E, which is itself recruited into a large preinitiation assembly based on eIF4G and containing eIF3 and eIF4A, an RNA helicase. The eIF3 complex, which consists of at least nine subunits, associates with the 40S ribosomal subunit and prevents premature binding of the 60S subunit. The initiator AUG is then identified by 5′ to 3′ scanning of the mRNA; once this is located, recruitment of the 60S subunit allows protein synthesis to begin.
Internal ribosome entry is an alternative mode of translation initiation used by a subset of cellular (and several viral) mRNAs (Jang et al., 1988; Pelletier and Sonenberg, 1988; Macejak and Sarnow, 1991; Bernstein et al., 1997; Keiper and Rhoads, 1997; Gan et al., 1998; Huez et al., 1998; Stein et al., 1998). This process is similar to that used in prokaryotes, in that there is no requirement for a 5′ mRNA cap. Instead, some internal aspect of the mRNA structure is recognized directly by the translational machinery in order to direct initiation from a specific AUG triplet. Internal ribosome entry is less well understood than cap-dependent initiation, but recent evidence suggests a role for eIF3 in the recognition of internal ribosome entry sites (IRESs) in the 5′ untranslated regions of viral mRNAs (Sizova et al., 1998). If there is an analogous role for eIF3 in the recognition of IRES elements in cellular mRNAs, this initiation factor could play a regulatory role in determining the balance between cap-dependent and cap-independent translation. Alteration of this balance to favor internal ribosome entry under conditions of stress, such as hypoxia, may be an important mechanism by which the synthesis of stress-related proteins is favored (Vagner et al., 1996; Stein et al., 1998).
Comparison of the predicted amino acid sequences of eIF3 components in human cells and the budding yeast Saccharomyces cerevisiae has shown that these species share a common set of five ‘core’ subunits, while the remaining ‘accessory’ subunits are not conserved (Asano et al., 1997a; Asano et al., 1997c; Asano et al., 1998). One of the human accessory subunits of eIF3 was found to be identical to the Int-6 protein (Asano et al., 1997b). The murine Int-6 gene, the product of which is identical in its amino acid sequence to the human Int-6 protein, was first identified as a site of MMTV provirus insertion in mammary tumors and a preneoplastic outgrowth (Marchetti et al., 1995). In each case, the MMTV integration is within an Int-6 intron and in the opposite transcriptional orientation, such that a hybrid Int-6/MMTV mRNA is transcribed that would encode a C-terminally truncated Int-6 protein. The role of this truncated protein in tumorigenesis has not yet been described.
We report here the identification in the fission yeast, Schizosaccharomyces pombe, of a functional homolog of the mammalian Int-6 protein, in a screen designed to identify drug resistance determinants. This finding has allowed us to conduct the first study of Int-6 function in a simple, genetically amenable model organism.
MATERIALS AND METHODS
General Fission Yeast Methods and cDNA Library Screen
General fission yeast manipulations were carried out as described in detail elsewhere (Moreno et al., 1991; Norbury and Moreno, 1997) using EMM2 (Edinburgh minimal medium), containing where necessary leucine and uracil at 225 μg/ml. An h− leu1–32 strain of S. pombe was transformed by electroporation (Bio-Rad Gene Pulser, Richmond, CA) with a fission yeast cDNA library (Bruce Edgar and C. N., unpublished data) constructed in the vector pREP3X (Forsburg, 1993). In this vector, which carries a Saccharomyces cerevisiae LEU2 selectable marker that can complement the leu1–32 mutation, cDNA expression is under the control of the thiamine-repressible fission yeast nmt1 promoter (Maundrell, 1989). Approximately 200,000 leu+ transformants were screened directly for their ability to grow on EMM2 agar containing 20 μg/ml methyl benzimidazole-2-yl carbamate (MBC). The number of transformants screened was estimated by plating a small aliquot of the transformation mix onto EMM2 agar plates without drug. Plates were incubated at 30°C for 5–7 days, and plasmids were recovered from drug resistant S. pombe colonies by small-scale DNA preparation followed by transformation of Escherichia coli DH10β. Induction by individual plasmids of resistance to 20 μg/ml MBC, 17.5 mM caffeine, 0.5 mM CdCl2 or 1 μg/ml staurosporine was tested following retransformation of the S. pombe leu1–32 strain, and the pap1-dependence of drug resistance was tested by transformation of pREP3X-based plasmids into an h− pap1::ura4+ ura4-D18 leu1-32 (pap1Δ) strain (Toda et al., 1991) kindly provided by Takashi Toda (ICRF, London, United Kingdom). Sensitivity to 254 nm UV (UV) irradiation was measured by plating 500 cells (in triplicate) onto multiple EMM2 agar plates, exposing each plate to a known dose of UV (Stratalinker, Stratagene, La Jolla, CA), and counting viable colonies after 3 d growth at 30°C. Sensitivity to H2O2 was measured in exponentially growing cultures 16 h after washing out thiamine; cells were diluted to 10,000/ml in EMM2, exposed to H2O2 for 3 h, and viability was measured as described for UV above. The DNA sequence of the plasmid inserts was determined on both strands by primer walking using an ABI 377 DNA sequencer and ABI PRISM dRhodamine dye terminator reagents (Perkin Elmer-Cetus, Norwalk, CT) and was confirmed in part by database (BLAST) searching against the partial S. pombe genomic database held at the Sanger Center, Hinxton, United Kingdom (http://www.sanger.ac.uk/Projects/S_pombe/blast_server.shtml). The GenBank accession number for the S. pombe int6 sequence is AF117648. The protein sequence alignment was created using the PILEUP program (University of Wisconsin Genetics Computer Group, Madison, WI) and displayed using MacBoxshade (written by Michael Baron: michael.baron@bbsrc.ac.uk). With the exception of Int6 (described here) and Sum1 (Humphrey and Enoch, 1998), fission yeast homologs of additional putative eIF3 subunits were identified by BLAST searches of the S. pombe genome sequence database. The comparison between human and budding yeast eIF3 components shown in Table 1 is based on that described previously by Hershey and colleagues (Asano et al., 1997c). The int6-HA tagged strain used in the experiment shown in Figure 2F was constructed by the one-step PCR-based technique (Bahler et al., 1998). GFP-Pap1 was expressed from pREP42 (Maundrell, 1993) after excision from pREP41-GFP-Pap1 (Toone et al., 1998). Green fluorescence was visualized in live cells growing in EMM2 using a confocal laser scanning microscope (Zeiss LSM 510). Cell number was determined using an automated cell analyser (Sysmex F-820). Processing of cells for DAPI, calcofluor, rhodamine-phalloidin, and antitubulin (TAT-1; kindly provided by K. Gull, University of Manchester) staining was performed using standard methods (Moreno et al., 1991).
Table 1.
Comparison of the possible subunit compositions of eIF3 in budding yeast, fission yeast, and human cells
| H. sapiens | S. pombe | S. cerevisiae | Protein familya |
|---|---|---|---|
| p170 | c17D11b | Tif32 | PCI |
| p116 | eIF3x | Prt1 | RRM |
| p110 | SPAC4A8b | Nip1 | PCI |
| p66 | Moe1/c637b | (absent) | |
| p48/Int-6 | Int6 | (absent) | PCI |
| p47 | SPBC4C3b | (absent) | MPN |
| p44 | eIF35 | Tif35 | RRM |
| p40 | p40 | (absent) | MPN |
| p36/TRIP1 | Sum1 | Tif34 | WD40 |
Protein family to which the subunit belongs: RRM, RNA recognition motif; PCI, proteasome/COP9/Int-6; MPN, related to proteasome subunit Mov-34/Pad1.
Denotes cosmid sequence designation in the Sanger Centre database.
Figure 2.
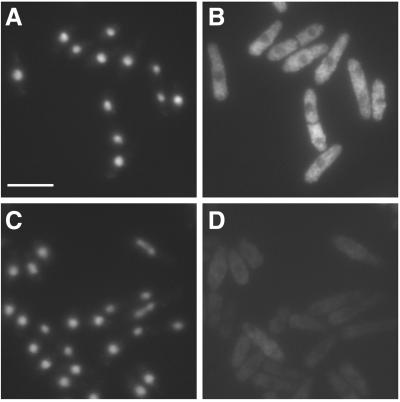
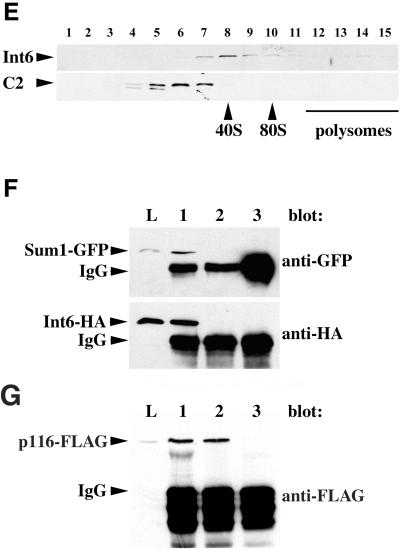
Fission yeast Int6 has several properties expected of an eIF3 component. (A-D) Indirect immunofluorescence of endogenous Int6. Methanol fixed cells of a wild-type (int6+) strain (A, B) or a strain deleted for int6 (int6Δ, see MATERIALS AND METHODS; (C, D) were processed for immunofluorescence after incubation with affinity-purified anti-Int6 antibodies and mounted in DAPI. For each strain a single representative field was used to collect images of DAPI (DNA) staining (A, C) and Cy3 antirabbit (Int6) staining (B, D). Scale bar, 10 μm. (E) Sucrose density gradient separation of whole cell extracts. After lysis of wild-type (int6+) cells and separation as described in MATERIALS AND METHODS, aliquots of sucrose gradient fractions were subjected to western blotting and were probed with antibodies against Int6 (upper panel) or the proteasome subunit C2 (lower panel). C2 is present in both 20S and 26S proteasomes. The positions at which 40S and 80S ribosomes and polysomes migrated, as determined by monitoring A254 during collection of the fractions, are indicated. (F) Coprecipitation of tagged Int6 and Sum1 proteins. Whole cell lysate (L) or immunoprecipitated proteins from a gene replacement strain expressing Int6-HA and Sum1-GFP (lanes 1 and 3) or a control strain expressing Sum1-GFP and untagged Int6 (lane 2) were subjected to western blotting using antibodies against GFP or HA, as indicated. Immunoprecipitating antibodies were anti-HA (lanes 1,2) or an isotype control (OX34 antirat CD2; lane 3). The immunoprecipitating antibody heavy chain (IgG) was also detected by the HRP-conjugated secondary antibody. (G) Coprecipitation of tagged Int6 and p116 proteins. Whole cell lysate (L) or immunoprecipitated proteins from a strain expressing Int6-HA and p116-FLAG were subjected to western blotting using anti-FLAG antibodies. Immunoprecipitating antibodies were anti-FLAG (lane 1), anti-HA (lane 2) or OX34 (antirat CD2; lane 3).
Antibodies, Western Blotting and Indirect Immunofluorescence
Recombinant Int6 was expressed in E. coli BL-21 as a glutathione S-transferase (GST) fusion protein after cloning the int6 open reading frame into the expression vector pGEX4T-2 (Amersham Pharmacia, Amersham, United Kingdom). Rabbit polyclonal antibodies against the gel-purified fusion protein were prepared by standard procedures (Harlow and Lane, 1988), were affinity purified using the GST-Int6 fusion protein, and were used at 1:2000 for Western blotting or 1:500 for immunofluorescence. Antibodies against the C2 proteasome subunit were prepared using the same procedure. Western blotting was performed essentially as described elsewhere (Ausubel et al., 1995) using Hybond ECL membranes (Amersham Pharmacia). Proteins were detected (ECL, Amersham) after 1-h incubations at room temperature with the respective primary and, where necessary, secondary antibodies. The rat anti-influenza hemagglutinin (HA) monoclonal 3F10 (Roche Diagnostics, UK) was used as a horseradish peroxidase (HRP) conjugate at a final concentration of 1 μg/ml for direct detection of HA-tagged proteins. Cdc2 was detected using the mouse monoclonal antibody Y100 (kindly provided by Dr J Gannon, ICRF Laboratories, South Mimms, United Kingdom). Polyclonal antibodies against Pap1 and p25 were generously provided by Mark Toone and Takashi Toda, respectively. Monoclonal anti-GFP (BAbCO) was used at a 1:500 dilution. Monoclonal anti-FLAG (clone M2, Sigma, St. Louis, MO) was used at 8.8 μg/ml. HRP-conjugated secondary antibodies (Sigma) were used at 1:1000. Immunofluorescence microscopy was performed on methanol-fixed cells using Cy3-conjugated goat antirabbit secondary antibodies (Sigma). Images were acquired using a Hamamatsu-cooled CCD camera (Hamamatsu Photonics UK, Welwyn Garden City, United Kingdom) attached to a Zeiss Axioskop (Thornwood, NY) microscope and Kromascan software (Kinetic Imaging) and were assembled using Adobe Photoshop.
Sucrose Density Gradient Fractionation and Immunoprecipitation
For the experiment shown in Figure 2E, a 200-ml culture of an h− leu1–32 strain was grown to midlog phase and lysed in 500 μl chilled lysis buffer A (10 mM Tris pH 7.5, 100 mM NaCl, 30 mM MgCl2, 50 μg/ml cycloheximide, 5 mM ATP) by vortexing with acid-washed glass beads. The lysate was clarified by centrifugation (14,000 x g, 10 min, 4°C) and then applied to a 13.5-ml 7–37% sucrose gradient prepared in 50 mM Tris acetate, 50 mM NH4Cl, 12 mM MgCl2, 5 mM ATP. After centrifugation for 1 h (Beckman, Fullerton, CA; SW41 rotor, 40,000 r.p.m., 4°C), 0.5-ml fractions were collected by displacement of the gradient from below. From each fraction, 20-μl aliquots were subjected to analysis by SDS-PAGE and Western blotting.
For the experiments shown in Figures 2F and 2G, 50-ml cultures of int6::int6-HA-kanMX sum1::ura4 leu1::sum1-GFP, sum1::ura4 leu1::sum1-GFP and int6::int6-HA-kanMX leu1–32 (pREP3X-p116FLAG) strains were grown to midlog phase and lysed in 200 μl chilled lysis buffer B (20% glycerol, 20 mM Tris pH 7.5, 1 mM β-mercaptoethanol, 0.1 mM EDTA, 5 mM ATP) by vortexing with acid-washed glass beads. The lysate was clarified by centrifugation (14000 x g, 10 min, 4°C), and 50-μl aliquots were incubated with 2 μg anti-HA or anti-CD2 (OX34, ICRF Research Monoclonal Antibody Services) antibody in lysis buffer B for 1 h. Antibody complexes were retrieved by incubation with protein G sepharose beads (Sigma) and washed four times in lysis buffer B before denaturation in SDS sample buffer and analysis by Western blotting.
Northern Blotting
RNA was prepared according to the manufacturer's instructions using RNAzol B (BioGenesis, Poole, United Kingdom) from midexponential cultures of appropriate fission yeast strains grown in EMM2 medium. Total RNA (20 μg per lane) was separated by formaldehyde/agarose gel electrophoresis and transferred to Hybond-N+ (Amersham Pharmacia), as described elsewhere (Ausubel et al., 1995). The apt1 probe was amplified from total genomic S. pombe DNA using the oligonucleotide primer pair GCAAACACCGTCGCTATTGTG and TCGGCTCCAGCATAGGAACC, the actin probe with the pair GATTTGGCATCACACTTTCTACAACGAGC and GATAGTGATAACTTGACCATCAGGAAGC, and the trr1 probe with the pair TCAGCTTACTACTACCACCG and ACGGTGTTCCACAAAACTTCC (all sequences 5′ to 3′). Probes were radiolabeled using [α32P]-dCTP and the Rediprime II random prime labeling kit (Amersham Pharmacia) and were hybridized to the membrane in ExpressHyb solution (Clontech, Palo Alto, CA) at 60°C for 12 h.
Disruption of the int6+ Gene
A diploid strain h−/h+ leu1–32/leu1–32 ura4-D18/ura4-D18 his7/his+ ade6-M210/ade6-M210 was transformed to uracil prototrophy using a linear DNA fragment amplified using the Expand high fidelity PCR system (Roche Diagnostics) from the ura4+ -containing template pREP4 (Maundrell, 1993) using the following oligonucleotides: 5′: G C G T G A A A C A T A T C A G A T A T G G G A T C C G A G C T T A A G A G T A C A A G C C C T T T A G C G G T C A A G T A T G A T T T G T C G C A A A A A A T T A T G C A A C A C C T T G A C C G C C C A A A T C C C A C T G G C T A T A T G T A T G C. This oligonucleotide contains 100 nt of the int6 gene spanning the ATG initiator codon (underlined) followed by 24 nt of the 5′UTR of the ura4+ gene (italicized).
3′: C C G A A A T G C T T T T A G C A A G A G A T T G C T C C A A G T T T T G A C T T T C A A A A C T T A A T G A C T T T G T A C G G T C T A T A A T C T G T T G G A A A G C T G A A T A A G T A G G G T G A A T T C T A A A T G C C T T C T G A C. This oligonucleotide consists of a sequence complementary to 100 nt of the coding strand of the int6 open reading frame beginning 21 nt upstream from the TAG stop codon, followed by 24 nt complementary to the 3′UTR of the ura4+ gene (italicized). PCR amplification of the ura4+ template using these primers generated a version of the int6 gene in which almost the entire open reading frame is replaced by the ura4+ selectable marker. The 100 bp of homology to int6+ at either end of this gene disruption cassette allowed targeted integration at one of the int6 loci in the diploid strain. Following transformation, individual ura+ colonies were tested for disruption of the int6 gene by PCR reactions using the following primer pair: GTCTAAACAGTAGCATGCTTTAACTCC (complementary to 27 nt immediately downstream from the putative integration site) and CGGGCTGGGACAGCAATATCG (internal to the ura4+ sequence). The desired gene disruption gave rise to a PCR product of ∼ 400 bp that was absent in reactions using DNA from the parental strain as template. The h−/h+ leu1–32/leu1–32 ura4-D18/ura4-D18 his7/his+ ade6-M210/ade6-M210 int6::ura4+/int6+ strain thus generated is referred to as int6+/int6− in the text.
Expression of p47, p116 and Human Int-6 in S. pombe
A p116 cDNA was amplified with a 3′ FLAG epitope tag using the primer pair GCTACGCTCGAGATCATGTCGGAAATCCTAATTG-AGG and GCTACGCCCGGGCTACTTGTCGTCATCGTCCT-TGTAGTCATCTTCAACGGGTTCTATCTCTTCAGAG and was cloned into pREP3X after digestion with XhoI and SmaI. A full-length human Int-6 cDNA was constructed by PCR amplification of the Int-6 open reading frame from a human embryonic fibroblast cDNA library, using the primer pair CCATGTCGACACCATGGCGGAGTACGACTTGACT and ATAAGATAGCGGCCGCTCAGTAGAAGCCAGAATCTTGAGT, followed by digestion with SalI and NotI and subsequent ligation into SalI and NotI-cleaved pREP3-HA3, a derivative of pREP3X encoding a triple HA tag beginning with an ATG codon between the XhoI and SalI sites. Analogous constructions were performed to generate the short and medium-length truncated Int-6 open reading frames as indicated in Figure 1, using the same first primer and either ATAAGATAGCGGCCGATCACAGCACTCTAAAAAAATAAAGATATTC or ATA-AGATAGCGGCCGCTCATGATTCACATTCCCTCAGCTTTTTC, respectively. The same strategy was used to clone a p47 cDNA using the primers CTACTGTCGACATGGCTTTGGGGACTAAGCACG and CTACTGCGGCCGCTTAGGGAAGCAAATTAAGACGGG.
Figure 1.
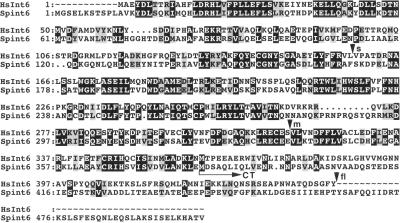
Alignment of the human Int-6 protein sequence (HsInt6) with that of the predicted product of the S. pombe int6 cDNA described here (Spint6). Sequence identities are boxed in black, and conservative substitutions are shaded. The C-terminal fragment (Int6CT) predicted to be expressed from the partial cDNA identified in the drug resistance screen would extend from the methionine residue indicated by the horizontal arrow to the carboxyl terminus. Arrowheads indicate the positions of the carboxyl termini of the short (s), medium (m), and full-length (fl) versions of the human protein expressed in fission yeast in this study.
RESULTS
A Fission Yeast Int-6-related Protein Confers Drug Resistance
To identify fission yeast genes capable of conferring multi-drug resistance, an S. pombe leu1–32 strain transformed with a regulatable cDNA library was plated onto agar containing the spindle poison MBC. Plasmids were recovered from transformants able to grow in the presence of 20 μg/ml MBC and were tested for their ability to confer resistance both to MBC and, separately, to 17.5 mM caffeine, 0.5 mM CdCl2, or 1 μg/ml staurosporine after retransformation into S. pombe leu1–32. After DNA sequencing, one of the cDNA inserts identified in this way was found to encode a 57 kDa fission yeast relative of the mammalian Int-6 protein (Figure 1). In recognition of this sequence conservation, the fission yeast protein was named Int6. A second cDNA isolated in the same screen was found to comprise a 3′ int6 fragment, presumably generated by incomplete reverse transcription during cDNA library construction. This partial cDNA encodes a 13 kDa C-terminal fragment of the Int6 protein (Figure 1), which we term Int6CT. A complete account describing other cDNAs isolated in this screen will be presented elsewhere.
Although the complete genomic sequence of S. pombe is not yet available, database searches revealed that, in addition to Int-6, a further eight components of human eIF3 are clearly conserved in fission yeast, although budding yeast lack counterparts of four of these (Int-6, p40, p47, and p66; Table 1). Overall amino acid identity/similarity in comparisons of the fission yeast and human Int6, p40, p47, and p66 sequences was 38/56%, 27/48%, 35/58%, and 41/58%, respectively.
Fission Yeast Int6 Is a Cytoplasmic Protein that Associates with Other Presumptive eIF3 Subunits
Antibodies raised against recombinant Int6 expressed in E. coli were used for indirect immunofluorescence microscopy of wild-type S. pombe (Figure 2A-D). The data indicate that Int6 is dispersed fairly evenly throughout the cytoplasm, in line with a possible role in the regulation of protein synthesis. Fractionation by sucrose density gradient centrifugation of fission yeast lysates, prepared under nondenaturing conditions, showed that the majority of Int6 has a sedimentation coefficient of ∼ 40S (Figure 2E). This is consistent with the value of 43S expected for a component of eIF3 associated with the 40S ribosomal subunit in the translation preinitiation complex.
The association of Int6 with other putative eIF3 components was investigated by co-immunoprecipitation experiments. A strain was constructed that expresses a GFP-tagged form of Sum1, an essential fission yeast gene product analogous to the human eIF3 subunit p36/TRIP1 (Humphrey and Enoch, 1998), and HA-tagged Int6. Each of the tagged genes in this strain replaces the corresponding endogenous gene, is present as a single copy expressed from its own chromosomal promoter, and appears to be fully functional, as judged by the growth rate of the strain. Immunoprecipitates of whole cell lysates prepared from this strain were subjected to immunoblotting analysis with anti-HA and anti-GFP antibodies (Figure 2F). Int6-HA and Sum1-GFP were both readily detectable in anti-HA immunoprecipitates, whereas control precipitations using an irrelevant antibody contained neither (Figure 2F). Further experiments showed that Int6-HA was associated with another presumptive eIF3 subunit, p116, which was expressed from a plasmid as a FLAG epitope-tagged form (Figure 2G). Taken together, these data show that Int6 has several of the properties expected of a bona fide component of eIF3.
Int6-induced Drug Resistance Is Pleiotropic and Pap1-dependent
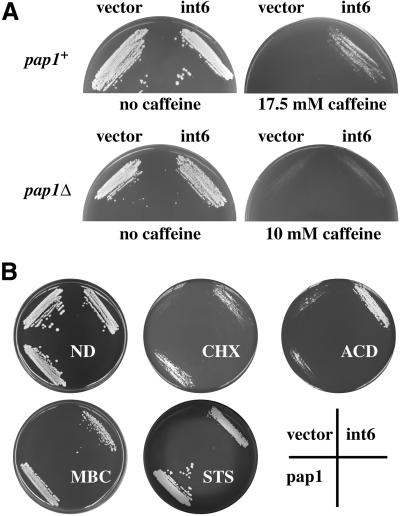
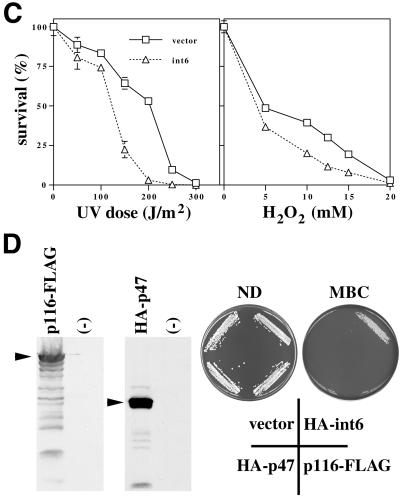
Drug resistance of the type induced by Int6 overexpression can also be conferred by overexpression of the nonessential fission yeast AP-1 transcription factor Pap1 (Toda et al., 1991). A number of Pap1-responsive genes have been identified that encode mediators of this drug resistance phenotype, such as ABC-type transporters, catalase, thioredoxin, and thioredoxin reductase (Toone et al., 1998). Expression of Int6 from the nmt1 promoter induced resistance to 17.5 mM caffeine in a wild-type (pap1+) fission yeast strain (Figure 3A). In a strain in which pap1 had been disrupted (pap1Δ), Int6-induced resistance was abolished (note that the pap1Δ strain is inherently more drug-sensitive than the pap1+ strain). Int6-mediated drug resistance is therefore dependent on Pap1 function. The range of drugs to which Int6 conferred resistance was similar to that seen on Pap1 overexpression and included the protein synthesis inhibitor, cycloheximide, and the RNA synthesis inhibitor, actinomycin D, as well as MBC, staurosporine, and caffeine (Figure 3B). In contrast to their drug resistance, cells overexpressing Int6 were slightly (but significantly) more sensitive to UV light or hydrogen peroxide than transformants containing the vector alone (Figure 3C).
Figure 3.
Overexpression of Int6 specifically induces Pap1-dependent drug resistance and sensitivity to UV or oxidative stress. (A) Transformants of S. pombe h- leu1–32 (pap1+; upper panels) or h- pap1::ura4+ ura4-D18 leu1–32 (pap1Δ; lower panels) containing either pREP3X (vector) or the pREP3X cDNA library plasmid directing expression of int6 (int6) were streaked onto minimal agar plates with or without caffeine at the concentrations indicated. Plates were photographed after five days incubation at 30°C. (B) Int6-induced multi-drug resistance. Transformants of S. pombe h- leu1–32 containing pREP3X (vector), or pREP3X cDNA library plasmids directing expression of pap1 (pap1) or int6 (int6) were streaked onto minimal agar plates containing no drug (ND), 20 μg/ml cycloheximide (CHX), 8 μg/ml actinomycin D (ACD), 15 μg/ml MBC or 3 μg/ml staurosporine (STS). Plates were photographed after 4–7 days incubation at 30°C. (C) The sensitivities to UV radiation or hydrogen peroxide exposure of transformants of S. pombe h- leu1–32 containing either pREP3X (vector) or pREP3X-int6 (int6) were determined as described in MATERIALS AND METHODS. Percentages of surviving cells were determined in triplicate. Error bars represent one SD; for several data points these are smaller than the plot symbols. (D) Overexpression of eIF3 subunits other than Int6 does not confer drug resistance. Expression of p116-FLAG and HA-p47 was confirmed by Western blotting with anti-FLAG and anti-HA antibodies (left hand panels). Strains lacking the tagged proteins (-) served as negative controls. Strains transformed with pREP3X (vector) or expressing HA-int6, HA-p47, or p116-FLAG, as indicated, were streaked onto minimal agar plates containing no drug (ND) or 15 μg/ml MBC. Plates were photographed after 5 days incubation at 30°C.
To address the specificity of the drug resistance phenotype seen on Int6 overexpression, genes encoding p116 and p47, two additional subunits of eIF3, were cloned separately as epitope-tagged cDNAs in pREP3X. Induction of expression of these tagged proteins was confirmed by Western blotting after removal of thiamine (Figure 3D). In comparison with HA-tagged Int6, neither p116-FLAG nor HA-p47 was able to induce resistance to MBC. The drug resistance phenotype is therefore not a general consequence of overproduction of individual eIF3 subunits.
Int6 Overexpression Up-regulates Pap1-responsive Gene Expression
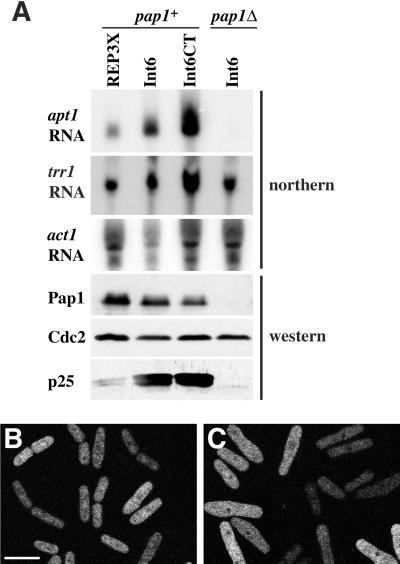
Pap1 up-regulation is accompanied by increased expression of a 25 kDa putative flavoprotein (p25), encoded by the apt1 gene, expression of which is almost entirely pap1-dependent (Toda et al., 1992) and, therefore, a useful marker of Pap1-dependent transcription. In line with the observed genetic dependence of Int6-induced drug resistance on pap1 integrity, Int6 or Int6CT overexpression was also associated with elevated levels of apt1 mRNA and p25 protein (Figure 4A). A second Pap1-responsive mRNA, that transcribed from the trr1 gene encoding thioredoxin reductase, was also up-regulated in response to overexpression of Int6 or Int6CT, although its expression was not absolutely Pap1-dependent. After normalization with respect to the loading controls for RNA (act1; actin) and protein (Cdc2), the level of up-regulation of apt1 RNA and p25 protein on Int6 or Int6CT overexpression was in each case found to be between 5- and 10-fold. These changes were Pap1-dependent but did not result from elevated Pap1 protein levels, as judged by immunoblotting (Figure 4A).
Figure 4.
Overexpression of Int6 causes elevated Pap1-responsive gene expression without Pap1 relocalization. (A) Transformants of S. pombe h- leu1–32 (pap1+) or h- pap1::ura4+ ura4-D18 leu1–32 (pap1Δ) containing either pREP3X, or pREP3X cDNA library plasmids directing expression of full length Int6 or Int6CT, as indicated, were grown in liquid culture for 16 h at 30°C in the absence of thiamine and were processed for northern or western blotting. A northern blot was probed to detect apt1 and trr1 RNA and subsequently stripped and reprobed to detect act1 RNA as a loading control, as indicated. A western blot of whole cell extracts prepared from the same cultures was probed with antibodies directed against Pap1, p25 and Cdc2 (loading control) as indicated. (B and C) An h- leu1–32 ura4-D18 strain transformed with pREP42-GFP-Pap1 and either pREP3X (B) or pREP3X-int6 (C) was grown in minimal medium without thiamine for 24 h in the dark at 25°C. GFP-Pap1 was visualized by confocal laser microscopy. Scale bar, 10 μm.
Pap1 is a substrate for the nuclear exportin Crm1, and one of the levels of regulation of Pap1 involves its relocalization to the nucleus (Toone et al., 1998). To address the possibility that Int6 overexpression influences Pap1-dependent gene expression through altering Pap1 localization, cells expressing GFP-Pap1 from the medium-strength nmt1 promoter in the vector pREP42 were examined by confocal microscopy. GFP-Pap1 was predominantly cytoplasmic, both in cells cotransformed with an empty pREP3X vector (Figure 4B) and in those overexpressing Int6, although the latter were somewhat swollen in appearance (Figure 4C). Int6-induced activation of Pap1-responsive gene expression would therefore seem to be independent of Pap1 relocalization.
Int6+ Is Required for Normal Growth Rate and Is Functionally Homologous to Human Int-6
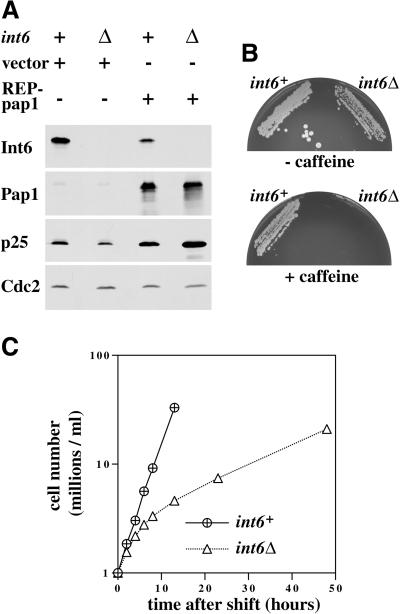
The absence of an Int-6 homolog in budding yeast might suggest that this eIF3 subunit is not essential for cell viability. To address this point, an int6+/int6− diploid fission yeast strain was constructed in which one of the two int6 alleles is disrupted by the ura4+ marker. After meiosis and tetrad dissection, it was found that all four haploid spores were viable. As expected, the int6::ura4+ progeny (Figure 5A, lanes 2 and 4) lacked detectable expression of Int6, as judged by Western blotting with the anti-Int6 antibody. The ura4+ meiotic products containing a disrupted int6 allele (int6Δ) contained slightly reduced p25 levels, but unchanged Pap1 levels (Figure 5A), and were found to be caffeine-sensitive (Figure 5B). Taken together with the overexpression data, this suggests that Int6 is a dosage-dependent regulator of drug resistance. However, Pap1 overexpression increased p25 protein levels regardless of int6 status (Figure 5A), indicating that Int6 is not an essential cofactor for Pap1.
Figure 5.
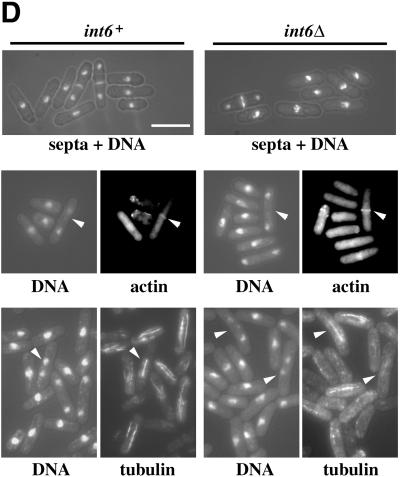
S. pombe cells lacking Int6 grow slowly, have reduced levels of p25, and are caffeine sensitive. (A) Haploid leu1–32 int6+ (+) and leu1–32 int6::ura4 (Δ) strains transformed either with pREP3X (vector) or pREP3X-pap1 (REP-pap1) were grown in EMM2 at 30°C for 16 h after removal of thiamine. Whole cell extracts prepared from these strains were subjected to Western blotting using antibodies directed against Int6, Pap1, p25, and Cdc2 (loading control), as indicated. (B) Two haploid segregants derived by sporulation of the int6+/int6− strain with genotypes h- int6+ leu1–32 ade- ura4-D18 (int6+) and h- int6::ura4+ leu1–32 ade- ura4-D18 (int6Δ) were streaked onto an EMM2 agar plate supplemented with adenine, leucine, and uracil, either without (upper plate) or with (lower plate) 10 mM caffeine. The plates were photographed after 6 days incubation at 30°C. (C) The int6+ and int6Δ strains were grown to midlog phase in rich (yeast extract) medium before being washed and resuspended in EMM2 medium at 106/ml and grown at 30°C. Cell number was determined for each culture at the times indicated after shifting to EMM2 medium. (D) Samples from each of the cultures used in C were taken at 18 h after shift to EMM2 medium, and were processed for staining with DAPI and calcofluor (visualized simultaneously using UV excitation; upper panels), with DAPI and rhodamine-phalloidin to determine DNA and actin distribution (visualized separately for each of the paired fields shown; middle panels), or with DAPI and an antitubulin antibody (lower panels). Arrowheads indicate cells with actin rings (middle panels) or anaphase cells with mitotic spindles (lower panels). Scale bar, 10 μm.
The int6Δ cells grew more slowly than those in which int6+ was intact (Figure 5C). This effect was most marked in cells growing in liquid minimal medium in which int6Δ cells grew with a doubling time of approximately 9 h at 30°C, compared with 2.5 h for an int6+ strain. Thus int6+ is required for maintenance of a normal growth rate in fission yeast. The slowly growing int6Δ cells retained an essentially normal morphology, as judged by staining of fixed cells to reveal distribution of DNA, septa or actin (Figure 5D). Antitubulin immunofluorescence revealed that the int6Δ cells were able to form apparently normal mitotic spindles, although interphase microtubules were less clearly defined than in the int6+ cells.
The growth defect in the int6Δ strain was complemented, as expected, by nmt1-driven expression of the cDNA encoding full-length Int6, but also by the Int6CT fragment isolated as a drug resistance determinant (Figure 6A). Transformants containing the empty vector were able to grow only slowly by comparison. The fact that Int6 is required for normal rates of fission yeast growth also allowed us to test whether human Int-6 can substitute functionally for its fission yeast relative. A derivative of the pREP3X vector containing an HA epitope-tagged full-length human Int-6 cDNA was constructed and transformed into the int6Δ strain. Expression of full-length human Int-6 was able to support growth at a rate comparable to that seen in an int6+ strain (Figure 6B). We conclude that the fission yeast int6+ and human Int-6 genes are functionally homologous.
Figure 6.
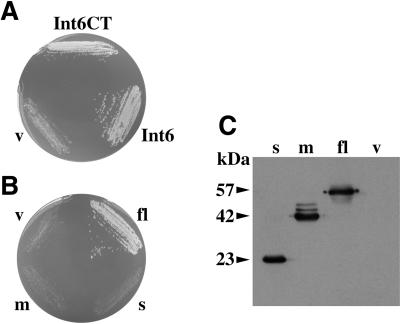
The growth-related function of fission yeast Int6 can be supplied by human Int-6. (A) Complementation of the int6Δ growth defect by Int6 or Int6CT. The int6Δ strain was transformed with pREP3X (v) or pREP3X cDNA library plasmids encoding full-length Int6 or Int6CT as indicated. Transformants were streaked onto an EMM2 agar plate supplemented with adenine and uracil, and were photographed after four days incubation at 30°C. (B and C) The int6Δ strain was transformed with pREP3X (v) or pREP3X derivatives containing full-length human Int-6 (fl), or either of the two C-terminally truncated Int-6 derivatives indicated in Figure 1 (m, s). Transformants were streaked onto an EMM2 agar plate supplemented with adenine and uracil, and were photographed after 4 days incubation at 30°C (B). Whole cell protein extracts prepared from liquid cultures of the same transformants were subjected to Western blotting using an anti-HA antibody (C). The molecular masses (in kDa) of the three tagged Int-6 proteins are indicated.
MMTV Integration Results in Loss of Normal Int-6 Function
One potential explanation for the apparent transforming activity of the truncated murine Int-6 proteins would be that they have lost a putative inhibitory C-terminal domain and, consequently, perform the normal Int-6 function constitutively (Marchetti et al., 1995). Alternatively, the truncated proteins might be nonfunctional and might contribute to transformation through dominant negative or gene dosage effects. The functional complementation of the int6 deletion by human Int-6 allowed us to distinguish between these possibilities. HA epitope-tagged versions of two truncated human Int-6 cDNAs analogous to two of the previously-described MMTV-disrupted alleles (Figure 1) were generated by PCR and cloned into pREP3X. Transformation of the resulting plasmids into the int6Δ strain followed by western blotting showed that the levels of truncated proteins produced were comparable to that produced by expression of the full-length Int-6 cDNA (Figure 6C). Despite these similar steady-state protein levels, neither of the truncated human cDNAs was able to support a normal growth rate in the int6Δ strain (Figure 6B). The MMTV-disrupted alleles therefore do not encode functional Int-6 proteins, as judged by this growth restoration assay.
DISCUSSION
While many aspects of the initiation of translation in mammals can be studied conveniently by genetic analysis of the corresponding components in budding yeast (Asano et al., 1997a; Asano et al., 1998), several human eIF3 components, including Int-6, do not have budding yeast counterparts (Asano et al., 1997c; Phan et al., 1998). The discovery of an Int-6 homolog in fission yeast was therefore unexpected, but the 38% amino acid identity (56% overall similarity) between the fission yeast and human proteins (Figure 1) suggests evolutionary conservation of an important function. This suggestion is given further weight by our findings that fission yeast Int6 functions to maintain normal rates of cell growth, and that this function can be performed by the human Int-6 protein (Figure 6). Given this degree of conservation, mammalian Int-6 probably also encodes an important growth-related function. It is unclear at this stage why normal rates of cell growth in S. cerevisiae do not require an Int-6-like eIF3 subunit.
In addition to Int-6, three further human eIF3 components without orthologs in S. cerevisiae appear to have been conserved in S. pombe. The S. pombe genome therefore includes genes encoding orthologs of all nine of the components of human eIF3 listed in Table 1. As the repertoire of genetic and biochemical techniques available in S. pombe is similar to that in budding yeast, a strong case can now be made for a more extensive investigation of eIF3 function in fission yeast. The effect of Int6 overexpression is specific, in that overexpression of the presumptive eIF3 components p47 or p116 in S. pombe did not cause drug resistance (Figure 3D).
Human Int-6 is a component of eIF3, and the preliminary characterization of fission yeast Int6 described here is consistent with a primary (and perhaps sole) role for this protein as a component of eIF3 in the cytoplasm under normal circumstances (Figure 2). One possible explanation for the Pap1-dependent drug resistance seen on Int6 overexpression in S. pombe (Figure 3) would therefore be an altered pattern of protein synthesis. This might be expected to result in accumulation of Pap1 itself or of gene products dependent for their synthesis on Pap1-mediated transcription. No alteration in the steady-state level of Pap1 was seen on Int6 overexpression, however, although the abundance of Pap1-responsive transcripts was increased (Figure 4A). The degree of up-regulation of p25 protein was similar to that of apt1 RNA, suggesting that in this case Int6 overexpression primarily influences the mRNA level, rather than the rate of protein synthesis.
Pap1 can be activated in response to a variety of cellular stresses through the activation of the StyI MAP kinase, but Int6-induced drug resistance was found to be sty1-independent (our unpublished results). We conclude that Int6 is unlikely to activate a stress response pathway upstream of Pap1. In line with this interpretation, Pap1 relocalization to the nucleus, which is StyI-dependent (Toone et al., 1998), was not observed on overexpression of Int6 (Figure 4B, C). Instead, elevated levels of Int6 might either increase Pap1 activity (for example by modulating the translation of an interacting transcription factor) or stabilize apt1 and trr1 mRNAs, as well as other transcripts that contribute to the drug resistance phenotype. It is quite conceivable that effects on translation and mRNA stabilization could be intimately linked, as some mutations affecting S. cerevisiae eIF3 components display defects in both processes (Schwartz and Parker, 1999). Interestingly, the Int6-induced drug resistance phenotype was accompanied by enhanced sensitivity to UV irradiation and hydrogen peroxide (Figure 3A, B). The reason for this sensitivity is not yet clear, but it is possible that Int6-mediated enhancement of Pap1-responsive, drug resistance-associated gene expression is achieved at the cost of suboptimal expression of other genes involved in the response to UV and oxidative stress. Although Pap1 is activated in response to oxidative stress, and pap1Δ cells are sensitive to pro-oxidants (Toone et al., 1998), overexpression of Pap1 has not been reported to confer resistance either to hydrogen peroxide or to UV radiation.
Deletion of int6 caused S. pombe cells to grow more slowly than int6+ controls, particularly when nutrients were limiting. The slowly growing int6Δ cells nonetheless exhibited balanced growth (Figure 5D), suggesting that multiple aspects of general biosynthesis were affected simultaneously. This could be taken as evidence for a role of Int6 in general translation, but there is a suggestion that, as in the case of Int6 overexpression, all gene products were not affected equally. Thus p25 was under-expressed relative to Pap1 and Cdc2 in the caffeine-sensitive int6Δ cells (Figure 5), and interphase microtubules were poorly defined (Figure 5D). A regulatory rather than essential role for Int6 in translation is also suggested by the absence of Int6 from S. cerevisiae.
The growth defect in the int6Δ strain could be complemented by Int6CT, the C-terminal fragment that was also found to induce drug resistance (Figure 6A). This fragment has only a limited degree of similarity to the corresponding region of the human protein, but appears to be functionally significant. It is possible that protein-protein interactions involving this C-terminal region are important for the normal function of Int6 within eIF3, and that mimicry of these interactions by overexpression of Int6CT is sufficient to induce drug resistance. Absence of the corresponding region in products of the MMTV-disrupted murine Int-6 alleles might also contribute to their putative oncogenic properties. Identification of proteins interacting with the Int6CT would therefore be a valuable objective for future work.
Expression of human Int-6 in fission yeast did not confer drug resistance (our unpublished results). This genetic separability of the cell growth-related and drug resistance-inducing functions of Int6 could suggest that these functions are also biochemically distinct. In this respect it is interesting that the Int6 homolog in Arabidopsis was shown recently to copurify with the COP9 complex (Karniol et al., 1998), the mammalian equivalent of which, the signalosome, appears to play a role in directing protein kinases to transcription factors (Wei et al., 1994; Seeger et al., 1998; Wei et al., 1998). The mammalian Jab1 protein, which binds the AP-1 factor c-Jun, is also a component of the signalosome complex and induces Pap1-dependent drug resistance when expressed in S. pombe (Claret et al., 1996; Seeger et al., 1998), and the COP9 signalosome is conserved in fission yeast (Mundt et al., 1999). It is therefore possible that the Int6-induced drug resistance described here is due to signalosome-mediated modulation of AP-1 activity, rather than any alteration in the pattern of translation. A caa1 deleted strain, in which signalosome assembly is defective (Mundt et al., 1999) was found to be generally drug-sensitive, but this sensitivity could not be overcome by overexpression of Int6 (our unpublished results). This might imply a role for the signalosome in Int6-induced drug resistance, but it seems unlikely that Int6 is itself a component of the S. pombe signalosome, as the size of the complex in which most of Int6 protein is found is considerably larger than the signalosome. Furthermore, the size of the Int6-containing complex was unaltered by caa1 deletion, which results in loss of signalosome integrity (Mundt et al., 1999).
Despite the lack of evidence linking Int6 with the signalosome in S. pombe, it is striking that the phenotype of Pap1-dependent drug resistance seen on Int6 overexpression was also seen on overexpression of Jab1, or of Pad1/POH1, a component of the 19S regulatory complex of the proteasome (Shimanuki et al., 1995; Claret et al., 1996; Spataro et al., 1997). An overall analogy among the subunit compositions of eIF3, the signalosome, and the 19S proteasome regulatory complex has been described. Each complex contains two members of the MPN (Pad1) family as well as multiple members of the PCI family, to which Int6 belongs (Hofmann and Bucher, 1998). The significance of this overall similarity in subunit composition is not yet clear, but is seems likely that the three complexes are derived from a common evolutionary ancestor, and it is conceivable that their activities are regulated in a coordinated manner.
The functional equivalence of the human and fission yeast Int-6/Int6 proteins has allowed us to investigate the biological activity of two truncated human Int-6 proteins corresponding to the products of MMTV-disrupted alleles found in mammary tumors. As neither truncated allele was able to suppress the slow growth phenotype resulting from int6 deletion in S. pombe, we suggest that the putative transforming activity of these alleles is likely to result from a loss-of-function or dominant negative effect. The reported retention of one wild-type Int-6 allele in each of the tumors containing an MMTV-disrupted Int-6 allele (Marchetti et al., 1995) would be consistent with this interpretation. The ability of human Int-6 to complement the growth defect in a fission yeast int6Δ strain can now be exploited as a straightforward means by which to examine more detailed structure-function relationships in the Int-6 protein.
ACKNOWLEDGMENTS
We are grateful to Tomohiro Matsumoto and Umadas Maitra for discussing results relating to the int6 deletion phenotype before publication. We thank Hiroto Okayama for providing the human embryonic fibroblast cDNA library, Kirsten Mundt and Tony Carr for providing the caa1 deletion strain before publication, Takashi Toda, Mark Toone, and Shao-Win Wang for providing strains, advice, and encouragement, Gracy Emilion for her invaluable help with northern blotting, Iain Goldsmith for excellent oligonucleotides, and Ian Hickson and other members of the Molecular Oncology Laboratory for their comments on the manuscript. This work was supported by the Imperial Cancer Research Fund.
Abbreviations used:
- eIF
eukaryotic initiation factor
- MMTV
mouse mammary tumor virus
- IRES
internal ribosome entry site
- EMM2
Edinburgh minimal medium
- MBC
methyl benzimidazole-2-yl carbamate
- HRP
horseradish peroxidase
REFERENCES
- Asano K, Kinzy TG, Merrick WC, Hershey JW. Conservation and diversity of eukaryotic translation initiation factor eIF3. J Biol Chem. 1997a;272:1101–1109. doi: 10.1074/jbc.272.2.1101. [DOI] [PubMed] [Google Scholar]
- Asano K, Merrick WC, Hershey JW. The translation initiation factor eIF3–p48 subunit is encoded by int-6, a site of frequent integration by the mouse mammary tumor virus genome. J Biol Chem. 1997b;272:23477–23480. doi: 10.1074/jbc.272.38.23477. [DOI] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologs of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem, 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K, Vornlocher HP, Richter-Cook NJ, Merrick WC, Hinnebusch AG, Hershey JW. Structure of cDNAs encoding human eukaryotic initiation factor 3 subunits. Possible roles in RNA binding and macromolecular assembly. J Biol Chem. 1997c;272:27042–27052. doi: 10.1074/jbc.272.43.27042. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: John Wiley & Sons, Inc; 1995. [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bernstein J, Sella O, Le SY, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J Biol Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature. 1996;383:453–457. doi: 10.1038/383453a0. [DOI] [PubMed] [Google Scholar]
- Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucl Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Celle ML, Rhoads RE. Functional characterization of the internal ribosome entry site of eIF4G mRNA. J Biol Chem. 1998;273:5006–5012. doi: 10.1074/jbc.273.9.5006. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Huez I, Creancier L, Audigier S, Gensac MC, Prats AC, Prats H. Two independent internal ribosome entry sites are involved in translation initiation of vascular endothelial growth factor mRNA. Mol Cell Biol. 1998;18:6178–6190. doi: 10.1128/mcb.18.11.6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T, Enoch T. Sum1, a highly conserved WD-repeat protein, suppresses S-M checkpoint mutants and inhibits the osmotic stress cell cycle response in fission yeast. Genetics. 1998;148:1731–1742. doi: 10.1093/genetics/148.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Wickens M. Translational controls impinging on the 5′-untranslated region and initiation factor proteins. Curr Opin Genet Dev. 1997;7:233–241. doi: 10.1016/s0959-437x(97)80133-5. [DOI] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karniol B, Yahalom A, Kwok S, Tsuge T, Matsui M, Deng XW, Chamovitz DA. The Arabidopsis homologue of an eIF3 complex subunit associates with the COP9 complex. FEBS Lett. 1998;439:173–179. doi: 10.1016/s0014-5793(98)01367-2. [DOI] [PubMed] [Google Scholar]
- Keiper BD, Rhoads RE. Cap-independent translation initiation in Xenopus oocytes. Nucl Acids Res. 1997;25:395–402. doi: 10.1093/nar/25.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macejak DG, Sarnow P. Internal initiation of translation mediated by the 5′ leader of a cellular mRNA. Nature. 1991;353:90–94. doi: 10.1038/353090a0. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Buttitta F, Miyazaki S, Gallahan D, Smith GH, Callahan R. Int-6, a highly conserved, widely expressed gene, is mutated by mouse mammary tumor virus in mammary preneoplasia. J Virol. 1995;69:1932–1938. doi: 10.1128/jvi.69.3.1932-1938.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. nmt1 of fission yeast: a highly transcribed gene completely repressed by thiamine. J Biol Chem. 1989;265:10857–10864. [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Mundt KE, Porte J, Murray JM, Brikos C, Christensen PU, Caspari T, Hagan IM, Millar JB, Simanis V, Hofmann K, Carr AM. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr Biol. 1999;9:1427–1430. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- Norbury C, Moreno S. Cloning cell cycle regulatory genes by transcomplementation in yeast. Meth Enzymol. 1997;283:44–59. doi: 10.1016/s0076-6879(97)83006-6. [DOI] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol, 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 1998;12:469–478. [PubMed] [Google Scholar]
- Shimanuki M, Saka Y, Yanagida M, Toda T. A novel essential fission yeast gene pad1+ positively regulates pap1+-dependent transcription and is implicated in the maintenance of chromosome structure. J Cell Sci. 1995;108:569–579. doi: 10.1242/jcs.108.2.569. [DOI] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C. Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26 S proteasome subunit. J Biol Chem. 1997;272:30470–30475. doi: 10.1074/jbc.272.48.30470. [DOI] [PubMed] [Google Scholar]
- Stein I, Itin A, Einat P, Skaliter R, Grossman Z, Keshet E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol Cell Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Yanagida M. Fission yeast genes that confer resistance to staurosporine encode an AP-1-like transcription factor and a protein kinase related to the mammalian ERK1/MAP2 and budding yeast FUS3 and KSS1 kinases. Genes Dev. 1991;5:60–73. doi: 10.1101/gad.5.1.60. [DOI] [PubMed] [Google Scholar]
- Toone WM, Kuge S, Samuels M, Morgan BA, Toda T, Jones N. Regulation of the fission yeast transcription factor Pap1 by oxidative stress: requirement for the nuclear export factor Crm1 (Exportin) and the stress-activated MAP kinase. Sty1/Spc1 Genes. Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Touriol C, Galy B, Audigier S, Gensac MC, Amalric F, Bayard F, Prats H, Prats AC. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Chamovitz DA, Deng XW. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78:117–124. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- Wei N, Tsuge T, Serino G, Dohmae N, Takio K, Matsui M, Deng XW. The COP9 complex is conserved between plants and mammals and is related to the 26S proteasome regulatory complex. Curr Biol. 1998;8:919–922. doi: 10.1016/s0960-9822(07)00372-7. [DOI] [PubMed] [Google Scholar]